Abstract
One of the primary challenges in applying deep learning approaches to medical imaging is the limited availability of data due to various factors. These factors include concerns about data privacy and the requirement for expert radiologists to perform the time-consuming and labor-intensive task of labeling data, particularly for tasks such as segmentation. Consequently, there is a critical need to develop novel approaches for few-shot learning tasks in this domain. In this work, we propose a Novel CNN-Transformer Fusion scheme to segment Multi-classes pneumonia infection from limited CT-scans data. In total, there are three main contributions: (i) CNN-Transformer encoders fusion, which allows to extract and fuse richer features in the encoding phase, which contains: local, global and long-range dependencies features, (ii) Multi-Branches Skip Connection (MBSC) is proposed to extract and fuse richer features from the encoder features then integrate them into the decoder layers, where MBSC blocks extract higher-level features related to the finer details of different infection types, and (iii) a Multi-classes Boundary Aware Cross-Entropy (MBA-CE) Loss function is proposed to deal with fuzzy boundaries, enhance the separability between classes and give more attention to the minority classes. The performance of the proposed approach is evaluated using two evaluation scenarios and compared with different baseline and state-of-the-art segmentation architectures for Multi-classes Covid-19 segmentation. The obtained results show that our approach outperforms the comparison methods in both Ground-Glass Opacity (GGO) and Consolidation segmentation. On the other hand, our approach shows consistent performance when the training data is reduced to half, which proves the efficiency of our approach in few-shot learning. In contrast, the performance of the comparison methods drops in this scenario. Moreover, our approach is able to deal with imbalanced data classes. These advantages prove the effectiveness and efficiency of the proposed EMB-TrAttUnet approach in a pandemic scenario where time is critical to save patient lives.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
Over the past three years, the world has been facing a global crisis due to the spread of the Covid-19 pandemic, which has affected all world’s countries (Amyar et al., 2020; Bougourzi et al., 2022). By November 2023, more than 770 million cases and about than 7 million deaths as declared by World Health Organization (WHO) WHO (2023). The Covid-19 pandemic is caused by the infectious SARS-CoV-2 virus, which mainly affects the lungs and spreads to other organs (Parasher 2021). Since the start of the pandemic, medical imaging has been widely used for Covid-19 analysis, including: Covid-19 diagnosis (Vantaggiato et al. 2021), segmentation (Pezzano et al. 2021; Bougourzi et al. 2022), and severity estimation and prediction (Li et al. 2021; Bougourzi et al. 2021). Medical scan modalities include: X-ray, Computerized Tomography (CT) and Ultrasound (Vantaggiato et al. 2021; Bougourzi et al. 2021; Roy et al. 2020. In fact, CT-scans are more efficient for Covid-19 analysis because they provide more details about the spread and severity of the infection, which allows to following-up the patient state and taking the right decision to save the patient’s life (Shi et al. 2020; Zhang et al. 2020). In particular, Covid-19 infection segmentation from CT-scans is a very important step in detecting, quantifying and evaluating the infection and predicting its severity (Shi et al. 2020; Zhang et al. 2020).
In the last decade, Deep Leaning (DL) approaches have become the dominant trend in many machine learning and computer vision tasks (Van der Velden et al. 2022; Bougourzi et al. Apr. 2022; Goceri 2023). In particular, DL approaches have been widely studied for automatically analysing various infections and diseases using different medical imaging modalities, which can assist or even replace experienced physicians and radiologists in diagnosing and analysing the diseases (Xie et al. 2023; Bougourzi et al. 2023; Soomro et al. 2022). However, In the case of the Covid-19 pandemic, the number of cases in a short period of time is enormous, which overwhelming the resources and personnel of hospitals, including physicians and radiologists. Therefore, it is necessary to develop automatic machine learning solutions to support the control of this disease and reduce the workload of medical staff (Vantaggiato et al. 2021; Roy et al. 2020; Rehman et al. 2023).
