Abstract
In motor imagery-based brain-computer interfaces (BCIs), the spatial covariance features of electroencephalography (EEG) signals that lie on Riemannian manifolds are used to enhance the classification performance of motor imagery BCIs. However, the problem of subject-specific bandpass frequency selection frequently arises in Riemannian manifold-based methods. In this study, we propose a multiple Riemannian graph fusion (MRGF) model to optimize the subject-specific frequency band for a Riemannian manifold. After constructing multiple Riemannian graphs corresponding to multiple bandpass frequency bands, graph embedding based on bilinear mapping and graph fusion based on mutual information were applied to simultaneously extract the spatial and spectral features of the EEG signals from Riemannian graphs. Furthermore, with a support vector machine (SVM) classifier performed on learned features, we obtained an efficient algorithm, which achieves higher classification performance on various datasets, such as BCI competition IIa and in-house BCI datasets. The proposed methods can also be used in other classification problems with sample data in the form of covariance matrices.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
The brain-computer interface (BCI) provides nonmuscular communication between human brains and external devices to aid people with motor impairments. It can be used to control a wheelchair, manipulator or mouse cursor on a computer screen, after decoding the electroencephalogram (EEG) signal from the cerebral scalp [1,2,3,4]. Different types of EEG modalities have been used to design multiple BCI systems [5,6,7,8]. Particularly, motor imagery BCI systems record EEG signals by imagining movements of different parts of the body, such as, the right hand, left hand, feet and tongue [9, 10]. The spatial and spectral information of motor imagery EEG signals can help identify the movement intention of the body. Therefore, the major challenge in motor imagery BCIs is the efficient extraction of the spatial and spectral features of EEG signals.
During the past decade, many methods to extract spatial and spectral features from motor imagery EEG signals have been proposed. In particular, the spatial covariance matrices of EEG signals are commonly used in feature extraction methods. The two types of feature extraction methods, based on covariance matrices, are the common spatial pattern (CSP)-based methods [11, 12] and Riemannian manifold-based methods [13, 14]. CSP-based methods, such as CSP, filter bankCSP (FBCSP) [15], sub-band CSP (SBCSP) [16], adaptive FBCSP (AFBCSP) [17], and the common sparse spectral-spatial pattern (CSSSP) [18], tend to extract features using by a spatial filter, which can maximize the variance in one class while minimizing the variance in the others. The original CSP obtains the spatial filter under a large bandpass frequency (8–30 Hz) that contains alpha and beta waves closely related to motor imagery. CSP strongly depends on the appropriate selection of subject-specific frequency bands. However, a large bandpass frequency band cannot distinguish the contributions of alpha or beta waves. Several CSP extensions have been proposed to address this problem. The FBCSP decomposes a larger frequency band into multiple sub-bands and learns the corresponding spatial filter on multiple sub-bands. In addition, it selects features from multiple sub-bands based on mutual information [15]. The SBCSP first obtains multiple after-filtering EEG signals by applying a Gabor filter to the multiple sub-frequency bands and then selecting discriminative features according to the sub-band score fusion techniques [16]. The AFBCSP designs the time-frequency map of the Fisher ratio to adaptively choose subject-specific frequency bands [17]. The CSSSP simultaneously optimizes the spatial filter and finite impulse response filter to learn the spectral-spatial features from the EEG signal [18]. Most CSP-based methods calculate the center of covariance matrices using the arithmetic mean. However, covariance matrices, with the symmetric positive definite form, lie in a Riemannian manifold in nature.
Riemannian manifold-based methods, such as Riemannian CSP [13], tangent space linear discriminant analysis (TSLDA) [14], bilinear sub-manifold learning (BSML) [19] and bilinear regularized locality preserving (BRLP) [20], attempt to project EEG signals from Euclidean space into Riemannian manifolds, where the relationship of samples is expressed by the Riemannian distance. Many efficient Riemannian manifold tools, such as the Riemannian mean and tangent space, can be applied to enhance the classification performance of motor imagery. Riemannian CSP recalculated the center of covariance matrices using the Riemannian mean and obtained the spatial filter through solving the joint diagonalization of mean covariance [13]. TSLDA extracted features by mapping the covariance matrices into tangent space, where the distance structure was consistent with the Riemannian manifold and the relationship between points was linear [14]. BSML designed a bilinear mapping framework for dimensionality reduction in covariance matrices. It learned low-dimensionality features by maximally preserving the global structure of the original manifold [19]. In contrast, BRLP is a locality-preserving dimensionality reduction method that attempts to preserve the similarities between vertex pairs on the Riemannian graph into embedding [20]. Although Riemannian manifold-based methods have been proposed to obtain efficient spatial features from EEG signals, they are primarily designed to project EEG signals into one Riemannian manifold corresponding to a large bandpass frequency, without considering frequency band selection in the Riemannian manifold mapping.
