Abstract
Early detection of diabetic retinopathy is a serious disease for diabetics to minimize their sightlessness risks. The different approaches take a much longer time for a very large training dataset. In classifying to better the accuracy of diabetic retinopathy, a novel technique called MAP Concordance Regressive Camargo’s Index-Based Deep Multilayer Perceptive Learning Classification (MAPCRCI-DMPLC) has been introduced with minimum time consumption. The novel model of MAPCRCI-DMPLC comprises the input layer, hidden layers, and output layer for detecting diabetic retinopathy at an early stage through high accuracy and less moment consumption. The proposed MAPCRCI-DMPLC model collected the retinal fundus images from the dataset as input. After that, we carried out image preprocessing using the MAP-estimated local region filtering-based preprocessing technique in the first hidden layer. In the second hidden layer, Camargo’s index-based ROI extraction is performed to identify the infected region. Then, Concordance Correlative Regression is applied for texture feature extraction. Then the color feature is extracted, beginning the image. The features extracted to the output layer to classify the different levels of DR using the swish activation function through higher accuracy. An investigational assessment using a retinal image dataset on factors such as peak signal-to-noise ratio (PSNR), disease detection accuracy (DDA), false-positive rate (FPR), and disease detection time (DDT), regarding the quantity of retinal fundus images and image dimension. The quantitative and qualitatively analyzed outcome shows a better presentation of our proposed MAPCRCI-DMPLC technique when compared through the five state-of-the-art approaches.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
People with diabetes are prone to an eye disease named “Diabetic retinopathy”.DR is a well-known reason for blindness and severe eye diseases in citizens who have a medical condition from diabetes. The study by International Diabetes Federation demonstrates that the prevalence of DR is increasing day by day. It is considered that DR has ranked in the first place in diseases threatening the eye health of human beings. One might not experience symptoms in diabetic retinopathy. The symptoms include such as poor night vision, fluctuating vision, unable to differentiate colors, and so on. The disease has no obvious symptoms at early stages and may lead to chronic destruction, thus causing permanent blindness if not detected at an early stage. It may lead to permanent loss of vision.
Diagnosing at an early stage can assist one self in saving their vision. Early detection of the disease plays a crucial role in preventing vision loss for treatment of patients with minimal time periods. The conventional automated system consists of many stages, like the detection and categorization of lesions starting with fundus images. Therefore, MLT based on top of automatic removal has been presented for DRD. However the existence of many small features in the images causes a complex grading of severity and a slow progression for clinicians. New developments like deep learning methods are used to provide better achievement than conventional MLT for DRD.
Diabetic retinopathy is a disease that damages the eyes and is caused as a result of diabetes. If blood sugar levels aren't maintained for an unlimited period of time, the disease can extend. It is mainly caused by the damage of blood vessels in the retina. Retinopathy is the major cause of blindness in the world. Doctors can diagnose blindness before it occurs using Deep Learning model. To prevent diabetic patients from becoming blind, early diagnosis and precise detection of DR are very important. Deep learning is employed for classification in DR detection.
Deep learning (DL) is a branch of machine learning techniques for detecting, predicting, and classifying diabetic retinopathy images. DL applications to medical image analysis comprise the classification, feature selection, and detection. The classification is the machine learning model that accurately classifies the image. In the proposed algorithm, we have designed MAPCRCI-DMPLC to accurately classify the detected fundus image into one of the five stages of DR. The goal of the study is to automatically discover and classify the DR diagnosis by using a novel deep neural network model named MAP Concordance Regressive Camargo’s Index-Based Deep Multilayer Perceptive Learning Classification (MAPCRCI-DMPLC) technique.
-
Most significant symptoms of the disease are considered and irrelevant symptoms are eliminated for accurate classification.
-
A novel deep learning model has proven to be time-saving and efficient as compared to the standard existing method.
-
The main reason for the early detection of DR disease is to treat and cure vision loss in medical practitioners.
The main novelty and contribution of this work are listed below:
-
To propose a novel method called, MAPCRCI-DMPLC technique has been developed based on Deep Multilayer Perceptive Learning Classification that includes the preprocessing, ROI extraction, feature extraction, and classification for the early stage of DR detection.
-
Contrary to existing methods, we present a novel preprocessing model using the MAP estimated local region filtering technique in a filtering window concept to remove the noise pixels and increase the image quality. Also, it is providing good results. As a result, minimizes the mean square error (MSE) and increases the peak signal-to-noise ratio (PSNR).
-
Contrary to existing methods, ROI extraction and feature extraction are performed in MAPCRCI-DMPLC. First, the innovation of the Camargo index is applied for ROI extraction to identify the infected region based on pixel similarity estimation. Moreover, the innovation of Concordance Correlative Regression is applied for texture feature extraction. Subsequently, color features are extricated from the input image. With this, disease detection time is said to be minimized.
-
Contrary to traditional methods, the Gradient function is applied to a novelty of the swish activation function to analyze the extracted and testing features for correctly and precisely classify the processed color fundus images. With this, the accuracy and specificity are said to be improved.
-
Contrary to conventional methods, a novel gradient descent function is employed in the proposed MAPCRCI-DMPLC technique for finding a minimal error. The weight gets updated as well as attains precise outcomes by lesser error, depending on the error value. With this, incorrect classifications are minimized.
-
A sequence of experiments was conducted to estimate the quantitative analysis and qualitative analysis along with different results in metrics.
The rest of the paper is structured as follows. Section 2 reviews the related works for diabetic retinopathy discovery. Section 3 the proposed MAPCRCI-DMPLC is described. Section 4 presents experimental settings with the dataset explanation. Section 5 results in the quantitative and qualitative analysis of the investigational results with different results in metrics. Section 6 discusses the proposed method with the state-of-the-art methods. Finally, Section 7 concludes and features work in this paper.
2 Related works
Effective feature extraction, (Luo et al. 2021) proposed Multi-View DRD by Combining DCNNS and Attention Mechanisms (MVDRNet). However, the designed mechanisms failed to train a network with the lesion explanation to get better results. The multi-scale shallow CNN-based incorporated model was introduced by Chen. (2020) for the early recognition of DR from end to end in the arrangement of retinal images. However, the designed model failed to further pick up the classification precision.
A CNN was designed in (Martinez-Murcia et al. 2021) for the regular diagnosis of DR. But, the designed network was not efficient enough to improve the automatic diabetic retinopathy grading systems in clinical use. Deep CNN (MPDCNN) was developed in (Deepa et al. 2022) for efficient recognition using fundus images. However, it failed to select more advanced neural network architectures for more accurate automatic diabetic retinopathy discovery. The deep learning architecture was designed in (Das et al. 2021) based scheduled segmented fundus image features for the categorization of DR. A reformed capsule network was introduced by (Kalyani et al. 2023) for the categorization of DR based on feature extraction from the fundus images. However, an early and better diagnosis was not performed to avoid vision loss.
A novel DRD method was developed (Oh et al. 2021) based on top fundus photography and deep learning techniques. However, the designed method failed to set an ROI from the entire captured area for retinopathy detection to minimize the complexity. A deep learning- technique was developed by (Erciyas and Barısci 2021) to automatically detect diabetic retinopathy lesions based on ROI extraction. However, the designed algorithms failed to use the minimum system resources.
