Abstract
Astyanax caucanus is an endemic fish species to the Magdalena-Cauca basin in Colombia. It is considered a Least Concern species by the International Union for Conservation of Nature, and currently, it is not a fishery resource. Its fertilized eggs may drift up to 4–5 days before hatching and can be carried up to 340 km given the water velocity of the river. Although A. caucanus is listed as short -migratory species (< 50 km), this study hypothesized that it exhibits gene flow along the middle and lower section of the Cauca River due to the great potential for larval dispersal. To test this hypothesis, we developed a set of species-specific microsatellite primers suitable for population genetic studies. Genetic structure analyses with 193 samples evidenced two genetic stocks that coexist, comigrate, and exhibit gene flow along the study area. Both stocks show high genetic diversity indices (Na and HE) and effective population sizes (Ne > 1000), but also show evidence of bottlenecked populations and high values of the inbreeding coefficient (FIS). Finally, these results are useful to understand the effects of other anthropic activities, besides fishing pressure on population bottlenecks found for other fish species cohabiting the area.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Astyanax, one of the 142 valid genera family Characidae (Fricke et al., 2023a), is widely distributed in the Neotropical region, from southern United States to northern Argentina (Nelson et al., 2016). These fishes commonly known as tetras are highly diverse, comprising more than 129 species (Fricke et al., 2023b) that exhibit diagnostic characters generalized within the family, which hampers the taxonomic classification based on their morphology (Mirande, 2019). The final phylogenetic arrangement of the genus remains to be elucidated, although many studies have aimed to disentangle the phylogenetic relationships of Astyanax (Ornelas-García et al., 2008; Mirande, 2009, 2019; Mello et al., 2015; Casane & Rétaux, 2016; Rossini et al., 2016; Schmitter-Soto, 2016, 2017; Piscor et al., 2019; Terán et al., 2020). Complementary information about morphology and DNA sequences has allowed to validate the identity of some Astyanax species and assign others to valid or resurrected genera (Terán et al., 2020); however, such information is absent for other members of this genus limiting their validation or redescription (Terán et al., 2020). This is the case for Astyanax caucanus (Steindachner, 1879), an endemic fish species to the Magdalena-Cauca hydrographic region in Colombia, often confused with A. magdalenae Eigenmann & Henn, 1916 because their similar morphology and shared geographic distribution (Ruiz-C. et al., 2011; García-Alzate et al., 2021).
Astyanax caucanus is an omnivorous species that plays an important ecological role for the Magdalena-Cauca River basin (Gutiérrez-Moreno & de la Parra-Guerra, 2021). Due to its small size and high population number, A. caucanus among other species is considered an important part of the diet for larger species, not only fishes but also birds, turtles, and otters (Gutiérrez-Moreno & de la Parra-Guerra, 2021). This is a potamodromous and short-distance migratory species (< 50 km) with early maturity and periodic reproduction (Jiménez-Segura et al., 2021). Astyanax caucanus is included in the IUCN red list as a least concern (LC) species, due to its wide distribution and that there are no major threats for its populations (Villa-Navarro & Sanchez-Duarte, 2014). However, the genetic status of this species remains unknown, although their natural habitat is constantly exposed to anthropic impacts due to the Magdalena-Cauca River basin, Colombia's primary developmental area, housing 77% of its inhabitants and contributing to 80% of its Gross Domestic Product (Restrepo et al., 2021). This has resulted in a significant anthropic impact on aquatic biodiversity in the region, including sewage discharge, irresponsible mining, the desiccation of floodplain lakes for livestock and agricultural expansion, and dam construction, among other factors (Angarita et al., 2021; Gutiérrez-Moreno & de la Parra-Guerra, 2021). In February 2019, the closure of the gates during an emergency at the Ituango dam construction on the Cauca River's main channel caused the complete disruption of the river's flow, resulting in the death of thousands of low-migration-range fish located 10 km downstream of the dam. Consequently, it is essential to generate information about the structure and genetic diversity of the populations of A. caucanus to estimate the risk category and design conservation strategies for the species.
Colombia lacks population genetic studies for Astyanax species, in contrast to Mexico (Strecker et al., 2003; Panaram & Borowsky, 2005; Hausdorf et al., 2011; Bradic et al., 2012; Herman et al., 2018; Pérez-Rodríguez et al., 2021) and Brazil (Leuzzi et al., 2004; Peres et al., 2005; Sofia et al., 2006; Ferreira et al., 2016; Freitas-Lidani et al., 2018; Limeira et al., 2019). In Mexico, the studies are focused mainly on analyzing surface versus cave populations using different molecular markers that include microsatellite loci (Strecker et al., 2003; Panaram & Borowsky, 2005; Hausdorf et al., 2011; Bradic et al., 2012; Pérez-Rodríguez et al., 2021). In Brazil, different molecular approaches such as RAPDs (Leuzzi et al., 2004; Sofia et al., 2006; Ferreira et al., 2016; Freitas-Lidani et al., 2018), allozymes (Peres et al., 2005), and microsatellite loci (Ferreira et al., 2016; Limeira et al., 2019) are used to analyze the diversity and structure of Astyanax populations. In terms of genetic diversity, these studies show that high allelic richness and heterozygosity is generalized within the genus. So far, only one study found evidence of gene flow among cave and surface populations in geographically close locations (Panaram & Borowsky, 2005), whereas other studies reveal that Astyanax populations are frequently structured and may exhibit structure in population separated by only 5 km (Sofia et al., 2006). These works suggest that such structure may be related to an isolation by distance model (Strecker et al., 2003; Leuzzi et al., 2004; Peres et al., 2005; Sofia et al., 2006; Ferreira et al., 2016; Freitas-Lidani et al., 2018; Garita-Alvarado et al., 2021; Pérez-Rodríguez et al., 2021), phylogenetic distance (Hausdorf et al., 2011; Garita-Alvarado et al., 2021), or assortative mating (Bradic et al., 2012). Based on the coancestry analysis, Ferreira et al. (2016) and Garita-Alvarado et al. (2021) show the coexistence of different genetic groups of Astyanax in the same studied localities.
Aiming to generate information useful for developing management and conservation policies for this species, this study assessed the genetic status and tested the hypothesis that A. caucanus exhibit gene flow along middle and lower sections of the Cauca River. If A. caucanus fish fry exhibits a similar behavior as some congeners that become free swimming and ready to feed on plankton at 4–5 days post fertilization (Axelrod et al., 1971; Hinaux et al., 2011; Stevanato & Ostrensky, 2018; dos Santos et al., 2020), in the absence of physical barriers, it is expected that this short-distance migrator may exhibit gene flow in an area of 325 km. Since the annual average water velocity in the area is 1.18 ± 0.52 m/s (middle section: S2-S3; Fig. 1) and 0.50 ± 0.18 m/s (lower section: S4-S8) (unpublished data), the larvae may drift up to 340 km along the main channel of the river until going into the floodplain lakes. To test this hypothesis, this work developed a set of species-specific microsatellite loci in population genetic studies of A. caucanus, which were able to measure the genetic diversity of this tetra fish in the middle and lower sections of the Cauca River.
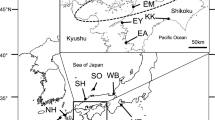
Geographical sections for collection of Astyanax caucanus along the Cauca River, Colombia, as described by Landínez-García & Márquez (2016). Triangle represents Ituango hydropower project (PHI). Samples of A. caucanus were collected from S2/3 to S7/8, in an area of 325 km
Materials and methods
Sample collection and DNA extraction
For population genetic purposes, this study used 193 ethanol-preserved muscle or tail tissues from individuals of A. caucanus collected in the middle and lower basin of the Cauca River to extract genomic DNA. The tissues were collected during the same fish rise period (subienda; December–March) in 2019 and 2020 by Grupo de Ictiología de la Universidad de Antioquia (GIUA). The specimens were collected along the main channel of the river in different environments including small affluents (middle basin) and floodplain lakes (middle and lower basin). The collection area was divided into nine sections, S1 to S8 as previously described by Landínez-García & Márquez (2016) and PHI corresponding to the current site of the Ituango hydropower project reservoir. Individuals were caught in seven of these sections; S1 and PHI were excluded due to lack of samples despite collection efforts. Sample number per site was as follows: S2/3: 30, S4: 31; S5: 47, S6: 37; S7/8: 48 (Fig. 1). Due to the extremely low number of individuals in those sections, S2 was merged with S3 (mouth of the Espiritu Santo River on the east margin of the Cauca River), and S7 (floodplain lakes located near the mouth of the Cauca River in the Magdalena River) with S8 (main channel of the Cauca River near the mouth of the Cauca River in the Magdalena River). Extraction of DNA was carried out using the PureLink™ Genomic DNA Mini Kit (Invitrogen), following manufacturer’s instructions and performing overnight digestions.