To address the issue-faced when using Riemannian manifold-based method, we propose a novel multiple Riemannian graph fusion method to combine multiple Riemannian manifolds corresponding to multiple bandpass frequency bands. As covariance matrices contain the spatial information of EEG signals, the proposed method attempts to obtain more spectral information and merge the spatial and spectral feature extraction into a unique framework. This unique framework is mainly composed of three parts: Riemannian graph construction on multiple frequency bands, graph embedding, and graph fusion. Many related works on graph fusion and motor imagery classification have been recently proposed. In [21], convolutional neural networks and graph convolutional networks were used to extract image-level features and relation-aware features from the images. Deep feature fusion was developed to fuse two types features to enhance the classifier performance. In [22], an adaptive spatiotemporal graph convolutional network was proposed to fully exploit the characteristics of EEG signals in the time domain and channel correlations in the spatial domain. In [23], a clustering based on a residual graph convolutional network was proposed to infer the possibility of a connection between a given node and its neighbors and achieve high clustering performance. However, the above methods fuse graphs on Euclidean space and ignore that the covariance matrices lie on the Riemannian manifold. The contributions of this study are threefold:
-
1) A novel framework of multiple graph fusion based on Riemannian geometry is proposed to extract the spatial and spectral features in motor imagery BCIs simultaneously. The proposed framework can be considered an extension of Riemannian manifold-based methods.
-
2) Insightful research on graph processing is proposed. Our method designs a fusion technique for the parallel processing of multiple graph embeddings. This is a significantly improved version of the traditional graph-embedding method.
-
3) The proposed method can efficiently alleviate the overfitting problem in the processing of motor imagery EEG signals using graph embedding and graph fusion.
The remainder of this paper is organized as follows. In Section 2, we provide more details on the multiple Riemannian graph fusion methods. In Section 3, we present extensive experimental results and discuss the findings. Finally, in Section 4, the conclusions are presented.
2 Materials and methods
In this section, some fundamental concepts of the space of symmetric positive-definite matrices and Riemannian geometry are briefly reviewed. In addition, the multiple Riemannian graph fusion method was proposed to learn the discriminative spectral-spatial features from motor imagery EEG signals.
2.1 Riemannian geometry
The spatial covariance matrix of the N-channel EEG signal \(\mathbf {X} \in \mathbb {R}^{N \times L}\) is represented by
where L is the number of sampled points in EEG trial X. The covariance matrix \(\mathbf {P} \in \mathbb {R}^{N \times N}\) lies in a space of symmetric positive-definite matrices, defined as
where \({\mathcal {S} }(N) = \left \{ \mathbf {P} \in \mathbb {R}^{N \times N},\mathbf {P} = {\mathbf {P}^{T}} \right \}\) is the space of positive-definite matrices and \({\mathcal {P} }(N)=\left \{ {\mathbf {P} \in {\mathbb {R}^{N \times N}},{\mathbf {u}^{T}}\mathbf {P}\mathbf {u} > 0,\forall \mathbf {u} \in {\mathbb {R}^{N}}} \right \}\) is the space of positive-definite matrices.
The space of symmetric positive-definite matrices endowed with the Riemannian metric is a differentiable Riemannian manifold \({{\mathscr{M}}}\)[24]. The concepts of Riemannian distance and tangent space play an important role in the application of Riemannian manifolds. Denoted by two symmetric positive-definite matrices \( {\mathbf {P}_{1}},{\mathbf {P}_{2}} \in {\mathcal {S}\mathcal {P}\mathcal {D}}(N) \), the Riemannian distance is defined as:
where ||⋅||F is the Frobenius norm of a matrix, and βi is the i-th real eigenvalue of \( {\mathbf {P}_{1}}^{- 1}{\mathbf {P}_{2}} \). The Riemannian distance is the minimum length of the curve connecting two points on a Riemannian manifold [25]. It satisfies three fundamental properties of the metric space: positivity, symmetry, and triangle inequality [24].