In the last decade, semantic segmentation tasks have been extensively studied and numerous deep learning segmentation approaches have been proposed (Ghosh et al. 2019; Minaee et al. 2021). In particular, the segmentation of Covid-19 infections has been widely investigated, which is a very challenging task due to two main factors. First, the available labelled data are very limited due to the significant time and effort required for the labelling process, with physicians and radiologists being overwhelmed due to the pandemic (Wang et al. 2020; Pezzano et al. 2021; Yao et al. 2021). The second factor is related to the nature of Covid-19 infection, which progress differently from one case to case. Moreover, Covid-19 infection exhibits high variability in terms of intensity, shape, position and type depending on the stage of infection (early vs. advanced), symptoms (asymptomatic vs. symptomatic patients), and severity (Shi et al. 2020; Zhang et al. 2020). In fact, the literature works have concentrated on segmenting Covid-19 infection as a binary task (background or infection) Pezzano et al. (2021); Paluru et al. (2021); Cong et al. (2022a, 2022b); Yao et al. (2021), as the available labelled data for Covid-19 infection types (Multi-classes) are very limited. However, segmentation of Covid-19 infections into multiple classes provides more details about infection progression, stage, and severity (Hefeda 2020; Salehi et al. 2020).
In this paper, a new CNN-Transformer based approach is proposed for segmenting Covid-19 infections into multiple classes from limited data. In detail, the proposed EMB-TrAttUnet consists of two main components: (i) CNN-Transformer architecture (MB-TrAttUnet) and (ii) Multi-classes Boundary Aware Cross-Entropy (MBA-CE) Loss function. The MB-TrAttUnet is an Encoder-Decoder architecture that follows the Unet architecture family (Ronneberger et al. 2015). The encoder of the proposed MB-TrAttUnet architecture combines CNN and Transformer layers to extract local, global, and sequential features in the encoding phase. Instead of the traditional Unet skip connection, a Multi Branches Skip Connection (MBSC) is proposed to extract high-level features, interpret the encoder features, and select the most important features. The decoder of the proposed MB-TrAttUnet is similar to the encoder of the Att-Unet architecture (Oktay et al. 2018), where the attention gates are better exploited because the encoder layers and the MBSC extract higher and richer features for Multi-classes Covid-19 Segmentation. The MBA-CE Loss function is designed to give more attention to the classes boundaries to enhance boundaries segmentation, the separability between classes and minority classes decision. The main contributions can be summarized as follows:
-
We propose a novel hybrid Transformer-CNN architecture (MB-TrAttUnet) for Multi-classes Covid-19 infection segmentation from CT-scans. The Encoder of TrAttUnet architecture combines CNN and Transformer blocks using Encoders Fusion Block (EFB). On the other hand, the Decoder of MB-TrAttUnet is AttUnet-like decoder.
-
Furthermore, Multi Branches Skip Connection is introduced to extract more advanced features and to interpret and select the most significant ones from the encoder features and pass them to the decoder block.
-
We propose a Multi-classes Boundary Aware Cross-Entropy (MBA-CE) Loss function to cope with fuzzy boundaries, enhance the separability between classes and give more attention to the minority classes. The trained MB-TrAttUnet with MBA-CE is called EMB-TrAttUnet.
-
The comparison between the proposed approach and both baseline and state-of-the-art segmentation architectures clearly demonstrates the superior performance of our method, particularly when faced with limited training data availability. The proposed EMB-TrAttUnet architecture is publicly available at: https://github.com/faresbougourzi/EMB-TrAttUnet.
The remainder of this paper is organized as follows: Sect. 2 summarizes the segmentation architectures for medical image segmentation and related work on Covid-19 infection segmentation. Section 3 explains the proposed approach. The datasets and evaluation metrics used are described in Sect. 4. Section 5 presents and discusses the experiments and results. Section 6 provides a visual analysis of the segmentation results of the proposed approach and the comparison methods. Finally, Section 7 concludes the paper.