Phylogenetic analysis
To avoid confusions with other phylogenetically related species of Astyanax cohabiting the area, the mitochondrial gene cytochrome c oxidase subunit 1 (MT-CO1) of 209 samples was partially sequenced using a four primers mix: VF2_t1 5′-TCAACCAACCACAAAGACATTGGCAC-3′, FishF2_t1 5′-TCGACTAATCATAAAGATATCGGCAC-3′, FishR2_t1 5′-ACTTCAGGGTGACCGAAGAATCAGAA-3′ (Ward et al., 2005) and FR1d_t1 5′-CACCTCAGGGTGTCCGAARAAYCARAA-3′ (Ivanova et al., 2007). Sequencing included samples from individuals tentatively identified as A. caucanus, Astyanax aff. fasciatus, and Astyanax microlepis Eigenmann, 1913 that were identified through traditional morphometrics following original descriptions of species (Steindachner, 1879; Eigenmann, 1913; Eigenmann & Henn, 1916); samples of A. caucanus collected in the type locality (Tarazá River), and A. aff. fasciatus (Colosó-Sucre: IAvH-CT-17785, Cimitarra- Santander: IAvH-CT-24679; Barrancabermeja- Santander: IAvH-CT-24548), were included as reference for validated nominal species. The amplification was performed following Rangel-Medrano et al. (2020) with some modifications to the final concentrations of DNA and primers: 1.0–1.5 ng/µL of DNA and 0.2 pmol/µL of the four-primer mix. The thermal profile for PCR comprised an initial denaturation of DNA at 90 °C for 3 min, followed by 35 temperature cycles of denaturing step at 90 °C for 30 s, annealing step at 56 °C for 60 s, and extension step at 72 °C for 30 s, and a final extension step at 72 °C for 2 min.
Sequences were purified, cut, aligned, and translated into amino acids to confirm absence of stop codons using GeneiousPrime® v2022.1.1 (www.geneious.com), and the software DnaSP v6.12.03 was used to obtain the list of haplotypes for further analyses. All the new sequences for A. caucanus and other species generated in this study are available on GenBank (accession numbers: OR064539—OR064743). Partial sequences MT-CO1 of A. microlepis (UdeA; BOLD: CIUA979), Psalidodon fasciatus (Cuvier, 1819) from Venezuela (GenBank KY267682), Psalidodon fasciatus from Brazil (GenBank KY268274), and the outgroup Brycon henni Eigenmann, 1913 (GenBank KP027535) were also included for phylogenetic tree reconstruction (Fig. 2). The software jModelTest v2.1.10 (Darriba et al., 2012) was used to select the best-fit nucleotide substitution model of the complete dataset. The tree was constructed using the Beauti v2.6.7 application of Beast2 (Bouckaert et al., 2014) with the following parameters for the run: HKY + G; Gamma category count: 4; 400,000,000 MCMC; 10% as burn-in; and storeEvery 40,000. The other parameters of the run were left as default. The run was carried out using the CIPRES platform (https://www.phylo.org/). Then, convergence of parameters (ESS > 200) in the run was checked with Tracer v1.7.1 application of Beast2. To summarize the 10,000 trees, we used TreeAnnotator v2.6.0 application of Beast2; finally, FigTree v1.4.4 application was used to visualize the final tree.
Phylogenetic relationships between haplotypes in this study and some Astyanax species. Names in blue, red, and green represent individuals identified, through traditional morphometrics, as Astyanax caucanus, Astyanax aff. fasciatus and Astyanax microlepis, respectively. Names in purple represent sequences used as reference
Microsatellite loci development
Aiming to identify microsatellite loci and develop a species-specific set of primers, the shotgun genomic library from one individual of the most frequent haplotype was sequenced with the Miseq Illumina platform. The subsequent analysis of the genomic data was performed as described previously by Landínez-García & Márquez (2016, 2018). Briefly, the reads with less than 50 bp, low-quality regions or duplicated were eliminated from analysis using Prinseq-lite v0.20.4 (Schmieder & Edwards, 2011), the cleaned reads were analyzed with PAL_FINDER v0.02.03 to extract potentially amplifiable loci (PAL), the primers were designed using PRIMER3 v.2.0 (Rozen & Skaletsky, 2000) and the correct primer alignment was verified by electronic PCR (Rotmistrovsky et al., 2004).
A set of 30 microsatellite loci were tested experimentally to check amplification consistency and level of polymorphism in 30 random samples. The 5´end of the forward primers was attached covalently to adapters as described in Table 1 complementary to a sequence fluorescently labeled with 6-FAM, VIC, NED, and PET (Blacket et al., 2012; Table 1). Concentrations and conditions for PCR amplifications were as proposed by Landínez-García & Márquez (2018) with some modifications described hereunder. The following final concentrations were used for the 10 µL amplification reactions: 5–10 ng/µL of DNA, 1 X of Platinum Mix and 2.5% of Enhancer GC, both from the Platinum™ Multiplex PCR Master Mix kit (Invitrogen), 0.50 pmol/μL of each forward primer, 1 pmol/μL reverse primer, and 0.50 pmol/μL of the universal-labeled primer. Additionally, amplifications were carried out in a T100 thermal cycler (BioRad) using the following profile that did not include both, elongation step and final extension: 3 min at 94 °C for initial denaturation, 35 cycles consisting of 35 s at 90 °C for denaturation step, and 35 s at 56 °C for annealing step. Finally, amplicons were separated using a SeqStudio Genetic Analyzer (Applied Biosystems) and LIZ600® (Applied Biosystems) as internal molecular size marker. The size of each fragment was annotated using the software GeneMarker v3.0.0 (SoftGenetics LLC®). Those microsatellite loci that satisfied the selection criteria described by Landínez-García & Márquez (2016, 2018) (polymorphism level, band resolution, specificity, desired fragment size, and heterozygosity) were used for further population genetic analyses.
Population genetics
The genetic study included 193 samples of A. caucanus genotyped using 12 primer pairs selected as described above. Micro-Checker v2.2.3 (van Oosterhout et al., 2004) was run to detect potential genotyping errors. The software GenAlEx v6.503 (Peakall & Smouse, 2012) was used to measure the average number of alleles per locus and the software Arlequin v3.5.2.2 (Excoffier & Lischer, 2010) to estimate the observed (HO) and expected (HE) heterozygosities, inbreeding coefficient (FIS), and departures of Hardy–Weinberg (HWE) and Linkage (LE) equilibria. In addition, we used Cervus v3.0 (Kalinowski et al., 2007) to calculate the polymorphic information content (PIC) for each microsatellite locus. For all analyses with multiple comparisons, sequential Bonferroni correction (Rice, 1989) was applied to adjust for statistical significance. Additionally, BayeScan v2.01 (Foll & Gaggiotti, 2008) with default parameters was used to explore nonneutral selection acting on microsatellite loci. Loci were ranked according to their estimated posterior probability or posterior odds (equivalent to Bayes factor), a statistical criterion to test the model (Foll, 2012). Positive alpha values were then used to distinguish microsatellites under diversifying selection, while negative alpha values were used to detect balancing selection.