The tangent space of a Riemannian manifold is a linear space, that can often be used to study the nonlinearity of manifolds. The tangent space \({\mathcal {T}}(N) \) at P is defined as [26]
where P is a tangent point, and the upper(⋅) operator maintains the upper triangular part of the matrix and vectorizes it. The logarithmic mapping operator is denoted by \(Log_{\mathbf {P}}({\mathbf {P}_{i}})= {\mathbf {P}^{\frac {1}{2}}}\log \left ({\mathbf {P}^{- \frac {1}{2}}}{\mathbf {P}_{i}}{\mathbf {P}^{- \frac {1}{2}}}\right ){\mathbf {P}^{\frac {1}{2}}}\). In the neighborhood of P, the Riemannian distance between P and the nearby point Pi is almost identical to the Euclidean distance between the corresponding points on tangent space s,si[14]:
However, the neighborhood of P is a vague area. Generally, all samples from the dataset can be considered to be neighbors, whereas the mean of all samples is regarded as the tangent point P. The relationship between the Riemannian manifold and the tangent space is shown in Fig. 1.
2.2 Multiple Riemannian graph fusion
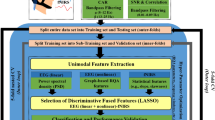
The framework of multiple Riemannian graph fusion algorithms is presented in Fig. 2. The overall framework includes a multiple Riemannian graph construction based on multiple frequency bands, multiple graph embedding for dimensionality reduction and graph fusion for feature selection.
2.2.1 Multiple Riemannian graph construction
The selection of an appropriate bandpass frequency band plays an important role in motor imagery classification. In this study, the EEG signal X was first bandpass filtered by three frequency bands–alpha band, beta band and total band, and the frequency components in the alpha and beta bands provided the best discrimination between the left and right-hand movement imagination [27]. In addition, to capture more information, the EEG signal was filtered by a large total frequency band that covered the alpha and beta bands. Three filtered signals \(\tilde {\mathbf {X}}^{(1)},\tilde {\mathbf {X}}^{(2)}, and \tilde {\mathbf {X}}^{(3)}\) were projected into three subsets of the Riemannian manifold (\({{\mathscr{M}}}^{(1)},{{\mathscr{M}}}^{(2)},{{\mathscr{M}}}^{(3)}\)). To learn the low-dimensional embedding of the Riemannian manifold, we constructed three Riemannian graphs (\({\mathcal {G}}_{l}^{(1)}, {\mathcal {G}}_{l}^{(2)}, {\mathcal {G}}_{l}^{(3)}\)) corresponding to three subsets on the Riemannian manifold. For each Riemannian graph \({\mathcal {G}}_{l}=({\mathcal {V}}, {\mathcal {E}})\), the vertices \(\mathcal {V}\) comprise all SPD matrices Pi in the l-th subset, and the edges \(\mathcal {E}\) contain adjacency and weights uij. The adjacency on \({\mathcal {G}}_{l}\) was designed using k-nearest neighbors with the Riemannian distance. The weight between two adjacent points Pi and \(\mathbf {P}_{j} \in \mathcal {V}\) is given by:
where \(d_{ij}=\delta _{R}\left (\mathbf {P}_{i},\mathbf {P}_{j} \right )\) and σ is a scaling factor.
2.2.2 Multiple graph embedding
For each Riemannian graph \({\mathcal {G}}_{l}\), we expect to design a bilinear mapping \(\mathbf {W} \in \mathbb {R}^{M \times N}\) and \(\mathbf {W}^{T} \in \mathbb {R}^{N \times M}\) to learn a low-dimensional embedding from a subset of Riemannian manifold. The learned low-dimensional embedding can be expressed as \({\mathbf {E}_{p}} = \mathbf {W}\mathbf {P}{{\mathbf {W}}^{T}} \in {\mathcal {S}\mathcal {P}\mathcal {D} }(M)\), where \(\mathbf {P} \in \mathbb {R}^{N \times N}\). This embedding is also a Riemannian sub-manifold.
The bilinear mapping matrices have many variations with different types of property preservation, such as distance preservation and locality preservation. In this study, we aim to learn bilinear mapping matrices by preserving the distance structure between a high-dimensional manifold and low-dimensional embedding. A reasonable bilinear mapping W, with respect to the minimum distance loss, can be obtained by solving the following objective function:
where C is the experimental dataset of matrices in the \({\mathcal {S}\mathcal {P}\mathcal {D}}(N)\). Eq. (6) can achieve an isometric mapping between the original Riemannian manifold and the low-dimensional sub-manifold. δR(Pi,Pj) represents the Riemannian distance of points (I,j) on the original Riemannian manifold, and \({\delta _{R}}(\mathbf {W}{\mathbf {P}_{i}}{{\mathbf {W}}^{T}},\mathbf {W}{\mathbf {P}_{j}}{{\mathbf {W}}^{T}})\) is the Riemannian distance of the mapped points on the low-dimensional sub-manifold. The mapping matrix, learned using by (6), can best preserve the distance structure between the manifold and its sub-manifold. The solution of (6) is a nonconvex problem that is difficult to solve. In previous works [19], we showed that the optimal mapping W of (6) is equivalent to the solution of the joint diagonalization of the mean covariance in the CSP algorithm. For the two-class classification problem, the solution to (6) is equivalent to the mapping error among the between-class and within-class points. In [19], we proved that the between-class distance can be approximated as the distance between the means of two classes set, particularly when the within-class variance is much smaller than the between-class distance. Therefore, we approximate optimization (6) as the minimum loss of distance between the mean covariance of the two classes. The solution can be obtained by joint diagonalization of the mean covariance.