Automatic recognition of retinopathy detection was developed by (Qureshi et al. 2021) based on the ML architecture of dynamic deep learning technique. However, the designed architecture failed to enlarge the multi-layer architecture to a variety of multimedia applications. In (Liu et al. 2021), the deep symmetric CNN method was developed to detect diabetic retinopathy. However, the designed method did not accurately detect diabetic retinopathy from the images.
A multitask deep learning model was introduced by (Majumder and Kehtarnavaz 2021) to detect different types of diabetic retinalopathy. However, it failed to apply the real-time accomplishment of this model. They developed a multi-scale attention network for diabetic retinopathy categorization (Al-Antary and Arafa 2021). A bio-inspired approach was developed by (Vives-Boix and Ruiz-Fernández 2021) based on convolutional neural networks. However, the designed approach was not efficient for using large data sets for diabetic retinopathy detection. The CNN-singular value decomposition (CNN-SVD) was introduced for binary and multiclass diabetic retinopathy classification (Nahiduzzaman et al. 2021). However, the time consumption for detecting the different stages of diabetic retinopathy diagnosis was not reduced.
A VGG-network-in-network (NiN) model was developed by (Khan et al. 2021) for diabetic retinopathy images at an earlier stage. However, it failed to apply the preprocessing techniques for improving the categorization of diabetic retinopathy stages. In (Sugeno et al. 2021), a transfer learning system based on a recent CNN was developed to identify the severity grading system for diabetic retinopathy. A convolutional neural network (CNN)-based move toward diagnosing DR and its severity from fundus images was described in (Sridhar et al. 2021). However, the designed approach failed to use additional accessible algorithms for feature extraction and classification to enhance accuracy and efficiency (Muthusamy and Sathyamoorthy 2022). They developed a LEQHF with an LSTMN for detecting diabetic retinopathy (Ashir et al. 2021). Deep DR, in deep learning technique, was developed by (Dai et al. 2021) for the early detection of diabetic retinopathy. However the error rate during the diabetic retinopathy detection was not reduced. Deep CNN was introduced (Hemanth et al. 2020) for diagnosing DR beginning retinal fundus images. However, the performance of accurately diagnosing diabetic retinopathy with minimum complexity was not achieved.
Deep Feature Extraction CNN was introduced by (Sungheetha and Sharma 2021) to improve the accuracy of image classification. Trilateral filterative hermitian feature are perfectly extracted (Muthusamy and Rakkimuthu 2022). A hierarchical multi-task deep learning structure was developed by (Wang et al. 2020) for obtaining precise outcomes in DR detection. Enhanced automated detection was introduced in (Lin et al. 2018) with higher machinery detection accuracy. An automatic diabetic retinopathy screening method was designed (Zhu et al. 2019) with the aid of color fundus images (Ozbay 2023). However, the noise was not removed. For automatically determining the DR, the Deep Learning Ensemble Approach was discussed in (Qummar et al. 2019). However, the detection accuracy was not enhanced. The DL-based method was introduced by (Erciyas and Barısci 2021) to find diabetic retinopathy disease.
A novel automated Hyperparameter Tuning Inception-v4 (HPTI-v4) model was developed by (Shankar et al. 2020) to classify the fundus images. But, the detection time was advanced. A hybrid DL model called DRISTI was discussed by (Kumar et al. 2021) for discovering DR. An automatic DR detection model was designed by (Hacisoftaoglua et al. 2020) for enhancing the performance of DR detection. Deep learning-based techniques were introduced by (Bhuiyan et al. 2021) to perform screening with maximum accuracy. The designed technique failed to perform feature extraction. The diagnostic efficiency was enhanced in (Chen et al. 2021) by using the General deep learning model. However, the false-positive rate was not reduced. Automated detection based on machine intelligence was introduced in (Nawaz et al. 2021) for precise diagnostic decisions. However, if failed to eliminate the noisy pixel. The hybrid DL approach was introduced by (Menaouer et al. 2022) to automatically discover DR. The preprocessing was applied to eradicate the noisy images. But, the image quality was better (Muthusamy and Sathyamoorthy 2023). An automatic deep learning-based hemorrhage detection method was developed by (Aziz et al. 2023) to minimize the time. The designed method performance outcomes were more sufficient (Muthusamy and Rakkimuthu 2022). Deep recurrent neural networks were examined by (Gunasekaran et al. 2022) for diabetic retinopathy recognition. However, the specificity was not improved.
Generative adversarial network (GAN) was employed in (Mithun et al. 2022) for precise retinal vessel identification. A deep ensemble model was introduced by (Poonguzhal et al. 2022) with a stacking ensemble learning technique for achieving optimum performance. An optimal configuration was chosen by a two-stage ensemble selection method. Nevertheless, it failed to lessen time. To address the issue, deep learning based 19 layer U-net architecture was developed (Yellamelli et al. 2022) for enhancing the image quality. Deep learning-based semantic segmentation techniques were developed by (Mithun et al. 2021) employing medical image. Multiscale Independent component analysis was utilized in (Malaya et al. 2013) for feature identification. However, the accuracy was not enhanced.
A hierarchical severity level grading (HSG) system was investigated (Bhardwaj et al. 2021a, b, c, d) for classifying the retinal images. However, time was not enhanced. Transfer learning-based automated DR detection system was discussed by (Bhardwaj et al. 2021a, b, c,d) with higher accuracy. However, it failed to classify the different diabetic retinopathy levels such as mild, moderate, and severe. Quadrant-based model employing the Inception-V3 CNN model (Quadrant-based Inception-V3) was introduced by (Bhardwaj et al. 2021a, b, c, d) to offer DR diagnosis. But, the accuracy was not enhanced. The quadrant ensemble automated DR grading approach was analyzed by (Bhardwaj et al. 2021a, b, c, d) with enhancing network performance. Various image pre-processing techniques were developed by (Bhardwaj et al. 2021a, b, c, d) for determining the objects. However, feature extraction and classification were not performed.
Diabetic retinopathy is vital to automatically find diabetic disease. A traditional DL method is clinically helpful for medical applications and diabetic disease identification. Several DL diabetic disease detection methods were developed to determine the DR detection. However, the accuracy was not enough for the classification process. In the existing method, the preprocessing was unable to eradicate the noisy pixel. The classification approaches for DR extracts the complex feature attributes which increases the time complexity. In addition, feature extraction was not performed for dissimilar features.
To address this issue, a novel deep learning technique named MAPCRCI-DMPLC technique is needed to perform three different steps such as preprocessing, feature extraction, and classification. The MAPCRCI-DMPLC is designed with the innovation of the MAP estimated local region filter, Camargo’s index, Concordance Correlative Regression and swish activation function. This innovation of the proposed method is very helpful in reducing the computational complexity of the network and provides precise classification outcomes. In addition, the preprocessing is performed by MAP-estimated local region filtering to eliminate noisy pixels. Feature extraction is carried out on ROI, texture, and color features. The classification is employed for classifying the image with higher accuracy and less time. The DR classification issue is addressed through deep learning methods that automatically categorize the DR levels. The performance of the proposed method is done over the retinal image dataset. Comparative analysis with other deep learning and classification algorithms shows that our proposed MAPCRCI-DMPLC outperforms the state-of-the-art approaches.