Exploration of recent genetic bottlenecks was performed calculating the levels of heterozygosity using the Wilcoxon sign-rank test (Luikart & Cornuet, 1998) with Bottleneck v1.2.02 (Piry & Luikart, 1999) and M-ratio using Arlequin v3.5.2.2 (Excoffier & Lischer, 2010). Values of M-ratio smaller than 0.680 indicates that the population experienced recent reductions in the population size (Garza & Williamson, 2001). The software Migrate v5.0.4 (Beerli & Palczewski, 2010) was used to estimate effective population size (Ne), setting the following parameters: Brownian model as it is recommended for Microsatellite data, θ: 0–120 (uniform), heating: 1.0, 1.5, 3.0, 1,000,000 (static), long chains: 6, number recorded steps: 20,000, number sampling increment: 200, and burn-in: 2000 for every chain. The runs was performed in CIPRES platform (https://www.phylo.org/). To transform the θ parameter in Ne, θ = 4 × Ne × µ (Gilbert & Whitlock, 2015), where µ = 5.56 × 10–4 changes/locus/generation (Yue et al., 2007).
The Geneclass2 software (Piry et al., 2004) was employed to assess migration events between different sites, encompassing both the overall sample and each distinct stock. The approach entailed assigning each individual to its most probable section of origin based on the collected samples. This assignment was performed using a Bayesian criterion, specifically the maximum likelihood estimator L_home/L_max, which represents the relationship between an individual's maximum likelihood of originating from the sampled section and the maximum likelihood across all sections. In addition, a statistical Monte Carlo resampling method (Paetkau et al., 2004) was applied, setting the simulation of 10,000 individuals and an error probability of 0.01.
Spatial structure of A. caucanus was calculated with an analysis of molecular variance (AMOVA) with the standardized statistics F′ST (Meirmans, 2006) and Jost’s D’est (Jost, 2008), and setting 10,000 permutations, all included in GenAlEx v6.503 (Peakall & Smouse, 2012). To test if there is correlation of genetic distance and geographical distance a Mantel test included in GenAlEx v6.503 (Peakall & Smouse, 2012) was performed with 9999 permutations. In addition, structure was explored with a discriminant analysis of principal components (DAPC) with the R package adegenet (Jombart, 2008) and using the diploid genotypes of 12 loci (24 variables) in 193 individuals. Furthermore, genetic structure was explored in R-based Parallel Structure (implemented from Structure v2.3.4; Pritchard et al., 2000) in CIPRES (https://www.phylo.org/) using 1,000,000 MCMC and an additional 10% as burn-in; admixture and non-admixture assumptions, and the LocPrior option. Each number of clusters (K) simulated ranged from K = 1 to 8 and each was simulated 20 times. After runs, the best K value was selected using the methods described by Evanno et al. (2005) and Puechmaille (2016) in the web-based software StructureSelector (Li & Liu, 2018). The coancestry plot was generated using the program Clumpak (Kopelman et al., 2015) included in StructureSelector.
Results
Phylogenetic analysis
After edition and alignment, 193 sequences of 492 bp of length (37 haplotypes) showed no start or stop codons after translation, suggesting that they are part of a functional gene. The phylogenetic tree showed three main groups (Fig. 2): (1) samples from the middle and lower Cauca River [25 haplotypes] clustered with A. caucanus from Tarazá. This haplogroup was considered as A. caucanus due to inclusion of topotype sequences of this nominal species, having priority over remaining names used to identify samples analyzed here; (2) samples of A. aff. fasciatus [4 haplotypes] clustered with samples of A. aff. fasciatus from Colosó—Sucre (IAvH-CT-17785), Cimitarra—Santander (IAvH-CT-24679) and Barrancabermeja–Santander (IAvH-CT-24548); and (3) samples of A. aff. fasciatus [6] and A. microlepis [1 haplotype] clustered with A. microlepis from the Porce River (BOLD: CIUA979), and Psalidodon fasciatus from Venezuela (GenBank: KY267682). All samples named A. aff. fasciatus were phylogenetically different from Psalidodon fasciatus from Brazil (GenBank KY268274).
Microsatellite loci development
The analysis of genomic sequencing of one individual showed that 76,206 of the total reads contained microsatellite loci, and the 5-mer and 6-mer were the most common repeat motifs with 42.72% and 32.25%, respectively, followed by 3-mer (10.81%), 2-mer (7.12%), and 4-mer (7.10%). Of the total microsatellite loci, 55,119 were potentially amplifiable, and 24,967 were validated through electronic PCR. Sixteen of the 30 loci tested experimentally satisfied the selection criteria described above (Table 1), amplified in the expected size range (122–335) and were highly polymorphic (average PIC: 0.904). All loci showed significant departures from Hardy–Weinberg equilibrium except in a few cases in the genetic stocks and four showed linkage disequilibrium (Aca01–Aca15 and Aca06Aca16) in the subgroup of 30 individuals. Both the average number of alleles per locus (Na) and the allelic richness (AR) showed values greater than 18 (Table 2).
Population genetics
Four loci were excluded from population genetic analyses since they showed outlier data according to BayeScan v2.01 (Aca07, Aca14, Aca21; Supplementary Table 1) or showed inconsistent amplification (Aca16; Table 2). There is no evidence of scoring errors detected by Micro-Checker; additionally, all loci showed evidence for a null allele and the overall sample showed departures from Hardy–Weinberg equilibrium and significant heterozygosity deficit. According to the different analyses of genetic differentiation, A. caucanus does not exhibit geographic structure due to the AMOVA was not statistically significant, which agrees with the values of F′ST and Jost’s D’est that were low and non-significant for all sections of the river under analysis (Table 3). The Mantel test showed a weak and statistically non-significant signal of spatial correlation of genetic distances (R2 = 0.055; P = 0.291). Similarly, DAPC indicated that all geographical demes constitute only one group (Fig. 3A). Furthermore, the analysis in Structure showed that all demes exhibit gene flow, although it was clear the existence of two genetic stocks (K = 2) distributed along the study area (Fig. 3B).
The diversity of genetic stocks was similar to the pooled sample (Table 4) particularly, Stock1 and Stock2 showed high values of AR. Additionally, A. caucanus exhibits significant values of FIS coefficient > 0.380 indicating high inbreeding levels in the species. Furthermore, bottleneck analysis showed that this fish species experienced recent and severe reduction of population size given the values of M-ratio smaller than 0.680 in addition to significant values found in the I.A.M. and T.P.M. tests (Table 5). In addition, values of Ne were higher than 1000 in the two stocks (Stock1: 24,730, 95% CI: 22,878–26,439, autocorrelation value: 0.119; Stock2: 25,234, 95% CI: 23,417–26,978, autocorrelation value: 0.084). In the sample, 32 individuals were identified as likely migrants (P < 0.01) among river sections according to the results of the first-generation migrant detection test. Specifically, 17 individuals collected downstream of collection sectors were detected as likely immigrants from upstream sections (Stock1: 8; Stock2:9), while 15 individuals collected upstream was identified as likely immigrant from downstream sections (Stock1: 11; Stock2: 4).
Discussion
This study assessed the genetic status and structure of the short-distance migratory species Astyanax caucanus in the middle and lower sections of the Cauca River in Colombia. Due to the morphological similarities with other Astyanax species distributed in the studied area, species identification was supported by a phylogenetic analysis based on partial sequences of MT-COI that revealed three main groups. The haplogroup clustered with A. caucanus from Tarazá, hence, was considered as A. caucanus due to the inclusion of sequences from specimens unequivocally assigned to this species, coming from its type locality, also having nomenclatural priority over remaining species names applied to analyzed samples from the Magdalena-Cauca River hydrographic region. The two haplogroups that contain samples morphologically identified as A. aff. fasciatus are phylogenetically distant from Psalidodon fasciatus from Brazil supporting the restricted distribution recently proposed for this last species (Terán et al., 2020). Since some limitations of MT-COI have been reported for discrimination of some morphologically distinguishable species or with high intraspecific variation (Rosini et al., 2016; Terán et al., 2020), our outcomes indicate that an integrative revision is required to clarify the taxonomy of this genus in Colombia.