2.2.3 Graph fusion
After learning multiple low-dimensional distance-preserving embeddings from multiple subsets, we constructed three new Riemannian graphs (\({\mathcal {G}}_{E}^{(1)},{\mathcal {G}}_{E}^{(2)},{\mathcal {G}}_{E}^{(3)}\)) corresponding to three embeddings. The vertices of \({\mathcal {G}}_{E}\) are comprise Ep, and the adjacency and weight are calculated using the Riemannian distance between two points on the embedding. Evidently, \({\mathcal {G}}_{E}\) is close to \({\mathcal {G}}_{l}\). However, multiple graphs include considerable redundant information, which leads to high computational costs and low classification performance. Thus, we propose a multiple graph fusion method to fuse multiple graphs \({\mathcal {G}}_{E}\) into a unique graph, that contains the most discriminative information from multiple embeddings.
In this study, multiple graph fusion refers to the fusion of the corresponding nodes on different graphs. As the SPD matrix form of the node on \({\mathcal {G}}_{E}\) is difficult to merge directly, we proposed vectorization processing for node Ep on \(\mathcal {G}_{E}\) by
where E is the Riemannian mean of the embedding. Notably, vectorization processing is a tangent space mapping in (4). Thus, such vectorization processing can maximally preserve the structure of the \({\mathcal {G}}_{E}\) using (7).
Next, we used mutual information to fuse the corresponding nodes on different graphs [15]. As shown in (7), a node on the Riemannian graph is represented by a tangent vector. In this study, we regarded the multiple-node fusion problem as an element selection from multiple tangent vectors. Because mutual information can measure arbitrary relations between variables and does not depend on transformations acting on the different variables, we calculated the mutual information of each element and selected the top k elements as the final fused nodes. Assume V(1), V(2) and V(3) are the node matrices of \({\mathcal {G}}_{E}^{(1)}\), \({\mathcal {G}}_{E}^{(2)}\), and \({\mathcal {G}}_{E}^{(3)}\), corresponding to the EEG signal X. The total matrix is formed as V = [V(1),V(2),V(3)]. The i-th column of V is the concatenation of the i-th node on \({\mathcal {G}}_{E}^{(1)}\), \({\mathcal {G}}_{E}^{(2)}\), and \({\mathcal {G}}_{E}^{(3)}\), and the j-th row on V is the jth element of the EEG signal. The mutual information of the j-th element can be computed as
where H(⋅) is the entropy calculation [15] and y is the label of the EEG signal X. Finally, we fuse the corresponding node by retaining elements with a high value of mutual information and removing elements with a low value. The nodes of the fusion graph can be regarded as spatial and spectral features for motor imagery classification. The pseudocode of the proposed algorithm is presented in Algorithm 1.

3 Results and discussion
In this section, to evaluate the effectiveness of the proposed MRGF method, the proposed algorithm was tested on two motor imagery datasets and compared against three competing methods.
3.1 Experimental setup
3.1.1 Data description
The EEG data used in this study were come from two motor imagery datasets, that is, the BCI competition IV dataset and an in-house dataset. The experimental settings of the two datasets were as follows.
-
1) 1) Dataset IIa of BCI competition IV included four types of motor imagery tasks (right hand, left hand, foot, and tongue imagined movements), which were performed on nine different subjects (S01-S09). The experimental protocol for dataset IIa is as follows. At the beginning of 0-2 s, the computer presented a short acoustic warning tone. After the sound, the screen shows an arrow pointing left, right, down, or up for a period of 1.25 seconds (2-3.25 s). In the period 3.25-6 s, the subjects were asked to perform a motor imagery task corresponding to the arrow. Finally, a short break of 1.5 s was given. This dataset consisted of 576 trials, recorded by 22 EEG channels. For one mental task, there were 72 training and 72 test trials. The sampling rate was set at 250 Hz.