3 Proposal methodology
The proposed model of diabetic retinopathy is a significant retinal disease to causes failure of sight in diabetic patients. Appropriate screening of retinal images decreases the chance of vision failure. However, physical screening of retinal images in the early hour’s stages of diabetic retinopathy is time-consuming. The proposed model of the deep neural network is designed for accurate detection and measurement of diabetic retinopathy using fundus images. A novel proposed MAPCRCI-DMPLC technique for early detection of diabetic retinopathy disease by means of retinal colored images has been developed. This MAPCRCI-DMPLC technique focuses on classifying diabetic retinopathy in different stages with the minimum possible learnable features to speed up the training and model convergence. The MAPCRCI-DMPLC also provides an accurate computer-aided diagnosis technique to maintain the clinical diagnosis with a minimum amount of time.
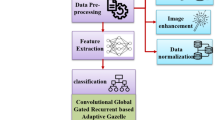
Overall architecture of the MAPCRCI-DMPLC technique step-by-step procedure to identify diabetic retinopathy disease at earlier stages is shown in Fig. 1. The deep multilayer perceptive neural learning classifier consists of an input layer, hidden layers, and an output layer. The proposed multilayer perceptron is a type of deep neural network that consists of various layers of perceptrons. The multilayer consists of three or more layers with nonlinearly-activating nodes. Every node in one layer is fully connected to another layer with a certain weight. The proposed MAPCRCI-DMPLC technique is designed with three dissimilar steps, are preprocessing, feature extraction, and classification. Initially, the Retinal fundus image dataset is considered. The number of retinal images \({FI}_{1},F{I}_{2},{FI}_{3},\dots .{FI}_{n}\) are collected from the large dataset and given to the input layer of a deep multilayer perceptive neural learning classifier. The input is sent to the first hidden layer.
After, the MAP estimated local region filtering-based preprocessing technique is employed to perform preprocessing in the first hidden layer for eradicating the noisy pixel and enhancing the image quality with higher PSNR and lesser MSE. Followed by feature extraction is said to be performed in the second hidden layer. The infected region is determined by means of Camargo’s index-based ROI extraction. Then, Texture feature extraction is executed with Concordance Correlative Regression. The color feature is extracted with decreased dimensionality of the image and time complexity. Finally, the classification is performed using the swish activation function technique in the output layer for investigating extracted features and tested disease features. The dissimilar levels of diabetic retinopathy namely normal, mild, moderate, severe, and proliferative correctly determined. As a diabetic retinopathy detection is performed with maximum accuracy and minimum time.
Then the retinal images are given to the input layer. The activity of the neuron in the input layer ‘z \(\left(t\right)\)’ is expressed as follows,
From the above Eq. (1), Where ‘\(c\)’ indicates a bias that stored the value is ‘1’,\({FI}_{i}\left(t\right)\) indicates input images, \({\tau }_{ih}\)’ symbolizes the weight between input at the input layer. Then the input layer is transferred into the first hidden layer.
3.1 MAP estimated local region filtering-based preprocessing
The first process of the proposed MAPCRCI-DMPLC technique is to perform image preprocessing to enhance the image quality. In general, there are various types of noise that appear in the input fundus images. Noise may occur due to different factors, such as image acquisition, transforming images, and so on. This kind of noise affects the image illustration and decreases the classification accuracy. Hence, it is necessary to remove the high-frequency noise and provide a better visual interpretation. An efficient technique called MAP estimated local region filtering is introduced in this section to enhance the image quality by minimizing the mean square error and increasing the peak signal to noise ratio.
Figure 2 illustrates the different steps of image preprocessing using the MAP estimated local region filtering technique. Let us consider the input fundus images \({FI}_{1},F{I}_{2},{FI}_{3},\dots .{FI}_{n}\) and the pixels are denoted by \({\varphi }_{1,}{\varphi }_{2},{\varphi }_{3},\dots {\varphi }_{m}\). The pixels are arranged and class the pixels in a filtering window. The center assessment of the pixels is selected from the window by organizing the pixels in ascending order. Then the maximum likelihood between the center pixels and the neighboring pixels (i.e. local region) is measured as given below,
From the above Eqs. (2) and (3), Where, \(L \left( { \varphi }_{i}|{ \varphi }_{c}\right)\) indicates a likelihood between the center pixel ‘\({\varphi }_{c}\)’ and the neighboring pixels ‘\({\varphi }_{i}\)’, \(\sigma\) indicates a deviation between the pixels, \(m\) denotes the number of pixels. \(F\) denotes a filtered output, \(arg max\) denotes an argument of maximum function. Based on estimation, the pixels that deviate from the center pixels are filtered to obtain quality enhanced images.
3.2 Camargo’s index-based ROI extraction
After the image preprocessing, ROI (region of interest) extraction is performed to visualize the object details more clearly. Instead of extracting the features from the whole image, the infected regions are extracted beginning of the image. This process helps to get better the accurate in disease diagnosis with minimum time complexity.
Figure 3 shows the ROI extraction and Feature extraction process. The ROI extraction is performed using pixel-level segmentation based on Camargo’s index. The Camargo’s index is used to find the pixel similarity for identifying the infected region. The pixels are denoted by \({\varphi }_{1,}{\varphi }_{2},{\varphi }_{3},\dots {\varphi }_{m}\) and Camargo’s index-based similarity is measured as follows,
From the above Eq. 4, Where ‘\(S\)’ denotes a similarity output, \({\varphi }_{i}\) denotes pixels, \({\varphi }_{j}\) indicates the neighboring pixels, \(k\) indicates the total number of pixels. Based on the similarity measure, similar pixels are identified and extract the region of interest.
3.3 Concordance correlative regression-based texture feature extraction
After extracting the ROI, the features are extracted from the particular region. The texture feature classification of extraction is performed in the third hidden layer to minimize the complexity of disease classification. The feature plays a significant role in image processing. Feature extraction is a significant step in the construction of disease classification, with the aim of extracting the relevant information from objects to form the feature vectors. These extracted feature vectors are then applied for further processing. The proposed technique first extracts the texture feature from the infected regions using Concordance Correlative Gaussian Process Regression. Regression is a machine learning technique that involves dependent variables (i.e., image pixels) and predictor variables (i.e., the mean of the pixel intensity). The regression worked on the basis of Gaussian distributive parameters such as mean and deviation with the help of Concordance Correlation. The texture feature provides information about the spatial representation.
From the above mathematical Eq. 5, ‘\(T\)’ indicates the texture feature, \({m}_{i}\) and \({m}_{j}\) indicates a mean of the pixels \({\varphi }_{i}, { \varphi }_{j}\) and deviation of the pixels are \({\sigma }_{i}\) and \({\sigma }_{j}\).
3.4 Color feature extraction
Color is the significant visual feature extracted from the image. The color feature classification of extraction is performed in the fourth hidden layer by transforming the given RGB color image \({\beta }_{r},{\beta }_{g},{\beta }_{b}\) into the hue (\({\delta }_{h}\)), saturation (\({\delta }_{s})\), value (\({\delta }_{v})\) as given below,
From the above Eqs. (6), (7) and (8), \({m}_{x}\) denotes a maximum pixel, \({m}_{n}\) indicates a minimum pixel. The hue color is obtained in degree (\({\delta }_{h}\epsilon \left[\text{0,360}\right]\)),\({\delta }_{s}\epsilon \left[\text{0,1}\right]\),\({\delta }_{v}\epsilon \left[\text{0,1}\right]\). By using (6), (7) and (8), the color features are extracted at the hidden layer. The hidden layer output is given by,
From the above Eq. (9), where, ‘\(R\left(t\right)\)’ denotes the hidden layer result, ‘\({\tau }_{ih}\)’ denotes the weight between the input layer and the hidden layer,\({\tau }_{h}\) denotes the weight of the hidden layer, \({r}_{i-1}\) and denotes an output layer of the previously hidden layer. After the feature extraction, the disease severity level is estimated at the output layer.