Although there are microsatellite loci developed for other Astyanax members (Strecker, 2003; Zaganini et al., 2012), this study developed a set of species-specific microsatellite loci for Astyanax caucanus aiming avoid problems associated to the use of heterologous microsatellite loci and the type of repeat motifs (see review: Guichoux et al., 2011). The newly developed set of 16 loci is characterized by high polymorphism level, good band resolution, specificity, desired fragment size, and high levels of expected heterozygosity. In addition, the loci show departures of Hardy–Weinberg equilibrium and deficit of observed heterozygosity in the analyzed sample, which may be explained by the high number of alleles per locus that demand a great sample size for analysis (see review: Abdul-Muneer, 2014), and biological causes that are discussed below such as Wahlund effect, inbreeding, and assortative mating.
Results reveal that geographic demes are not genetically differentiated supporting the hypothesis that A. caucanus exhibits gene flow along the analyzed area (325 km). This outcome is congruent with the detection of 32 individuals identified as likely migrants among river sections, as indicated by the results of the Bayesian analysis with Geneclass2 (Piry et al., 2004) that suggest the potential movement of individuals in both upstream and downstream directions, as well as the drift of larvae from upstream sections. This result differs from the pattern found in other congeners that show populations geographically structured (Strecker et al., 2003; Leuzzi et al., 2004; Peres et al., 2005; Hausdorf et al., 2011; Bradic et al., 2012; Freitas-Lidani et al., 2018; Limeira et al., 2022) even when they were separated by only 5 km (Sofia et al., 2006). If larval duration in A. caucanus reaches five days post fertilization as observed in related congeners (Axelrod et al., 1971; Hinaux et al., 2011; Stevanato & Ostrensky, 2018; dos Santos et al., 2020), A. caucanus larvae may travel distances even longer (up to 340 km) than the adults in the migration events (50 km; Jiménez-Segura et al., 2021), favoring gene flow. Among marine species, this explanation is usual since the gene flow is favored via larval dispersal (Hellberg, 1996; Matschiner et al., 2009; Pascual et al., 2017).
Despite the absence of spatial structure, A. caucanus exhibits two stocks that coexist in the middle and lower sections of the Cauca River. Due to the limitation of MT-COI and morphology for Astyanax species discrimination (Rosini et al., 2016; Terán et al., 2020), the presence of two genetic stocks may be explained by morphological differences recognized in the names A. caucanus and A. magdalenae. However, there are other possible aspects in the life history of A. caucanus that could explain the presence of both stocks. One explanation considers the two annual spawning periods of this species (Jiménez-Segura et al., 2016), related to the bimodal hydrological cycle of the basin (López-Casas et al., 2016), which may reflect temporal reproductive isolation between the two genetic groups (Wahlund effect). Although the presence of sexually mature individuals from both stocks at the same hydrological period may not provide support to this explanation, other aspects such as assortative mating by preference of environmental factors (e.g., temperature, light intensity, and/or timing), asynchronous spawning, or the temporal presence of hooks in the anal fin as was shown in Psalidodon scabripinnis (Jenyns, 1842) (Castro et al., 2014), might explain the reproductive isolation of both stocks. These hypotheses must be tested in further studies with more information regarding the reproductive characteristics of A. caucanus.
The coexistence of different genetic groups has been recorded for the genus, as it was also found in Astyanax altiparanae Garutti & Britski, 2000 in several sections of the Penacho stream (Ferreira et al., 2016) and in sympatric Astyanax morphs from Central America (Garita-Alvarado et al., 2021), although its causes remain to be clearly established. This genetic structure has been also found in other species that inhabit the Cauca River, such as Prochilodus magdalenae Steindachner, 1879 (Landínez-García et al., 2020), Megaleporinus muyscorum (Steindachner, 1900) (Márquez et al., 2021), Pimelodus grosskopfii Steindachner, 1879 (Restrepo-Escobar et al., 2021), and Pimelodus yuma Villa-Navarro & Acero P., 2017 (Joya et al., 2021). The coexistence of genetic groups in the Cauca River is explained by spatial or temporal reproductive isolation; nevertheless, both explanations require further research to clarify the probable causes of these patterns of genetic structuring (Márquez et al., 2021).
The two stocks in this study show high genetic diversity levels, specifically the average number of alleles/locus and expected heterozygosity are higher than the average values for Neotropical Characiformes (Na: 10.920; HE: 0.675; Hilsdorf & Hallerman, 2017). Particularly for A. caucanus, values of HE (0.827–0.840) are higher than those found in Astyanax sp. (0.550; Strecker et al., 2012), A. mexicanus (De Philippi, 1853) (0.487; Strecker et al., 2003) and P. scabripinnis (0.487; Limeira et al., 2019), and similar to those found in A. altiparanae (0.833; Ferreira et al., 2016), and A. bransfordii (Gill, 1877), A. nicaraguensis Eigenmann & Ogle, 1907 and A. nasutus Meek, 1907 (0.840; Garita-Alvarado et al., 2021). Compared to species of the Cauca River, A. caucanus shows higher values of HE than those found in B. henni (0.604–0.662; Landínez-García & Márquez, 2020), Ichthyoelephas longirostris (Steindachner, 1879) (0.771–0.798; Landínez-García & Márquez, 2016), Curimata mivartii Steindachner, 1878 (0.793–0.810; Landínez-García & Marquez, 2018), Sorubim cuspicaudus Littmann, Burr & Nass, 2000 (0.771–0.785; Restrepo-Escobar et al., 2021), and Pseudoplatystoma magdaleniatum Buitrago-Suárez & Burr, 2007 (0.770–0.798; García-Castro et al., 2021), similar to those found in Ageneiosus pardalis Lütken, 1874 (0.832–0.839; Restrepo-Escobar et al., 2021), Pimelodus yuma (0.825–0.875; Joya et al., 2021), Pseudopimelodus atricaudus Restrepo-Gómez, Rangel-Medrano, Márquez & Ortega-Lara, 2020 (0.804–0.840; Rangel-Medrano & Márquez, 2021), and lower than those found in Prochilodus magdalenae (0.898; Landínez-García et al., 2020) and Pimelodus grosskopfii (0.849–0.866; Restrepo-Escobar et al., 2021).
The high levels of inbreeding of A. caucanus exceed by more than three times the threshold to avoid short-term decreases in reproductive performance in captive populations: 0.100 (Soulé, 1980). Although this is a threshold for captive populations, Frankham et al. (2014) indicate that any sign of inbreeding reduces population viability in wild populations. Due to the scarce information about the life cycle of A. caucanus, causes for such values of inbreeding remain unclear; however, Keller & Waller (2002) indicate that inbreeding occurs in wild populations regularly and can be severe but do not exclude the increase of environmentally inflicted mortality. Specifically, the elevated levels of inbreeding may stem from a nonrandom contribution by parents, possibly influenced by variations in their fertility, the viability of their offspring, or differences in their reproductive behavior (see: Caballero, 1994).
Alternatively, the heightened levels of inbreeding may be attributed to a recent and severe reduction in population size as estimated by Bottleneck analysis, a common characteristic showed by other species present in the Cauca River even before the construction of Ituango Hydropower (Landínez-García & Marquez, 2018; Landínez-García et al., 2020; García-Castro et al., 2021; Restrepo-Escobar et al., 2021). Since A. caucanus is not a fishery resource, anthropic activities different to the fishery and likely climate events may explain these results. Among the remaining anthropic harmful activities are mining, oil refinery, hydropower generation, urban water supply and dumping, agricultural crops, and livestock production (Angarita et al., 2021; Gutiérrez-Moreno & de la Parra-Guerra, 2021). They lead to chemical dumping (e.g., heavy metals, pesticides, drugs), oil spill (accidental or terrorism related), sedimentation (30,000–300,000 m3/day), hydrological disconnection, deforestation, among others (Angarita et al., 2021; Gutiérrez-Moreno & de la Parra-Guerra, 2021; Hernández-Barrero et al., 2021; Lasso et al., 2021).
It is important to recall that A. caucanus along with other congeners represent the dominant biomass of the river (Jiménez-Segura et al., 2021; Valderrama-Barco et al., 2021), hence, its population is still large, even after experiencing bottlenecks. This last idea is supported by the estimated value of the effective population size (Ne > 1000) that indicates little or no genetic threats to its evolutionary potential (Frankham et al., 2014). Nevertheless, consequently with the signals of genetic bottleneck and levels of inbreeding above discussed, careful conservation strategies focused on habitat preservation are required to lower risk of future reductions in population size.