-
2) Our in-house EEG data only included two types of mental tasks (left/right and imagined movements) that were performed on seven subjects (A01-A07) with 32 EEG channels. The experimental protocol for the in-house dataset was set as follows. At the initial stage 0-2.25 s, the screen remained blank. From 2.25-4 s, the screen shows a cross to attract the subject’s visual fixation. In the time period 4-8 s, a left/right arrow appears and prompts the subject to perform the required task. This dataset consisted of 234 trials. On one subject, 117 training and test trials each were conducted. The sampling rate was set at 250 Hz.
3.1.2 Algorithms evaluated
The MRGF was compared against the following competing algorithms:
-
1) A shrinkage estimator-based CSP was used to extract highly discriminative spatial features, and an enhanced one versus one structure was used to classify the EEG signals [28].
-
2) DPLM: Low-dimensional features, learned by distance preserving to local means (DPLM), were used to improve the performance of motor imagery [29].
-
3) MEMDBF: Multivariate empirical mode decomposition-based filtering (MEMDBF) was used to classify EEG signals into multiple classes [30].
-
4) ESVL: Ensemble support vector learning (ESVL) was used for feature combinations to improve classification performance [31].
-
5) LDA+TSSM: The LDA classifier was applied in the tangent space of the submanifold (TSSM) learned by the distance-preserving dimensionality reduction method [19].
-
6) Hybrid learning of transductive and inductive models was used to handle non-stationarities in motor imagery classification [32].
-
7) FBCSP: The 1st winner method for BCI competition IV. The recorded EEG signal was band-pass filtered by multiple sub-frequency bands of 4-8 Hz and 8-12 Hz..., 36-40 Hz. Then, the CSP algorithm was used to extract the spatial features from each sub-band. In addition, discriminative features were selected from spatial features based on mutual information. Finally, the naive Bayes Parzen window was used for classification [15].
-
8) CSP+LDA+Bayes: The 2nd winner method for BCI competition IV. The recorded EEG signal was band-pass filtered at 8-30 Hz. Then, the CSP algorithm was used to extract spatial features, and Fisher LDA was used to select features. Finally, a Bayesian classifier was applied for classification.
-
9) CSP+SVM: The 3rd winner method on BCI competition IV. The recorded EEG signal was band-pass filtered at 8-25 Hz. Standard CSP was applied to learn spatial features, and an ensemble support vector machine was used as a classifier to classify the features.
3.1.3 Parameters setting
The dimensions of embedding were set to 10 for the BCI competition dataset and 6 for the in-house dataset based on cross-validation. The number of selected features was set to 25 and 12. SVM is a built-in function of MATLAB, the parameters of the SVM classifier are set as linear kernels, and the penalty factor is set to 1. An analysis of the parameter settings is included in the following section.
3.2 Results and discussion
3.2.1 Classification results
As the nodes on the fusion graph have capture the spatial and spectral information of the motor imagery EEG signal, we regarded the nodes on the fusion graph as the feature vectors and applied SVM to classify it. To evaluate the classification performance, we tested the MRGF-SVM on the BCI competition and in-house datasets. Table 1 shows the kappa value of the MRGF-SVM and the nine competing algorithms on the BCI competition dataset. The kappa value is commonly adopted to evaluate the classification performance of the four-class problem in dataset IIa of competition IV because the kappa value considers the misclassification of multi-class problems. As shown in Table 1, MRGF-SVM achieved a mean kappa value of 0.616, which is the highest result in Table 1. More specifically, the MRGF-SVM was significantly higher than 2nd (p= 0.0012) and 3rd (p= 0.00041). There was no significant difference between the performance of the MRGF-SVM method and FBCSP (p = 0.072). However, the value of p is close to 0.05.
Furthermore, we compared the classification performance of the MRGF-SVM with the three competing methods on an in-house dataset. Because the in-house motor imagery BCI classifies right and left imagined movements (two-class problem), for simplicity, we used classification accuracy as a performance measure for the in-house dataset. As shown in Table 2, the accuracy of the MRGF-SVM method is higher than that of FBCSP, CSP+LDA+Bayes, and CSP+SVM by 8.4 %, 9.49 % and 10.9 %, respectively. Upon examination, all p< 0.05, and the results in Table 2 were statistically significant.
From the comparison of methods in Tables 1 and 2, the high performance of the proposed method might be attributable, in part, to the highly discriminative features learned by MRGF as the SVM classifier is also commonly used in other competing methods.