From the above Eq. (10), ‘\(Y\left(t\right)\)’ denotes the output result, ‘\({\tau }_{ho}\)’ represents the weight allocated between the hidden layer and output layer, \(h\left(t\right)\) denotes a hidden layer output, \(\omega\) and indicates an activation function. The swish activation function of a node generates the output of that node according to the set of inputs (i.e. extracted features). By applying the swish activation function, the sigmoid function is multiplied by its input.
From the above Eq. (11), Where, \({E}_{f}\) denotes an extracted feature, \({t}_{f}\) denotes a testing feature, \(\theta\) denotes a trainable parameter \(( \theta =1)\). The output of the swish activation function returns the value range from 0 to 1.
From the above Eq. (12), in this way, diabetic retinopathy is correctly identified. After the classification, the degree of error in an output node is measured based on target value and predicted results observed from the deep learning classifier.
From the above Eq. (13), Where,\({e}_{r}\) denotes an error, \({t}_{r}\) indicates a target results, \({p}_{r}\) represents the predicted results from the deep learning classifier. Based on the error value, the weight gets updated and computes the error rate. Finally, the proposed MAPCRCI-DMPLC technique uses the steepest descent function to find the minimum error.
From the above Eq. (14), Where, \(f(x)\) represents the steepest descent function, \(\text{arg}\;min\) denotes an argument of the minimum function, \({e}_{r}\) indicates an error. In this way, diabetic retinopathy (DR) is correctly identified with minimum error. The work flow chart of the MAPCRCI-DMPLC technique is shown in Fig. 4.
Figure 4 demonstrates the work flow chart of the MAPCRCI-DMPLC technique to get higher accuracy. The MAPCRCI-DMPLC algorithmic process is described as given below,
Algorithm. 1 above illustrates the step-by-step procedure of diabetic retinopathy through higher accuracy and lowest time consumption. The input images are collected and given to the input layer. First, the MAP estimated local region filtering-based preprocessing is performed within the first hidden layer to improve the image quality. In the second hidden layer, Camargo’s index is functional for ROI extraction toward distinguishing the infected region. Then the texture feature and color feature are extracted from the input image. Finally, the extracted features are sent to the output layer to find the disease. In the output layer, the swish activation function is applied to identify the different levels of disease, such as normal, mild, moderate, and severe. After the classification, the minimum error rate is identified to get better the classification accuracy at the output layer.
4 Experimental setup
In this section, we implemented six proposed MAPCRCI-DMPLC techniques and five state-of-the-art approaches such as MVDRNet, Multi-Scale Shallow CNN-based integrated model, Deep Residual CNN, MPDCNN, and Hybrid Deep Learning Approach compared their performances on the retinal image dataset with using the MATLAB R2022b simulator. The implementation is conducted by using hardware specification of Windows 10, Operating system, core i3-4130 3.40GHZ Processor, 8 GB RAM, 1 TB (1000 GB) Hard disk.
4.1 Dataset
The retinal image dataset is used in to evaluate the experiment. The dataset is taken from the https://www.kaggle.com/amanneo/diabetic-retinopathy-resized-arranged. The number of face images is gathered from the dataset. The high-resolution retina images are gathered in a variety of imaging circumstances that influence the visual look of the left and right eye. The dataset comprises five dissimilar image files and the clinician has rated the attendance of diabetic retinopathy in each file on a scale of 0 to 4, based on the following scale: No DR, Mild, Moderate, Severe, and Proliferative DR. The dataset includes 35,126 images for the classification of disease. The dataset is divided into training and testing. The majority of the images are used for training, and a smaller portion of the images is applied for testing. From the collected images, the training and testing input images are taken for conducting the simulation. The hyper-parameters and their description utilized in the proposed method are in Table 1.
5 Results
In this section, the proposed MAPCRCI-DMPLC technique and five state-of-the-art approaches namely MVDRNet (Luo et al. 2021), Multi-Scale Shallow CNN-based integrated model (Chen et al. 2020), Deep Residual CNN (Martinez-Murcia et al. 2021), MPDCNN (Deepa et al. 2022), and Hybrid Deep Learning Approach (Menaouer, B et al. 2022) are discussed based on quantitative and qualitative manner.
6 Quantitative analysis
The quantitative analysis in performance of the proposed MAPCRCI-DMPLC technique and five state-of-the-art approaches, namely the MVDRNet (Luo et al. 2021), Multi-Scale Shallow CNN-based Integrated Model (Chen et al. 2020), Deep Residual CNN (Martinez-Murcia et al. 2021), MPDCNN (Deepa et al. 2022), and Hybrid Deep Learning Approach (Menaouer et al. 2022) are discussed in this section with different results of performance metrics such as peak signal-to-noise ratio (PSNR), disease detection accuracy (DDA), false-positive rate (FPR), and disease detection time (DDT).
6.1 Impact of peak signal to noise ratio
A PSNR is estimated depending on MSE based on the preprocessed image and the original image. The formula for calculating the PSNR is given below,
From the above equations are (15) and (16), Where, ‘\({SE}_{M}\)’ refers a mean square error, ‘\({FI}_{p}\)’ denotes a preprocessing image size and original input image size ‘\({FI}_{o}\)’, ‘\({Ratio}_{pk}\)’ represents the PSNR and ‘\(M\)’ denotes the maximum probable pixel range i.e. 255. The PSNR is measured in decibels (dB).
The above Table 2 represents the comparison of PSNR based on six different techniques, namely the MAPCRCI-DMPLC technique, MVDRNet (Xiaoling Luo et al. 2021), Multi-Scale Shallow CNN-based integrated model (Chen et al. 2020), Deep Residual CNN (Martinez-Murcia et al. 2021), MPDCNN (Deepa et al. 2022) and Hybrid Deep Learning Approach (Menaouer et al. 2022). The PSNR is measured based on the sizes of the eye’s retina images in terms of KB. For each method, ten different runs are carried out versus different sizes of images. The results reported that the performance of the PSNR is comparatively higher when compared to existing methods. This is established through numerical calculation. Let us consider the input eye retina images with the size and the performance of PSNR is and the MSE is using the MAPCRCI-DMPLC technique. By applying the six existing MVDRNet (Xiaoling Luo et al. 2021), Multi-Scale Shallow CNN-based integrated model (Chen et al. 2020), Deep Residual CNN (Martinez-Murcia et al. 2021), MPDCNN (Deepa et al. 2022), and Hybrid Deep Learning Approach (Menaouer et al. 2022), the observed MSE and the PSNR are respectively. Likewise, different results are observed for all six methods. The observed results of the proposed technique are compared to the conventional methods. The reason for higher PSNR is to apply a novel application of the MAP estimated local region filtering-based preprocessing technique in the first hidden layer. First, the retina image is taken from the dataset. The pixels of the input retina image are arranged and the pixels are in ascending classified. The center significance is selected from the window. Then, the maximum likelihood between the center pixels and the neighbouring pixels in the filtering window is estimated. Finally, the pixels that deviate from the center value are identified as “noisy pixels”. These pixels are removed, which enhances the image feature. This process minimizes the MSE and improves the PSNR. Then the average of ten comparison results indicates that the performance of PSNR is considerably using MAPCRCI-DMPLC improved by 7% when compared to (Xiaoling Luo et al. 2021), 14% when compared to (Chen 2020), 18% when compared to (Martinez Murcia et al. 2021), and 22% when compared to (Deepa et al. 2022), and 3% when compared to (Menaouer et al. 2022).