In summary, this work found two genetic stocks that coexist, comigrate and exhibit gene flow along the middle and lower sections of the Cauca River. Each stock show high levels of expected heterozygosity and effective population numbers, in addition to evidence of recent and severe reductions in population size and high values of inbreeding coefficients. Likewise, this study developed a set of species-specific microsatellite loci that may help monitor the genetic diversity, structure, and demography of A. caucanus, information relevant for designing short-term regulations that are crucial to preserve the natural habitat of this tetra fish and support the food chain for commercially important species. Finally, this work provides insights on the genetic status of a species that is not influenced by fishing pressure.
References
Abdul-Muneer, P. M., 2014. Application of microsatellite markers in conservation genetics and fisheries management: recent advances in population structure analysis and conservation strategies. Genetics Research International 2014: 1–11. https://doi.org/10.1155/2014/691759.
Angarita, H., A. Santos-Fleischmann, C. Regéliz, G. Narváez-Campo, J. Delgado, T. Santos, A. Santos, G. Herrera-R, & L. F. Jiménez-Segura, 2021. Capítulo 7 - Modificación del hábitat para los peces de la cuenca del río Magdalena, Colombia. In Jiménez-Segura, L.F., & C.A. Lasso (eds), Peces de la cuenca del río Magdalena, Colombia: diversidad, conservación y uso sostenible. Instituto de Investigación de Recursos Biológicos Alexander von Humboldt. https://doi.org/10.21068/A2020RRHHXIX.
Axelrod, H., C. Emmens, D. Sculthorpe, W. Vonderwinkler & N. Pronek, 1971. Exotic Tropical Fishes, TFH Publications, New Jersey:
Beerli, P. & M. Palczewski, 2010. Unified framework to evaluate panmixia and migration direction among multiple sampling locations. Genetics 185(1): 313–326. https://doi.org/10.1534/genetics.109.112532.
Blacket, M. J., C. Robin, R. T. Good, S. F. Lee & A. D. Miller, 2012. Universal primers for fluorescent labelling of PCR fragments—an efficient and cost-effective approach to genotyping by fluorescence. Molecular Ecology Resources 12: 456–463. https://doi.org/10.1111/j.1755-0998.2011.03104.x.
Bouckaert, R., J. Heled, D. Kühnert, T. Vaughan, C.-H. Wu, D. Xie, M. A. Suchard, A. Rambaut & A. J. Drummond, 2014. BEAST 2: A Software Platform for Bayesian Evolutionary Analysis. PLoS Computational Biology 10: e1003537. https://doi.org/10.1371/journal.pcbi.1003537.
Bradic, M., P. Beerli, F. J. García-de León, S. Esquivel-Bobadilla & R. L. Borowsky, 2012. Gene flow and population structure in the Mexican blind cavefish complex (Astyanax mexicanus). BMC Evolutionary Biology 12: 9. https://doi.org/10.1186/1471-2148-12-9.
Caballero, A., 1994. Developments in the prediction of effective population size. Heredity 73: 657–679. https://doi.org/10.1038/hdy.1994.174.
Casane, D., & S. Rétaux, 2016. Chapter Five - Evolutionary Genetics of the Cavefish Astyanax mexicanus In Foulkes, N. S. (ed), Advances in Genetics. Academic Press: 117–159, https://www.sciencedirect.com/science/article/pii/S0065266016300189.
Castro, J. P., M. O. Moura, O. Moreira-Filho, O. A. Shibatta, M. H. Santos, V. Nogaroto, M. R. Vicari, M. C. de Almeida & R. F. Artoni, 2014. Evidence of incipient speciation in Astyanax scabripinnis species complex (Teleostei: Characidae). Neotropical Ichthyology 12: 429–438. https://doi.org/10.1590/1982-0224-20130222.
Darriba, D., G. L. Taboada, R. Doallo & D. Posada, 2012. jModelTest 2: more models, new heuristics and parallel computing. Nature Methods 9: 772–772. https://doi.org/10.1038/nmeth.2109.
dos Santos, J. A., C. M. Soares & A. Bialetzki, 2020. Early ontogeny of yellowtail tetra fish Astyanax lacustris (Characiformes: Characidae). Aquaculture Research 51: 4030–4042. https://doi.org/10.1111/are.14746.
Eigenmann, C. H., 1913. Some results from an ichthyological reconnaissance of Colombia, South America—Part II, Indiana University Studies, Indiana:
Eigenmann, C. H. & A. Henn, 1916. Description of three new species of characid fishes. Annals of the Carnegie Museum 10: 87–90.
Evanno, G., S. Regnaut & J. Goudet, 2005. Detecting the number of clusters of individuals using the software structure: a simulation study. Molecular Ecology 14: 2611–2620. https://doi.org/10.1111/j.1365-294X.2005.02553.x.
Excoffier, L. & H. E. L. Lischer, 2010. Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Molecular Ecology Resources 10: 564–567. https://doi.org/10.1111/j.1755-0998.2010.02847.x.
Ferreira, D. G., S. C. Lima, W. Frantine-Silva, J. F. Silva, C. Apolinário-Silva, S. H. Sofia, S. Carvalho & B. A. Galindo, 2016. Fine-scale genetic structure patterns in two freshwater fish species, Geophagus brasiliensis (Osteichthyes, Cichlidae) and Astyanax altiparanae (Osteichthyes, Characidae) throughout a Neotropical stream. Genetics and Molecular Research. https://doi.org/10.4238/gmr15048124.
Foll, M., 2012. BayeScan v2.1: user manual. Ecology 20: 1450–1462.
Foll, M. & O. Gaggiotti, 2008. A genome-scan method to identify selected loci appropriate for both dominant and codominant markers: a Bayesian perspective. Genetics 180: 977–993. https://doi.org/10.1534/genetics.108.092221.
Frankham, R., C. J. A. Bradshaw & B. W. Brook, 2014. Genetics in conservation management: revised recommendations for the 50/500 rules, Red List criteria and population viability analyses. Biological Conservation 170: 56–63. https://doi.org/10.1016/j.biocon.2013.12.036.
Freitas-Lidani, K. C., R. A. Torres, H. M. F. Madeira, P. C. F. Carneiro, & J. E. Gabriel, 2018. Identificação de polimorfismos genéticos em populações nativas de lambaris Astyanax sp. B do Rio Iguaçu (estado do paraná, brasil) por análises de amplificação aleatória. Visão Acadêmica 19. https://revistas.ufpr.br/academica/article/view/61124.
Fricke, R., W. N. Eschmeyer, & J. D. Fong, 2023a. Eschmeyer’s catalog of fishes: Genera/Species by family/Subfamily. http://researcharchive.calacademy.org/research/ichthyology/catalog/SpeciesByFamily.asp.
Fricke, R., W. N. Eschmeyer, & R. Van Der Laan, 2023b. Eschmeyer’s catalog of fishes: Genera, Species, References. http://researcharchive.calacademy.org/research/ichthyology/catalog/fishcatmain.asp.
García-Alzate, C., C. DoNascimiento, F. A. Villa-Navarro, J. E. García-Melo, & G. A. Herrera-R., 2021. Capítulo 2 - Diversidad de peces de la cuenca del Rïo Magdalena, Colombia In Jiménez-Segura, L. F., & C. A. Lasso (eds), Peces de la cuenca del río Magdalena, Colombia: diversidad, conservación y uso sostenible. Instituto de Investigación de Recursos Biológicos Alexander von Humboldt. https://doi.org/10.15472/numrso.
García-Castro, K. L., J. D. Rangel-Medrano, R. M. Landínez-García & E. J. Márquez, 2021. Population genetics of the endangered catfish Pseudoplatystoma magdaleniatum (Siluriformes: Pimelodidae) based on species-specific microsatellite loci. Neotropical Ichthyology 19: e200120. https://doi.org/10.1590/1982-0224-2020-0120.