3.2.2 Discussion of graph structure
The proposed MRGF method constructs three graphs corresponding to three frequency sub-bands from a single dataset and fuses them into one unified graph. To reveal the principle of multiple graph fusion, we analyzed the changes in graph structures during the execution of the MRGF method. The structures of the graph can be expressed using the weight matrix of the graph U. The weight between the ith point and the ith point is calculated by
where dij is the distance of two points.
In Fig. 3, the trials of the left/right-hand imagined movements from the competition BCI dataset were selected to calculate the weight matrix. The abscissa of 1-72 represents the left-hand trials, and the abscissa of 73-144 represents the right-hand imagery trials. The ordinate is the same as the abscissa. Therefore, the high values in the top left and bottom right of the weight matrix indicate that the points of the graph have low within-class distances. The low values in the top right and bottom left lead to a high between-class distance. Figure 3 shows the weight matrix of the high-dimensional Riemannian graph, low-dimensional embedding graph, tangent space graph and fusion graph on the BCI competition datasets. The weight matrices of embedding (Fig. 3 (b)) and tangent space (Fig. 3 (c)) have higher values at the top left and bottom right than the weight matrix of the Riemannian graph (Fig. 3 (a)). Furthermore, the weight matrix of the fusion graph (Fig. 3 (d)) has the highest value at the top left and bottom right and the lowest value at the top right and bottom left. Figure 4 shows the weight matrix of the high-dimensional Riemannian graph, low-dimensional embedding graph, tangent space graph and fusion graph corresponding to the in-house datasets. The weight matrices in Fig. 4 are similar to those shown in Fig. 3. Based on the results of Figs. 3 and 4, we can infer that the graph embedding and graph fusion of MRGF can help obtain more discriminative features from EEG signals.
In addition, to provide more intuitive results (discriminative features), we calculate the distance of each point from two class-related means on a high-dimensional Riemannian graph, a low-dimensional embedding graph, a graph of tangent space, and a fusion graph. In Figs. 5 and 6, the distance from the right-hand mean is regarded as the abscissa, and the distance from the right and mean is regarded as the ordinate. Figures 5 (d) and 6 (d) have the most separability. Figures 5 (b,c) and 6 (b,c) are more separable than those in Figs. 5 (a) and 6(a). These results provide evidence for the higher discriminative graph structure observed in Figs. 3 and 4.
Separability of right/left hand trials on subject 3 on BCI competition dataset. a) separability of points on high-dimensional Riemannian graph, b) separability of points on low-dimensional embedding graph, c) separability of points on graph of tangent space, d) separability of points on fusion graph
3.2.3 Discussion of parameter influence
Finally, we analyze the influence of the parameters adopted within the MRGF method, such as the frequency of sub-bands, the number of selected features and the dimension of embedding.
- (I) Analysis of the frequency of sub-bands:
-
To find the optimal frequency of sub-bands, Figs. 7 and 8 show the short-term Fourier transform of the EEG signal from both the BCI competition and the in-house datasets. The time-frequency diagram of the short-time Fourier transform can be used to analyze the changes in the power spectrum during motor imagery, especially for event synchronization and desynchronization. After observing the time-frequency spectrum of the left/right-hand motor imagery modes in Figs. 7 and 8, we can clearly observe the phenomenon of synchronization and desynchronization, which appear in frequency bands of 7.5 Hz± 2.5 Hz\(\sim \)13.5 Hz± 2.5 Hz and 15.5 Hz± 2.5 Hz\(\sim \)25 Hz± 2.0 Hz In fact, these frequency bands are close to the μ and β rhythms. Therefore, the optimal frequency of the sub-band in the MRGF method depends on the frequency band, which can cause synchronization and desynchronization. In addition, to capture more information, we used a total band of 7-35 Hz as the sub-band frequency. Thus, three sub-bands of μ and β rhythms and the total band are used in the proposed method.
- (II) Analysis of selected features:
-
In graph fusion processing, we retain features with high mutual information values and remove the low-value features. The key problem that remains is how to determine the number of selected features. Figure 9 shows the mutual information entropy of the features and the ratio of the selected features to the total features. We rank the entropy value of the features from high to low. As shown in Fig. 9, a larger entropy ratio can be obtained when more features are selected. A large entropy ratio indicates that the selected features accurately represents the total features. However, if the number of selected features is too large, it will lead to high computational cost. Consequently, the number of selected features must be determined by achieving a trade-off between the degree of representation and computational costs. As shown in Fig. 9, we can obtain 25 for the BCI competition dataset and 12 for the in-house dataset.