6.2 Impact of disease detection accuracy
It is defined as the number of retinal images that are accurately detected as disease or average from the total number of retinal images. The disease detection accuracy is computed as given below,
From the above Eq. (17), Where, \(ACC\) indicates the disease detection accuracy, \({n}_{correctly\; identified}\) designates the number of images correctly detected as disease or normal, \(n\) denotes a number of images. The disease detection accurateness is calculated in terms of percentage (%).
Table 3 given above demonstrates the graphical demonstration of disease detection precision for 35,000 retina images. From Table 3, it is inferred that the disease detection accurateness was found to be comparatively higher using MAPCRCI-DMPLC upon comparison with MVDRNet (Xiaoling Luo et al. 2021) and the Multi-Scale Shallow CNN-based integrated model (Chen. 2020), Deep Residual CNN (Martinez-Murcia et al. 2021), MPDCNN (Deepa et al. 2022) and Hybrid Deep Learning Approach (Menaouer et al. 2022). The increase in the disease detection accuracy using the MAPCRCI-DMPLC technique was higher. Let us consider 3500 images in the first iteration to compute the accuracy. By applying the MAPCRCI-DMPLC, 3250 images are correctly classified and the accuracy is 92.85% whereas the accuracy percentage of the existing (Luo et al. 2021), (Chen et al. 2020), (Martinez-Murcia et al. 2021), (Deepa et al. 2022) and (Menaouer et al. 2022) are 89.14%, 87.14%, 84.42% 81.71% and 91.42% respectively. Following that, the nine remaining results are observed based on a different number of images. Finally, the performance of the MAPCRCI-DMPLC technique is compared to existing methods. The reason for higher accuracy is to apply innovation of the swish activation function in the output layer. By using this activation function, extracted features, and tested disease features are examined. Based on the swish activation function, the different levels of the disease are normal, mild, moderate, severe, and proliferative correctly identified. Therefore, the disease detection accuracy is improved using the MAPCRCI-DMPLC technique. The overall evaluation result showed that the recognition accuracy using the proposed MAPCRCI-DMPLC technique is significantly increased by 4%, 7%, 8%, and 9% when compared to state-of-the-art approaches (Xiaoling Luo et al. 2021), (Chen. 2020), (Martinez-Murcia et al. 2021), (Deepa et al. 2022), (Menaouer et al. 2022).
6.3 Impact of false-positive rate
Defined as the number of retinal images those are incorrectly detected as disease or normal from the total number of retinal images. The false-positive rate is computed as given below,
From the above Eqs. (18), Where, \({PR}_{fal}\) indicate the false positive rate, \({n}_{incorrectly\; identified}\) designates the number of images inaccurately detected as disease or normal, \(n\) denotes the number of images. The FPR is measured in terms of percentage (%).
Table 4 given above illustrates the performance of the FPR of a different quantity of retina images (3500–35000). The false-positive rate of six methods, namely the MAPCRCI-DMPLC technique, MVDRNet (Xiaoling Luo et al. 2021), Multi-Scale Shallow CNN-based integrated model (Chen et al. 2020), Deep Residual CNN (Martinez-Murcia et al. 2021), MPDCNN (Deepa et al. 2022), and Hybrid Deep Learning Approach (Menaouer et al. 2022) respectively. The performance of the MAPCRCI-DMPLC technique minimizes the FPR. Then the FPR of MAPCRCI-DMPLC is \(7.14\%\) whereas the FPR of MVDRNet (Xiaoling Luo et al. 2021), Multi-Scale Shallow CNN-based integrated model (Chen et al. 2020), Deep Residual CNN (Martinez-Murcia et al. 2021), MPDCNN (Deepa et al. 2022), Hybrid Deep Learning Approach (Menaouer et al. 2022) are \(10.85\%\), \(12.85\%,\) \(15.57\%,\) \(18.28\%\) and \(8.57\%\) by considering 3500 images. Followed by, the various estimation results are obtained for each method. The obtained results of the MAPCRCI-DMPLC technique are compared to existing methods. The main reason is to apply the innovation of the gradient descent function in the proposed MAPCRCI-DMPLC technique to discover the lowest error. Based on the error value, the weight gets updated and obtains accurate results with a minimum error. This helps to decrease the number of incorrect classifications. The overall performance of the FPR using the MAPCRCI-DMPLC technique is reduced by 35%, 46%, 50%,54% and 23% when compared to state-of-the-art approaches (Xiaoling Luo et al. 2021), (Chen et al. 2020), (Martinez-Murcia et al. 2021), (Deepa et al. 2022) (Menaouer et al. 2022), respectively.
6.4 Impact of time complexity
Computational complexity is measured as the amount of computing resources (time) that a particular algorithm consumes when it runs. Define the amount of time consumed by the algorithm to detect the disease or normal image. The overall time consumption is measured as follows,
From the above Eq. (19), Where,\(n\) denotes the number of images, \(DSI\)’ denotes the disease detection using a single image. The time complexity is computed in terms of milliseconds (ms).
Table 5 given above shows the graphical representation of time complexity results based on a number of retinal images. In the case of the collected retinal mages, the time complexity involved was analyzed and plotted in the above Table 5. As shown in Table 5, the performance of time complexity is increased linearly while increasing the count of retinal images for all five methods. However, the time complexity was found to be minimized using the MAPCRCI-DMPLC technique upon comparison with MVDRNet (Luo et al. 2021), Multi-Scale Shallow CNN-based integrated model (Chen et al. 2020), Deep Residual CNN (Martinez Murcia et al. 2021), MPDCNN (Deepa et al. 2022), and Hybrid Deep Learning Approach (Menaouer et al. 2022). Let us consider the ‘3500’ number of retinal images for testing, the time consumed by MAPCRCI-DMPLC to discover the disease or normal image is 63 \(ms\), where as \(68.25ms\), 70 ms, 73 ms, \(76ms\) and \(64.75ms\) of time consumed by existing techniques (Luo et al. 2021), (Chen et al. 2020), (Martinez-Murcia et al. 2021), (Deepa et al. 2022), and (Menaouer et al. 2022). As revealed in the table, the time complexity is gradually increased for all five classification methods while raising the number of retinal images since the counts of data get increased for each run. In other words, the prediction time of each method gets increased while increasing the number of retinal images. The main reason for the lesser time complexity is to use the ROI extraction as well as feature extraction in the proposed MAPCRCI-DMPLC technique. First, the innovation of Camargo’s index is applied for extracting the ROI from the preprocessed image based on pixel similarity measure. Following that, the innovation of Concordance Correlative Regression is applied for extracting the texture feature from the extracted ROI region instead of using the whole image. The color feature of the image is extracted by transforming the RGB into HSV from the image. With the selected features, the different severity levels are accurately identified with a minimum amount of time. From this analysis, the overall ten comparison analysis indicates that the time complexity using the MAPCRCI-DMPLC technique was reduced by 5%, 8%, 13%,17% and 2% when compared to (Xiaoling Luo et al. 2021), (Chen 2020), (Martinez-Murcia et al. 2021), (Deepa et al. 2022), and (Menaouer et al. 2022), respectively.