Garita-Alvarado, C. A., M. A. Garduño-Sánchez, M. Barluenga & C. P. Ornelas-García, 2021. Genetic and ecomorphological divergence between sympatric Astyanax morphs from Central America. Journal of Evolutionary Biology 34: 1752–1766. https://doi.org/10.1111/jeb.13933.
Garza, J. C. & E. G. Williamson, 2001. Detection of reduction in population size using data from microsatellite loci. Molecular Ecology 10: 305–318. https://doi.org/10.1046/j.1365-294X.2001.01190.x.
Gilbert, K. J. & M. C. Whitlock, 2015. Evaluating methods for estimating local effective population size with and without migration. Evolution 69(8): 2154–2166. https://doi.org/10.1111/evo.12713.
Guichoux, E., L. Lagache, S. Wagner, P. Chaumeil, P. Léger, O. Lepais, C. Lepoittevin, T. Malausa, E. Revardel, F. Salin & R. J. Petit, 2011. Current trends in microsatellite genotyping. Molecular Ecology Resources 11: 591–611. https://doi.org/10.1111/j.1755-0998.2011.03014.x.
Gutiérrez-Moreno, L. C., & A. C. de la Parra-Guerra, 2021. Capítulo 6 - Contaminación del agua de la cuenca del río Magdalena, Colombia y su relación con los peces In Jiménez-Segura, L. F., & C. A. Lasso (eds), Peces de la cuenca del río Magdalena, Colombia: diversidad, conservación y uso sostenible. Instituto de Investigación de Recursos Biológicos Alexander von Humboldt. https://doi.org/10.15472/numrso.
Hausdorf, B., H. Wilkens & U. Strecker, 2011. Population genetic patterns revealed by microsatellite data challenge the mitochondrial DNA based taxonomy of Astyanax in Mexico (Characidae, Teleostei). Molecular Phylogenetics and Evolution 60: 89–97. https://doi.org/10.1016/j.ympev.2011.03.009.
Hellberg, M. E., 1996. Dependence of gene flow on geographic distance in two solitary corals with different larval dispersal capabilities. Evolution 50: 1167–1175. https://doi.org/10.1111/j.1558-5646.1996.tb02357.x.
Herman, A., Y. Brandvain, J. Weagley, W. R. Jeffery, A. C. Keene, T. J. Y. Kono, H. Bilandžija, R. Borowsky, L. Espinasa, K. O’Quin, C. P. Ornelas-García, M. Yoshizawa, B. Carlson, E. Maldonado, J. B. Gross, R. A. Cartwright, N. Rohner, W. C. Warren & S. E. McGaugh, 2018. The role of gene flow in rapid and repeated evolution of cave-related traits in Mexican tetra, Astyanax mexicanus. Molecular Ecology 27: 4397–4416. https://doi.org/10.1111/mec.14877.
Hernández-Barrero, S., C. G. Barreto-Reyes, & M. Valderrama-Barco, 2021. Capítulo 9 - Presión de uso del recurso íctico por la pesca artesanal en la cuenca del río Magdalena, Colombia In Jiménez-Segura, L. F., & C. A. Lasso (eds), Peces de la cuenca del río Magdalena, Colombia: diversidad, conservación y uso sostenible. Instituto de Investigación de Recursos Biológicos Alexander von Humboldt. https://doi.org/10.15472/numrso.
Hill, W. G., 1981. Estimation of effective population size from data on linkage disequilibrium. Genetical Research 38: 209–216. https://doi.org/10.1017/S0016672300020553.
Hilsdorf, A. W. S., & E. M. Hallerman, 2017. Genetic Resources of Neotropical Fishes. Springer International Publishing, Cham, http://link.springer.com/https://doi.org/10.1007/978-3-319-55838-7.
Hinaux, H., K. Pottin, H. Chalhoub, S. Père, Y. Elipot, L. Legendre & S. Rétaux, 2011. A developmental staging table for Astyanax mexicanus surface fish and Pachón cavefish. Zebrafish 8: 155–165. https://doi.org/10.1089/zeb.2011.0713.
Ivanova, N. V., T. S. Zemlak, R. H. Hanner & P. D. N. Hebert, 2007. Universal primer cocktails for fish DNA barcoding. Molecular Ecology Notes 7: 544–548. https://doi.org/10.1111/j.1471-8286.2007.01748.x.
Jiménez-Segura, L. F., G. Galvis-Vergara, P. Cala-Cala, C. A. García-Alzate, S. López-Casas, M. I. Ríos-Pulgarín, G. A. Arango, N. J. Mancera-Rodríguez, F. Gutiérrez-Bonilla & R. Álvarez-León, 2016. Freshwater fish faunas, habitats and conservation challenges in the Caribbean river basins of north-western South America. Journal of Fish Biology 89: 65–101. https://doi.org/10.1111/jfb.13018.
Jiménez-Segura, L. F., J. Herrera-Pérez, D. Valencia-Rodríguez, I. Castaño-Tenorio, S. López-Casas, M. I. Ríos, Y. Rondón.Martínez, K. Rivera-Coley, J. Morales, M. F. Arboleda, S. Muñoz-Duque, & V. Atencio, 2021. Capítulo 4 - Ecología e historias de vida de los peces en la cuenca del río Magdalena, Colombia In Jimenez-Segura, & C. A. Lasso (eds), Peces de la cuenca del río Magdalena, Colombia: diversidad, conservación y uso sostenible. Instituto de Investigación de Recursos Biológicos Alexander von Humboldt. https://doi.org/10.15472/numrso.
Jombart, T., 2008. adegenet: a R package for the multivariate analysis of genetic markers. Bioinformatics 24: 1403–1405. https://doi.org/10.1093/bioinformatics/btn129.
Jost, L., 2008. GST and its relatives do not measure differentiation. Molecular Ecology 17: 4015–4026. https://doi.org/10.1111/j.1365-294X.2008.03887.x.
Joya, C. D., R. M. Landínez-García & E. J. Márquez, 2021. Development of microsatellite loci and population genetics of the catfish Pimelodus yuma (Siluriformes: Pimelodidae). Neotropical Ichthyology 19: e200114. https://doi.org/10.1590/1982-0224-2020-0114.
Kalinowski, S. T., M. L. Taper & T. C. Marshall, 2007. Revising how the computer program cervus accommodates genotyping error increases success in paternity assignment: Cervus likelihood model. Molecular Ecology 16: 1099–1106. https://doi.org/10.1111/j.1365-294X.2007.03089.x.
Keller, L. F. & D. M. Waller, 2002. Inbreeding effects in wild populations. Trends in Ecology & Evolution 17: 230–241. https://doi.org/10.1016/S0169-5347(02)02489-8.
Kopelman, N. M., J. Mayzel, M. Jakobsson, N. A. Rosenberg & I. Mayrose, 2015. Clumpak : a program for identifying clustering modes and packaging population structure inferences across K. Molecular Ecology Resources 15: 1179–1191. https://doi.org/10.1111/1755-0998.12387.
Landínez-García, R. M. & E. J. Márquez, 2016. Development and characterization of 24 polymorphic microsatellite loci for the freshwater fish Ichthyoelephas longirostris (Characiformes: Prochilodontidae). PeerJ 4: e2419. https://doi.org/10.7717/peerj.2419.
Landínez-García, R. M. & E. J. Marquez, 2018. Microsatellite loci development and population genetics in Neotropical fish Curimata mivartii (Characiformes: Curimatidae). PeerJ 6: e5959. https://doi.org/10.7717/peerj.5959.
Landínez-García, R. M. & E. J. Márquez, 2020. Population genetics of the fish Brycon henni (Characiformes: Bryconidae) using species-specific polymorphic microsatellite loci. Revista de Biología Tropical. https://doi.org/10.15517/rbt.v68i3.38405.
Landínez-García, R. M., J. C. Narváez & E. J. Márquez, 2020. Population genetics of the freshwater fish Prochilodus magdalenae (Characiformes: Prochilodontidae), using species-specific microsatellite loci. PeerJ 8: e10327. https://doi.org/10.7717/peerj.10327.