- (III) Selection of dimension of embedding:
-
After setting the frequency of the sub-band and the number of selected features, we could determine the dimensions of embedding using a cross-validation procedure. Tables 3 and 4 show the cross-validation results of the BCI competition dataset and in-house dataset, while the dimension of the embedding changes. In Table 3, the highest mean accuracy of 70.11 % is obtained when the embedding dimension is 10. In Table 4, the highest mean accuracy of 86.34 % is obtained when the embedding dimension is 6.
Time-frequency analysis for subject 3 on BCI competition dataset. a) Time-frequency spectrum of electrode C3 in the left hand motor imagery; b) Time-frequency spectrum of electrode C4 in the left hand motor imagery; c)Time-frequency spectrum of electrode C3 in the right hand motor imagery; (d)Time-frequency spectrum of electrode C4 in the right hand motor imagery
Time-frequency analysis for subject 2 on in-house dataset. a) Time-frequency spectrum of electrode C3 in the left hand motor imagery; b) Time-frequency spectrum of electrode C4in the left hand motor imagery; c)Time-frequency spectrum of electrode C3in the right hand motor imagery; (d)Time-frequency spectrum of electrode C4in the right hand motor imagery
4 Conclusions
To extract the spatial and spectral features from EEG signals, we construct multiple Riemannian graphs corresponding to multiple sub-frequency bands and fuse them into a unified graph. Experimental results on the BCI competition and an in-house dataset show that the proposed MRGF can capture discriminative features and lead to high classification performance. The proposed methods can also be applied to many other pattern-recognition problems with input data in the form of SPD matrices.
References
Rebsamen B, Burdet E, Guan C, Zhang H, Teo CL, Zeng Q, Laugier C, Ang MH (2007) Controlling a wheelchair indoors using thought. IEEE Intell Syst 22(2):18
Birbaumer N (2006) Brain-computer-interface research: Coming of age
Curran EA, Stokes MJ (2003) Learning to control brain activity: A review of the production and control of EEG components for driving brain–computer interface (BCI) systems. Brain Cogn 51(3):326
Wolpaw JR, Birbaumer N, Heetderks WJ, McFarland DJ, Peckham PH, Schalk G, Donchin E, Quatrano LA, Robinson CJ, Vaughan TM (2000) Brain-computer interface technology: a review of the first international meeting. IEEE Trans Rehabilitation Eng 8(2):164
Wolpaw JR, Birbaumer N, McFarland DJ, Pfurtscheller G, Vaughan TM (2002) Brain–computer interfaces for communication and control. Clinical neurophysiology 113(6):767
Sharma N, Pomeroy VM, Baron JC (2006) Motor imagery: a backdoor to the motor system after stroke?. Stroke 37(7):1941
Cai F, Wang T, Wu J, Zhang X (2020) Handheld four-dimensional optical sensor. Optik 164001:203
Blankertz B, Dornhege G, Krauledat M, Müller KR, Curio G (2007) The non-invasive Berlin brain–computer interface: fast acquisition of effective performance in untrained subjects. Neuroimage 37(2):539
Pfurtscheller G, Neuper C, Flotzinger D, Pregenzer M (1997) EEG-based discrimination between imagination of right and left hand movement. Electroenc Clin Neurophys 103(6):642
Zhang R , Yao D, Valdés-Sosa PA, Li F, Li P, Zhang T, Ma T, Li Y, Xu P (2015) Efficient resting-state EEG network facilitates motor imagery performance. J Neural Eng 12(6):066024
Ramoser H, Muller-Gerking J, Pfurtscheller G (2000) Optimal spatial filtering of single trial EEG during imagined hand movement. IEEE Trans Rehabilitation Eng 8(4):441
Guger C, Ramoser H, Pfurtscheller G (2000) Real-time EEG analysis with subject-specific spatial patterns for a brain-computer interface (BCI). IEEE Trans Rehabilitation Eng 8(4):447
Barachant A, Bonnet S, Congedo M, Jutten C (2010) Riemannian geometry applied to BCI classification. In: Vigneron V, Zarzoso V, Moreau E, Gribonval R, Vincent E (eds) Latent variable analysis and signal separation. Lecture Notes in Computer Science. Springer-Verlag, Berli, pp 629–636
Barachant A, Bonnet S, Congedo M, Jutten C, Trans IEEE (2012) Multiclass brain–computer interface classification by Riemannian geometry. Biomed Eng 59(4):920
Ang KK, Chin ZY, Zhang H, Guan C (2008) Filter bank common spatial pattern (FBCSP) in brain-computer interface. In: IEEE International joint conference on neural networks (IEEE world congress on computational intelligence), vol 2008. IEEE, pp 2390–2397