6.5 Specificity
Specificity is measured as a percentage of correctly detected healthy fundus images (TN) out of a total number of retinal images. It is represented in terms of percentage (%).
From Eq. (20), ‘\(S\)’ refers a specificity, \({t}_{n}\) refers a true negative, \({f}_{p}\) refers to the false positive (correctly detected as a healthy fundus image).
Table 6, it is observed that the show of specificity using MAPCRCI-DMPLC, MVDRNet (Xiaoling Luo et al. 2021), Multi-Scale Shallow CNN-based integrated model (Chen et al. 2020), Deep Residual CNN (Martinez-Murcia et al. 2021), MPDCNN (Deepa et al. 2022), and Hybrid Deep Learning Approach (Menaouer et al. 2022). However, simulations performed with 3500 images observed 90.44%, 86.65%, 84.78%, 82.12%, 80.85%, and 89.35% using the six methods respectively. From these results, the specificity rate is found to be improved using the MAPCRCI-DMPLC technique upon comparison with the state-of-the-art methods. The reason for higher specificity is to apply the swish activation function in the MAPCRCI-DMPLC technique for investigating extracted s as well as tested features. Also, dissimilar levels of the disease are classified with maximum specificity. As a result, the specificity rate using the MAPCRCI-DMPLC technique was said to be improved by 4%, 6%, 9%, 12% and 15% compared to state-of-the-art approaches (Luo et al. 2021), (Chen et al. 2020), (Martinez-Murcia et al. 2021), (Deepa et al. 2022), and (Menaouer et al. 2022), respectively
6.6 Impact of sensitivity
Sensitivity is also called recall. It is calculated to find the number of true positives as well as false negatives during the fault prediction. It is also known as sensitivity and is calculated as follows,
From Eq. (21), \({t}_{p}\) denotes a true positive, \({f}_{n}\) denotes the false negative. The recall is measured in percentage (%).
Table 7 demonstrate of sensitivity using MAPCRCI-DMPLC, MVDRNet (Xiaoling Luo et al. 2021), Multi-Scale Shallow CNN-based integrated model (Chen et al. 2020), Deep Residual CNN (Martinez-Murcia et al. 2021), MPDCNN (Deepa et al. 2022), and Hybrid Deep Learning Approach (Menaouer et al. 2022). However, simulations performed with 3500 images observed 95.36%, 93.45%, 92.66%, 91.91%, 89.1%, and 87.22% using the six methods respectively. From these results, the sensitivity is found to be improved using the MAPCRCI-DMPLC technique upon comparison with the state-of-the-art methods. The reason for higher sensitivity is to apply the swish activation function in the MAPCRCI-DMPLC technique to find various levels of the disease. As a result, the sensitivity using MAPCRCI-DMPLC technique was said to be improved by 5%, 7%, 10%, 11% and 3% compared to state-of-the-art approaches (Xiaoling Luo et al. 2021), (Chen et al. 2020), (Martinez-Murcia et al. 2021), (Deepa et al. 2022), and (Menaouer et al. 2022), respectively.
6.7 Cross-Entropy loss
The cost analysis of the MAPCRCI-DMPLC model is done in terms of cross-entropy loss to compute the performance of a classification model. The cross-entropy loss assesses the display of the characterization model, and it ranges from 0 to 1 where 0 is the value of cross-entropy loss observed for an accurate classification case.
From Eq. (22), M denotes the number of classes, t is the true classification for observation o, p is the likelihood prediction, and y is the binary indicator (either 0 or 1) for correct class label classification.
Table 8 demonstrates the cross-entropy loss with respect to number of retinal images. As shown in Table 8, the performance of cross-entropy loss is increased linearly while increasing the count of retinal images for all five methods. However, the cross-entropy loss was found to be minimized using the MAPCRCI-DMPLC technique compared to exiting methods. ‘3500’ number of retinal images considered for experimental, the cross-entropy loss is obtained as 0.554 using MAPCRCI-DMPLC. In addition, 0.583, 0.625, 0.663, 0.692 and 0.792 of cross-entropy loss consumed by existing techniques (Luo et al. 2021), (Chen et al. 2020), (Martinez-Murcia et al. 2021), (Deepa et al. 2022), and (Menaouer et al. 2022). As revealed in the table, the cross-entropy loss of each method gets increased while increasing the number of retinal images. The reason for lesser cross-entropy loss is to apply the ROI extraction, texture, and color feature extraction in the proposed MAPCRCI-DMPLC technique. As a result, cross-entropy loss of MAPCRCI-DMPLC technique was minimized by 13%, 18%, 20%, 23%, and 7% when compared to MVDRNet (Xiaoling Luo et al. 2021), Multi-Scale Shallow CNN-based integrated model (Chen et al. 2020), Deep Residual CNN (Martinez-Murcia et al. 2021), MPDCNN (Deepa et al. 2022), and Hybrid Deep Learning Approach (Menaouer et al. 2022) respectively.
6.7.1 Comparison of proposed MAPCRCI-DMPLC technique
The statistics of the proposed derivative of the proposed MAPCRCI-DMPLC technique implementation after preprocessing, feature extraction, and classification are tabulated in Table 9. The performance of the MAPCRCI-DMPLC technique is compared for the accuracy, time, and cross-entropy loss values.
Table 9 represents the proposed MAPCRCI-DMPLC technique derivative. In the 100th iteration, the accuracy of 92.28% is achieved at the 10th epoch. Also, a much reduced cross-entropy value of 0.693 is obtained for the proposed derivative model. The time taken by the model is 63.12 ms. The maximum accuracy of 94.28% is achieved at the 4th epoch and 40th iteration. The cross-entropy value of 0.594 is obtained for the proposed derivative model. The time taken by the model is 84.08 ms. Yet another, the maximum accuracy of 94.28% is achieved at the 6th epoch and 60th iteration. The cross-entropy value of 0.634 is obtained for the proposed derivative model. The time taken by the model is 96.61 ms. The comparative result reveals that the MAPCRCI-DMPLC technique and its derivatives provide better performance in terms of both the performance parameters involving a trade-off with the computational time.
6.7.2 Ablation experiments
Ablation experiments are the procedure of elimination section of input systematically in which elements are inputs are similar to output. In our work, we have used the MAP estimated local region filtering based preprocessing to eradicate the noisy pixel and offer a significant output. Initially, the retina image is gathered from the dataset. The pixels of the input retina image are prearranged and group the pixels are in the filtering window concept. The center assessment is chosen from the window. Followed by, maximum likelihood among the center pixels as well as neighboring pixels is estimated. Pixels that deviate from the middle value are discovered as “noisy pixels”. These pixels are eradicated. We have employed Camargo’s index and Concordance Correlative Regression for ROI extraction and feature extraction. After the selected features, the different classification results namely Normal, Mild, Moderate, Severe, and Proliferative are accurately identified with higher accuracy by using the swish activation function. Ablation experiments are demonstrated in Fig. 5.