Lasso, C. A., M. D. Escobar, M. C. Catellanos, D. Valencia-Rodríguez, J. Campuzano, F. García, & L. Jimenez-Segura, 2021. Capítulo 8 - Peces introducidos en el río Magdalena y cuencas vecinas, Colombia In Jiménez-Segura, L. F., & C. A. Lasso (eds), Peces de la cuenca del río Magdalena, Colombia: diversidad, conservación y uso sostenible. Instituto de Investigación de Recursos Biológicos Alexander von Humboldt. https://doi.org/10.15472/numrso.
Leuzzi, M. S. P., F. S. de Almeida, M. L. Orsi & L. M. K. Sodré, 2004. Analysis by RAPD of the genetic structure of Astyanax altiparanae (Pisces, Characiformes) in reservoirs on the Paranapanema River, Brazil. Genetics and Molecular Biology 27: 355–362. https://doi.org/10.1590/S1415-47572004000300009.
Li, Y.-L. & J.-X. Liu, 2018. StructureSelector: A web-based software to select and visualize the optimal number of clusters using multiple methods. Molecular Ecology Resources 18: 176–177. https://doi.org/10.1111/1755-0998.12719.
Limeira, D. M., M. H. Santos, R. P. Mateus, M. C. de Almeida & R. F. Artoni, 2019. Genetic variability in a population of Astyanax scabripinnis: recent bottleneck and the possible influence of individuals with B chromosomes. Acta Scientiarum. Biological Sciences 41: e47323. https://doi.org/10.4025/actascibiolsci.v41i1.47323.
Limeira, D. M., M. H. Santos, R. P. Mateus, C. F. Ruas, M. C. de Almeida, O. Moreira-Filho & R. F. Artoni, 2022. Molecular data reveal a complex population genetic structure for Psalidodon scabripinnis (Teleostei: Characidae) in the Atlantic Rainforest. Brazil. Genetics and Molecular Biology 45: e20210048. https://doi.org/10.1590/1678-4685-GMB-2021-0048.
López-Casas, S., L. F. Jiménez-Segura, A. A. Agostinho & C. M. Pérez, 2016. Potamodromous migrations in the Magdalena River basin: bimodal reproductive patterns in neotropical rivers. Journal of Fish Biology 89: 157–171. https://doi.org/10.1111/jfb.12941.
Luikart, G. & J.-M. Cornuet, 1998. Empirical evaluation of a test for identifying recently bottlenecked populations from allele frequency data. Conservation Biology [wiley, Society for Conservation Biology] 12: 228–237. https://doi.org/10.1111/j.1523-1739.1998.96388.x.
Márquez, E. J., C. Restrepo-Escobar, F. A. Yepes-Acevedo, & J. E. Narváez Barandica, 2021. Capítulo 3 - Diversidad y estructura genética de los peces de la cuencuas del río Magdalena, Colombia In Jiménez-Segura, L. F., & C. A. Lasso (eds), Peces de la cuenca del río Magdalena, Colombia: diversidad, conservación y uso sostenible. Instituto de Investigación de Recursos Biológicos Alexander von Humboldt. https://doi.org/10.15472/numrso.
Matschiner, M., R. Hanel & W. Salzburger, 2009. Gene flow by larval dispersal in the Antarctic notothenioid fish Gobionotothen gibberifrons. Molecular Ecology 18: 2574–2587. https://doi.org/10.1111/j.1365-294X.2009.04220.x.
Meirmans, P. G., 2006. Using the AMOVA framework to estimate a standardized genetic differentiation measure. Evolution 60: 2399–2402. https://doi.org/10.1111/j.0014-3820.2006.tb01874.x.
Mello, R., T. C. Maniglia, S. M. A. P. Prioli & A. J. Prioli, 2015. Genetic and biogeographical relationships among species of Astyanax (Teleostei, Characidae) in Brazilian river basins. Genetics and Molecular Research 14: 15356–15364. https://doi.org/10.4238/2015.november.30.13.
Mirande, J. M., 2009. Weighted parsimony phylogeny of the family Characidae (Teleostei: Characiformes). Cladistics 25: 574–613. https://doi.org/10.1111/j.1096-0031.2009.00262.x.
Mirande, J. M., 2019. Morphology, molecules and the phylogeny of Characidae (Teleostei, Characiformes). Cladistics 35: 282–300. https://doi.org/10.1111/cla.12345.
Nelson, J. S., T. C. Grande, & M. V. H. Wilson, 2016. Fishes of the World. Wiley, Hoboken, New Jersey, http://lccn.loc.gov/2015037522.
van Oosterhout, C., W. F. Hutchinson, D. P. M. Wills & P. Shipley, 2004. MICRO-CHECKER: software for identifying and correcting genotyping errors in microsatellite data. Molecular Ecology Notes 4: 535–538. https://doi.org/10.1111/j.1471-8286.2004.00684.x.
Ornelas-García, C., O. Domínguez-Domínguez, & I. Doadrio, 2008. Evolutionary historyof the fish genus Astyanax Baird & Girard (1854) (Actinopterygii, Characidae) in Mesoamerica reveals multiple morphological homoplasies. BMC Evolutionary Biology 8: 340.
Panaram, K. & R. Borowsky, 2005. Gene flow and genetic variability in cave and surface populations of the mexican tetra, Astyanax mexicanus (Teleostei: Characidae). Copeia 2005: 409–416. https://doi.org/10.1643/CG-04-068R1.
Pascual, M., B. Rives, C. Schunter & E. Macpherson, 2017. Impact of life history traits on gene flow: A multispecies systematic review across oceanographic barriers in the Mediterranean Sea. PLOS ONE Public Library of Science 12: e0176419. https://doi.org/10.1371/journal.pone.0176419.
Peakall, R. & P. E. Smouse, 2012. GenAlEx 6.5: genetic analysis in Excel. Population genetic software for teaching and research–an update. Bioinformatics 28: 2537–2539. https://doi.org/10.1093/bioinformatics/bts460.
Peres, M. D. & M. dos S. Vasconcelos, & E. Renesto, 2005. Genetic variability in Astyanax altiparanae Garutti & Britski, 2000 (Teleostei, Characidae) from the Upper Paraná River basin, Brazil. Genetics and Molecular Biology 28: 717–724. https://doi.org/10.1590/S1415-47572005000500011.
Pérez-Rodríguez, R., S. Esquivel-Bobadilla, A. M. Orozco-Ruíz, J. L. Olivas-Hernández & F. J. García-De León, 2021. Genetic structure and historical and contemporary gene flow of Astyanax mexicanus in the Gulf of Mexico slope: a microsatellite-based analysis. PeerJ 9: e10784. https://doi.org/10.7717/peerj.10784.
Piry, S. & G. Luikart, 1999. BOTTLENECK: A computer program for detecting recent reductions in the effective population size using allele frequency data. Journal of Heredity 90: 502–503. https://doi.org/10.1093/jhered/90.4.502.
Piscor, D., A. P. B. Pozzobon, C. A. Fernandes, L. Centofante & P. P. Parise-Maltempi, 2019. Molecular clock as insight to estimate the evolutionary history and times of divergence for 10 nominal Astyanax species (Characiformes, Characidae): An evolutionary approach in species with 2n = 36, 46, 48, and 50 chromosomes. Zebrafish 16: 98–105. https://doi.org/10.1089/zeb.2018.1647.
Pritchard, J. K., M. Stephens & P. Donnelly, 2000. Inference of population structure using multilocus genotype data. Genetics 155: 945–959. https://doi.org/10.1093/genetics/155.2.945.
Puechmaille, S. J., 2016. The program structure does not reliably recover the correct population structure when sampling is uneven: subsampling and new estimators alleviate the problem. Molecular Ecology Resources 16: 608–627. https://doi.org/10.1111/1755-0998.12512.
Rangel-Medrano, J. D. & E. J. Márquez, 2021. Development of microsatellite loci and population genetics in the bumblebee catfish species Pseudopimelodus atricaudus and Pseudopimelodus magnus (Siluriformes: Pseudopimelodidae). Neotropical Ichthyology 19: e200053. https://doi.org/10.1590/1982-0224-2020-0053.