Novi Q, Guan C, Dat TH, Xue P (2007) Sub-band common spatial pattern (SBCSP) for brain-computer interface. In: 2007 3rd International IEEE/EMBS Conference on Neural Engineering. IEEE, pp 204–207
Thomas KP, Guan C, Tong LC, Prasad VA (2008) An adaptive filter bank for motor imagery based brain computer interface. In: 2008 30th Annual international conference of the IEEE engineering in medicine and biology society. IEEE, pp 1104–1107
Dornhege G, Blankertz B, Krauledat M, Losch F, Curio G, Muller KR (2006) Combined optimization of spatial and temporal filters for improving brain-computer interfacing. IEEE Trans Biomedical Eng 53(11):2274
Xie X, Yu ZL, Lu H, Gu Z, Li Y (2017) Motor imagery classification based on bilinear sub-manifold learning of symmetric positive-definite matrices. IEEE Trans Neural Syst Rehabil Eng 25(6):504
Xie X, Yu ZL, Gu Z, Zhang J, Cen L, Li Y (2018) Bilinear regularized locality preserving learning on Riemannian graph for motor imagery bci. IEEE Trans Neural Syst Rehabil Eng 26(3):698
Wang S, Govindaraj V, Gorriz J, Zhang X, Zhang YD (2020) Covid-19 classification by FGCNet with deep feature fusion from graph convolutional network and convolutional neural network. Inf Fusion. https://doi.org/10.1016/j.inffus.2020.10.004
Sun B, Zhang H, Wu Z, Zhang Y, Li T (2021) Adaptive Spatiotemporal Graph Convolutional Networks for Motor Imagery Classification. IEEE Signal Process Lett PP(99):1
Qi C, Zhang J, Jia H, Mao Q, Song H (2021) Deep face clustering using residual graph convolutional network. Knowl-Based Syst 211:106561
Förstner W, Moonen B (2003) A metric for covariance matrices. In: Grafarend EW, Krumm FW, Schwarze VS (eds) Geodesy-the challenge of the 3rd millennium. Springer-Verlag, Berli, pp 299– 309
Moakher M (2005) A differential geometric approach to the geometric mean of symmetric positive-definite matrices. SIAM J Matrix Anal Appl 26(3):735
Tuzel O, Porikli F, Meer P (2008) Pedestrian detection via classification on Riemannian manifolds. IEEE Trans Pattern Anal Mach Intell 30(10):1713
Pfurtscheller G, Neuper C, Flotzinger D, Pregenzer M (1997) EEG-based discrimination between imagination of right and left hand movement. Electroencephalography Clin Neurophysiol 103(6):642
Sharbaf ME, Fallah A, Rashidi S (2017) Shrinkage estimator based common spatial pattern for multi-class motor imagery classification by hybrid classifier. In: 2017 3rd International conference on pattern recognition and image analysis (IPRIA)
Davoudi A, Ghidary SS, Sadatnejad K (2017) Dimensionality reduction based on distance preservation to local mean for symmetric positive definite matrices and its application in brain–computer interfaces. J Neural Eng 14(3):036019. https://doi.org/10.1088/1741-2552/aa61bb
Gaur P, Pachori RB, Wang H, Prasad G (2018) A multi-class EEG-based BCI classification using multivariate empirical mode decomposition based filtering and Riemannian geometry. Expert Syst Appl 95(APR.):201
Luo J, Gao X, Zhu X, Wang B, Lu N, Wang J (2020) Motor imagery EEG classification based on ensemble support vector learning. Computer Methods Programs Biomed 193:105464
Raza H, Cecotti H, Prasad G (2016) A combination of transductive and inductive learning for handling non-stationarities in motor imagery classification. In: International Joint Conference on Neural Networks
Acknowledgements
This work was supported in part by the Natural Science Foundation of Hainan under grant 2019RC165, the National Natural Science Foundation of China (No. 61906048), Guangdong Basic and Applied Basic Research Foundation (No. 2020A1515010350), and the Young Talents Science and Technology Innovation Project of Hainan Association for Science and Technology under grant QCXM202011.
Funding
This work was supported in part by the Natural Science Foundation of Hainan under grant 2019RC165, the National Natural Science Foundation of China (No. 61906048), Guangdong Basic and Applied Basic Research Foundation (No. 2020A1515010350), and the Young Talents Science and Technology Innovation Project of Hainan Association for Science and Technology under grant QCXM202011.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of Interests
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Xie, X., Zou, X., Yu, T. et al. Multiple graph fusion based on Riemannian geometry for motor imagery classification. Appl Intell 52, 9067–9079 (2022). https://doi.org/10.1007/s10489-021-02975-2
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10489-021-02975-2