Figure 5 illustrates ablation experiment results for graphical representations of classification. In the above figure, the x-axis denotes the different classification results and the y-axis gives the accuracy in percentage (%) for the proposed MAPCRCI-DMPLC technique. The performance of the MAPCRCI-DMPLC technique provides better performance for each class of DR disease. For the retinal image dataset, the classifier accurately identified 80% for normal, 82% for mild, 88% for moderate, 90% for severe, and 93% for Proliferative.
6.8 Qualitative analysis
Qualitative analysis of the proposed MAPCRCI-DMPLC technique is shown in Fig. 6.
7 Discussion
This study discusses the proposed MAPCRCI-DMPLC technique with the existing MVDRNet, Multi-Scale Shallow CNN-based integrated model, Deep Residual CNN, MPDCNN, and Hybrid Deep Learning Approach based on various parameters, such as PSNR, DDA, FPR, and DDT. Compared with standard existing methods, the proposed MAPCRCI-DMPLC technique is to get the best performance for the desired output. Contrary to existing works, the MAPCRCI-DMPLC technique is designed with the innovation of MAP estimated local region filtering technique, Camargo index, Concordance Correlative Regression, Gradient function, and swish activation function. At first, MAP estimated local region filtering is employed to execute preprocessing to reduce the noise and improve the image quality. In this manner, MSE is decreased and PSNR is enhanced. Second, the novelty of Camargo’s index is developed to extract ROI. The features are extracted by using the novelty of Concordance Correlative Regression with the lowest time. Next, the swish activation function is for examining extracted features and testing disease features to classify different levels with higher accuracy. Gradient descent functions are utilized for finding the lowest error. This helps to minimize the false positive rate. Table 10 provides a detailed comparison of the proposed technique with the state-of-the-art methods.
In above Table 10, the analysis of the overall results confirms that the proposed MAPCRCI-DMPLC technique improves PSNR, DDA, specificity, and sensitivity and reduces FPR, DDT, and cross-entropy loss by 56.00 dB, 93.30%, 91.11%, 96.35% and 6.77%, 94.36 ms compared to the conventional methods using retinal image dataset respectively.
8 Conclusion and future work
Diabetic retinopathy is one of the major challenges for people who are suffering from diabetes. Diabetic retinopathy is avoided for early detection. The conventional methods available for this purpose take more time and the prediction accuracy is low and failed to detect in the early stages. If the issue is determined in advanced stages, the chance of proficient treatment for recovery is low. Therefore, early detection with maximum accuracy plays a major task in diabetic retinopathy. Deep learning is one technology that is proficient outcomes in the health domain. Deep Multilayer Perceptive Learning is one of the well-known networks for image analysis. A novel deep learning model called the MAPCRCI-DMPLC technique is used to avoid the problem in early hours detection of diabetic retinopathy disease with multiple layers.
We preprocessed the input retinal images using the MAP estimated local region filtering technique to get better image superiority. Then the ROI extraction process is carried out to detect the infected region. Then, regarding the texture and color feature, extraction processes are to be performed. Finally, the activation function is used to analyze the feature level by testing disease features. Based on the feature analysis, the different levels of diabetic retinopathy are identified with higher accuracy. To authenticate the proposed method, we conducted the simulation with a diabetic retinopathy dataset.
When compared to that method, the proposed MAPCRCI-DMPLC achieves good results for early detection of retinopathy issues in diabetic patients. We derived the quantitative analysis of five methods in terms of PSNR, DDA, FPR, DDT, and Specificity. The comparative result shows that the aim of the proposed MAPCRCI-DMPLC technique has significantly enhanced the accuracy and reduced the time consumption compared to the conventional methods. In the future, we focus on the different multidimensional classifications of diabetic retinopathy disease detection time consumption. Future developments of the proposed method include selecting more advanced neural network architectures such as m deep convolute neural network. A complete and more precise automatic DR grading system would successfully assist the retinopathy screening.
Data availability
Data will be provided based on a request to the corresponding author.
References
Al-Antary MT, Arafa Y (2021) Multi-Scale Attention Network for Diabetic Retinopathy Classification. IEEE Access 9:54190–54200
Ashir AM, Ibrahim S, Abdulghani M, Ibrahim AA, Anwar MS, (2021) Diabetic Retinopathy Detection Using Local Extrema Quantized Haralick Features with Long Short-Term Memory Network. Int J Biomed Imaging. 2021, Article ID 6618666, 12. https://doi.org/10.1155/2021/6618666
Aziz T, Charoenlarpnopparut C, Mahapakulchai S (2023) Deep learning-based hemorrhage detection for diabetic retinopathy screening. Sci Rep 13(1):1479. https://doi.org/10.1038/s41598-023-28680-3
Bhardwaj C, Jain S, Sood M (2021a) Hierarchical severity grade classification of non-proliferative diabetic retinopathy. J Ambient Intell Humaniz Comput 12:2649–2670. https://doi.org/10.1007/s12652-020-02426-9
Bhardwaj C, Jain S, Sood M (2021b) Transfer learning based robust automatic detection system for diabetic retinopathy grading. Neural Comput Appl 33(20):13999–14019. https://doi.org/10.1007/s00521-021-06042-2volV)
Bhardwaj C, Jain S, Sood M (2021c) Diabetic retinopathy severity grading employing quadrant-based Inception-V3 convolution neural network architecture. Int J Imaging Syst Technol 31(2):592–608. https://doi.org/10.1002/ima.22510
Bhardwaj C, Jain S, Sood M (2021d) Deep learning–based diabetic retinopathy severity grading system employing quadrant ensemble model. J Digit Imaging 34:440–457. https://doi.org/10.1007/s10278-021-00418-5
Bhuiyan A, Govindaiah A, Deobhakta A, Hossain M, Rosen R, Smith T (2021) Automated diabetic retinopathy screening for primary care settings using deep learning. Intelligence-Based Med , Elsevier 5:1–9. https://doi.org/10.1016/j.ibmed.2021.100045
Chen W, Yang B, Li J, Wang J (2020) An Approach to Detecting Diabetic Retinopathy Based on Integrated Shallow Convolutional Neural Networks. IEEE Access 8:178552–178562
Chen PN, Lee CC, Liang CM, Pao SI, Huang KH, Lin KF (2021) General deep learning model for detecting diabetic retinopathy. BMC Bioinformatics 22(84):1–14
Dai L, Wu L, Li H, Cai C, Wu Q, Kong H, Liu R, Wang X, Hou X, Liu Y, Long X, Wen Y, Lu L, Shen Y, Chen Y, Shen D, Yang X, Zou H, Sheng B, Jia W (2021) A deep learning system for detecting diabetic retinopathy across the disease spectrum. Nat Commun 12(1):3242