Rangel-Medrano, J. D., A. Ortega-Lara & E. J. Márquez, 2020. Ancient genetic divergence in bumblebee catfish of the genus Pseudopimelodus (Pseudopimelodidae: Siluriformes) from northwestern South America. PeerJ 8: e9028. https://doi.org/10.7717/peerj.9028.
Restrepo, J.D., A. Cárdenas-Rozo, J.F. Paniagua-Arroyave, & L. Jiménez-Segura., 2021. Capítulo 1 – Aspectos físicos de la cuenca del río Magdalena, Colombia: geología, hidrología, sedimentos, conectividad, ecosistemas acuáticos e implicaciones para la biota. In Jiménez-Segura, L. F., & C. A. Lasso (eds), Peces de la cuenca del río Magdalena, Colombia: diversidad, conservación y uso sostenible. Instituto de Investigación de Recursos Biológicos Alexander von Humboldt.
Restrepo-Escobar, N., A. J. Yepes-Acevedo & E. J. Márquez, 2021. Population genetics of three threatened catfish species in heterogeneous environments of the Cauca River, Colombia. Neotrop Ichthyol 19: e200040. https://doi.org/10.1590/1982-0224-2020-0040.
Rice, W. R., 1989. Analyzing tables of statistical tests. Evolution 43: 223–225. https://doi.org/10.2307/2409177.
Rossini, B. C., C. A. M. Oliveira, F. A. G. de Melo, V. A. de Bertaco, J. M. D. de Astarloa, J. J. Rosso, F. Foresti & C. Oliveira, 2016. Highlighting Astyanax species diversity through DNA barcoding. PLoS ONE 11: 67203. https://doi.org/10.1371/journal.pone.0167203.
Rotmistrovsky, K., W. Jang & G. D. Schuler, 2004. A web server for performing electronic PCR. Nucleic Acids Research 32: W108–W112. https://doi.org/10.1093/nar/gkh450.
Rozen, S. & H. Skaletsky, 2000. Primer on the world wide web for general users and for biologist programmers. In Krawets, S. & S. Misener (eds), Bioinformatics Methods and Protocols: Methods in Molecular Biology Humana Press, New Jersey: 365–386.
Ruiz-C, R. I., C. Román-Valencia, B. E. Herrera-Murcia, O. E. Peláez & A. Ermakova-A, 2011. Variación morfológica de las especies de Astyanax, subgénero Zygogaster (Teleostei, Characidae). Animal Biodiversity and Conservation 34: 47–66. https://doi.org/10.32800/abc.2011.34.0047.
Schmieder, R. & R. Edwards, 2011. Quality control and preprocessing of metagenomic datasets. Bioinformatics 27: 863–864. https://doi.org/10.1093/bioinformatics/btr026.
Schmitter-Soto, J. J., 2016. A phylogeny of Astyanax (Characiformes: Characidae) in Central and North America. Zootaxa 4109: 101. https://doi.org/10.11646/zootaxa.4109.2.1.
Schmitter-Soto, J. J., 2017. A revision of Astyanax (Characiformes: Characidae) in Central and North America, with the description of nine new species. Journal of Natural History 51: 1331–1424. https://doi.org/10.1080/00222933.2017.1324050.
Sofia, S. H., C. R. M. Silva, B. A. Galindo, F. S. Almeida, L. M. K. Sodré & C. B. R. Martinez, 2006. Population genetic structure of Astyanax scabripinnis (Teleostei, Characidae) from an urban stream. Hydrobiologia 553: 245–254. https://doi.org/10.1007/s10750-005-1106-4.
Soulé, M. E., 1980. Thresholds for survival: maintaining fitness and evolutionary potential - Chapter 9 In Soulé, M. E., & B. Wilcox (eds), Conservation Biology: An Evolutionary-Ecological Perspective. Sinauer, Sunderland: 151–169, http://ybfwrb.org/wp-content/uploads/2017/10/Soule_1980.pdf.
Steindachner, F., 1879. Ichthyologische Beiträge (VIII). Mathematisch-Naturwissenschaftliche Classe.
Stevanato, D. J. & A. Ostrensky, 2018. Ontogenetic development of tetra Astyanax lacustris (Characiformes: Characidae). Neotropical Ichthyology. https://doi.org/10.1590/1982-0224-20170073.
Strecker, U., 2003. Polymorphic microsatellites isolated from the cave fish: Astyanax fasciatus. Molecular Ecology Notes 3: 150–151. https://doi.org/10.1046/j.1471-8286.2003.00386.x.
Strecker, U., L. Bernatchez & H. Wilkens, 2003. Genetic divergence between cave and surface populations of Astyanax in Mexico (Characidae, Teleostei). Molecular Ecology 12: 699–710. https://doi.org/10.1046/j.1365-294X.2003.01753.x.
Strecker, U., B. Hausdorf & H. Wilkens, 2012. Parallel speciation in Astyanax cave fish (Teleostei) in Northern Mexico. Molecular Phylogenetics and Evolution 62: 62–70. https://doi.org/10.1016/j.ympev.2011.09.005.
Terán, G. E., M. F. Benitez & J. M. Mirande, 2020. Opening the Trojan horse: phylogeny of Astyanax, two new genera and resurrection of Psalidodon (Teleostei: Characidae). Zoological Journal of the Linnean Society. https://doi.org/10.1093/zoolinnean/zlaa019.
Valderrama-Barco, M., J. L. Escobar-Cardona, R. Pardo, M. Toro, J. C. Gutierrez, & S. López-Casas, 2021. Capítulo 5 - Servicios ecosistémicos generados por los peces de la cuenca del río Magdalena, Colombia In Jiménez-Segura, L. F., & C. A. Lasso (eds), Peces de la cuenca del río Magdalena, Colombia: diversidad, conservación y uso sostenible. Instituto de Investigación de Recursos Biológicos Alexander von Humboldt. https://doi.org/10.15472/numrso.
Villa-Navarro, F. A. & P. Sanchez-Duarte, 2014. Astyanax caucanus. The IUCN Red List of Threatened Species 2016. International Union for Conservation of Nature, http://www.iucnredlist.org/details/49829978/0.
Waples, R. S., 2006. A bias correction for estimates of effective population size based on linkage disequilibrium at unlinked gene loci*. Conservation Genetics 7: 167–184. https://doi.org/10.1007/s10592-005-9100-y.
Waples, R. S. & C. Do, 2010. Linkage disequilibrium estimates of contemporary Ne using highly variable genetic markers: a largely untapped resource for applied conservation and evolution. Evolutionary Applications 3: 244–262. https://doi.org/10.1111/j.1752-4571.2009.00104.x.
Ward, R. D., T. S. Zemlak, B. H. Innes, P. R. Last & P. D. N. Hebert, 2005. DNA barcoding Australia’s fish species. Philosophical Transactions of the Royal Society B: Biological Sciences 360: 1847–1857. https://doi.org/10.1098/rstb.2005.1716.
Yue, G. H., L. David & L. Orban, 2007. Mutation rate and pattern of microsatellites in common carp (Cyprinus carpio L.). Genetica 129: 329–331. https://doi.org/10.1007/s10709-006-0003-8.
Zaganini, R. L., D. T. Hashimoto, L. H. G. Pereira, C. Oliveira, F. F. Mendonça, F. Foresti & F. Porto-Foresti, 2012. Isolation and characterization of microsatellite loci in the Neotropical fish Astyanax altiparanae (Teleostei: Characiformes) and cross-species amplification. Journal of Genetics 93: 24–27. https://doi.org/10.1007/s12041-012-0143-9.
Acknowledgements
This study was granted by Empresas Públicas de Medellín framed under the project “Variabilidad genética de un banco de peces de los sectores medio y bajo del río Cauca” (CT-2019-000661).
Funding
Open Access funding provided by Colombia Consortium.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflicts of interest.
Additional information
Handling editor: Nicholas R. Bond
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Velandia, R.A., Campo-Nieto, O. & Márquez, E.J. Astyanax caucanus: microsatellite loci development and population genetics in the Cauca River, Colombia. Hydrobiologia 851, 2007–2024 (2024). https://doi.org/10.1007/s10750-023-05434-w
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10750-023-05434-w