Das S, Kharbanda K, Suchetha M, Raman R, Dhas E (2021) Deep learning architecture based on segmented fundus image features for classification of diabetic retinopathy. Biomed Signal Processing Control, Elsevier 68:1–10. https://doi.org/10.1016/j.bspc.2021.102600
Deepa V, Kumar CS, Cherian T (2022) Ensemble of multi-stage deep convolutional neural networks for automated grading of diabetic retinopathy using image patches. J King Saud Univ- Computer Inform Sci, Elsevier 34(8):6255–6265. https://doi.org/10.1016/j.jksuci.2021.05.009
Erciyas A, Barısci N (2021) An Effective Method for Detecting and Classifying Diabetic Retinopathy Lesions Based on Deep Learning. Computational and Mathematical Methods in Medicine. 2021, Article ID 9928899, 13. https://doi.org/10.1155/2021/9928899
Gunasekaran K, Pitchai R, Chaitanya GK, Selvaraj D, Annie Sheryl S, Almoallim HS, Alharbi SA, Raghavan SS, Tesemma BG (2022) A Deep Learning Framework for Earlier Prediction of Diabetic Retinopathy from Fundus Photographs. Biomed Res Int 2022:3163496. https://doi.org/10.1155/2022/3163496
Hemanth DJ, Deperlioglu O, Kose U (2020) An enhanced diabetic retinopathy detection and classification approach using deep convolutional neural network. Neural Computing Applications, Springer 32:707–721
Kalyani G, Janakiramaiah B, Karuna A (2023) Diabetic retinopathy detection and classification using capsule networks. Complex Intell Syst 9:2651–2664. https://doi.org/10.1007/s40747-021-00318-9
Khan Z, Khan FG, Khan A, Rehman ZU, Shah S, Qummar S, Ali F, Pack S (2021) Diabetic Retinopathy Detection Using VGG-NIN a Deep Learning Architecture. IEEE Access 9:61408–61416
Kumar G, Chatterjee S, Chattopadhyay C (2021) DRISTI: a hybrid deep neural network for diabetic retinopathy diagnosis. Signal, Image and Video Processing, Springer 15:1679–1686
Lin GM, Chen MJ, Yeh CH, Lin YY, Kuo HY, Lin MH, Chen MC, Lin SD, Gao Y, Ran A, Cheung CY (2018) Transforming Retinal Photographs to Entropy Images in Deep Learning to Improve Automated Detection for Diabetic Retinopathy. Hindawi, J Ophthalmol 2018:1–18
Liu T, Chen Y, Shen H, Zhou R, Zhang M, Liu T, Liu J (2021) A Novel Diabetic Retinopathy Detection Approach Based on Deep Symmetric Convolutional Neural Network. IEEE Access 9:160552–160558
Luo X, Pu Z, Xu Y, Wong WK, Su J, Dou X, Ye B, Hu J, Mou L (2021) MVDRNet: Multi-view diabetic retinopathy detection by combining DCNNs and attention mechanisms. Pattern Recognition, Elsevier 120:1–12
Majumder S, Kehtarnavaz N (2021) Multitasking Deep Learning Model for Detection of Five Stages of Diabetic Retinopathy. IEEE Access 9:123220–123230
Martinez-Murcia FJ, Ortiz A, Ramírez J, Górriz JM, Cruz R (2021) Deep Residual Transfer learning for Automatic Diagnosis and Grading of Diabetic Retinopathy. Neurocomputing, Elsevier 452:424–434
Menaouer B, Dermane Z, El Houda Kebir N, Matta N. (2022) Diabetic Retinopathy Classification Using Hybrid Deep Learning Approach. SN Comput Sci. 3(357). https://doi.org/10.1007/s42979-022-01240-8
Muthusamy D, Rakkimuthu P (2022) Trilateral filterative hermitian feature transformed deep perceptive fuzzy neural network for finger vein verification. Expert Syst Appl 196:116678. https://doi.org/10.1016/j.eswa.2022.116678
Muthusamy D, Sathyamoorthy S (2022) Deep belief network for solving the image quality assessment in full reference and no reference model. Neural Comp Applications, Springer Nature 34(24):21809–21833
Muthusamy D, Sathyamoorthy S (2023) Feature Sampling based on Multilayer Perceptive Neural Network for image quality assessment, Engineering Applications of Artificial Intelligence. Elsevier 121:106015. https://doi.org/10.1016/j.engappai.2023.106015
Nahiduzzaman M, Islam MR, Islam SR, Goni MO, Anower MS, Kwak KS (2021) Hybrid CNN-SVD Based Prominent Feature Extraction and Selection for Grading Diabetic Retinopathy Using Extreme Learning Machine Algorithm. IEEE Access 9:152261–152274
Nawaz F, Ramzan M, Mehmood K, Khan HU, Khan SH, Bhutta MR (2021) Early Detection of Diabetic Retinopathy Using Machine Intelligence through Deep Transfer and Representational Learning. Computers, Mater Continua 66(2):1631–1645
Oh K, Kang HM, Leem D, Lee H, Seo KY, Yoon S, (2021) Early detection of diabetic retinopathy based on deep learning and ultra-wide-field fundus images, Scientific Reports. 11(1897). https://doi.org/10.1038/s41598-021-81539-3
Ozbay E (2023) An active deep learning method for diabetic retinopathy detection in segmented fundus images using artificial bee colony algorithm. Artif Intell Rev 56:3291–3318. https://doi.org/10.1007/s10462-022-10231-3
Qummar S, Khan FG, Shah S, Khan A, Shamshirband S, Rehman ZU, Khan IA (2019) A Deep Learning Ensemble Approach for Diabetic Retinopathy Detection. IEEE Access 7:150530–150539
Qureshi I, Ma J, Abbas Q (2021) Diabetic retinopathy detection and stage classification in eye fundus images using active deep learning. Multimedia Tools Applications, Springer 80:11691–11721
Shankar K, Zhang Y, Liu Y, Wu L, Chen CH (2020) Hyperparameter Tuning Deep Learning for Diabetic Retinopathy Fundus Image Classification. IEEE Access 8:118164–118173
Sridhar S, PradeepKandhasamy J, Sinthuja M, Minish TS, (2021) Diabetic retinopathy detection using convolutional nueral networks algorithm, Materials Today: In Proceedings, Elsevier, 1–3
Sugeno A, Ishikawa Y, Ohshima T, Muramatsu R (2021) Simple methods for the lesion detection and severity grading of diabetic retinopathy by image processing and transfer learning. Computers Biol Med, Elsevier 137:1–9. https://doi.org/10.1016/j.compbiomed.2021.104795
Sungheetha A, Sharma R (2021) Design an Early Detection and Classification for Diabetic Retinopathy by Deep Feature Extraction based Convolution Neural Network. J Trends Comp Sci Smart Technol 3(2):81–94
Vives-Boix V, Ruiz-Fernández D (2021) Diabetic retinopathy detection through convolutional neural networks with synaptic metaplasticity. Comput Methods Programs Biomed 206:1–8
Wang J, Bai Y, Xia B (2020) Simultaneous Diagnosis of Severity and Features of Diabetic Retinopathy in Fundus Photography Using Deep Learning. IEEE J Biomed Health Inform 24(12):3397–3407
Zhu CZ, Hu R, Zou BJ, Zhao RC, Chen CL, Xiao YL (2019) Automatic Diabetic Retinopathy Screening via Cascaded Framework Based on Image- and Lesion-Level Features Fusion. J Comput Sci Technol 34(6):1307–1318
Funding
The authors did not receive support from any organization for the submitted work.
Author information
Authors and Affiliations
Contributions
Dharmalingam Muthusamy: Conceptualization, Investigation, Writing—original draft, Writing—review & editing, Visualization, Formal analysis, Methodology, and Supervision. Parimala Palani: Writing—review & editing, Software, Supervision.
Corresponding author
Ethics declarations
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Competing interests
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Muthusamy, D., Palani, P. Deep learning model using classification for diabetic retinopathy detection: an overview. Artif Intell Rev 57, 185 (2024). https://doi.org/10.1007/s10462-024-10806-2
Accepted:
Published:
DOI: https://doi.org/10.1007/s10462-024-10806-2