2 Related work
This section is dedicated to describing the related works concerning state-of-the-art deep learning architectures for medical image segmentation, deep learning approaches for respiratory diseases, and deep learning approaches for COVID-19 infection segmentation.
2.1 DL architectures for medical imaging segmentation
In the last decade, Deep Learning approaches have proved their efficiency in many medical imaging tasks using different medical imaging modalities including but not limited to: Magnetic Resonance Imaging (MRI), X-rays, Computed Tomography (CT) and Ultrasound images (Litjens et al. 2017; Paladini et al. 2021). In particular, medical image segmentation is one of the most studied tasks in recent years. Since the development of Unet architecture in 2015, CNN-based architectures have become the dominant approaches in medical image segmentation. Consequently, plenty of the Unet Ronneberger et al. (2015) variants have been proposed such as Att-Unet Oktay et al. (2018), Unet++ Zhou et al. (2018), ARU-GD Maji et al. (2022), FANet Tomar et al. (2022), and KiU-Net Valanarasu et al. (2021).
In short, Unet Ronneberger et al. (2015) has an Encoder-Decoder structure with a“U”shape. The encoder of the Unet architecture consists of successive convolutional blocks followed by max-pooling operators to extract high-level features from the input. On the other side, Unet’s decoder exploits the extracted features of the encoder to reconstruct the segmentation mask through successive deconvolution layers. Moreover, the skip connections incorporate the feature maps of the encoder layers into the decoder layers to prevent the loss of detailed features due to the shrinking of the encoder spatial dimensionalty. In the Att-Unet architecture (Oktay et al. 2018), Attention Gates (AGs) are placed between the skip connections and the decoder layers to select the most important salient parts from the encoder features and pass them to the decoder.
In recent years, Transformers have shown great success in Natural Language Processing (NLP) domain (Vaswani et al. 2017; Khan et al. 2021). This inspired the computer vision community to exploit the transformer for vision tasks (Dosovitskiy et al. 2020; Liu et al. 2021; Touvron et al. 2021). In particular, transformers have shown promising performance for segmentation tasks in medical imaging (Li et al. 2023). Therefore, plenty of Transformer-based segmentation architectures have been proposed, such as UNETR Hatamizadeh et al. (2022), GT U-Net Li et al. (2021), and Swin-Unet Cao et al. (2021). In Li et al. (2021), a Group Transformer Unet (GT U-Net) architecture was proposed for tooth root segmentation. In more details, GT U-Net keeps the general structure of the Unet architecture, with the encoder and decoder consisting of Group Transformer blocks combining CNN and Transformer components. Also, SwinUnet Cao et al. (2021) architecture follows the same structure as Unet. However, the encoder and decoder of the Swin-Unet were purely constructed using Swin Transformer blocks (Liu et al. 2021) without any convolutional operations. The experimental results in Cao et al. (2021) proved the efficiency of the Swin-Unet architecture compared to CNN-based segmentation architectures for multi-organ and cardiac segmentation tasks.
In addition to pure Transformer architectures, hybrid Transformer-CNN architectures have been investigated in the last two years in the field of medical imaging segmentation (Wang et al. 2022; Huang et al. 2022; Wang et al. 2022). In Wang et al. (2022), the same structure as Unet was retained while proposing a Mixed Transformer Module (MTM), which aims to learn inter and intra-affinity features. MTM consists of a Local–Global Gaussian-Weighted Self-Attention (LGG-SA) and an External Attention (EA) block. In Huang et al. (2022), the aim of the MISSFormer architecture is to propose an Enhanced Transformer Block by using convolutional operations. Additionally, the authors introduced the Enhanced Transformer Context Bridge to extract long-range dependencies and local context from multi-scale features.
In contrast to these state-of-the-art hybrid approaches, our method involves two encoder paths. The first one is designed to extract global and long-range dependency features, while the second encoder aims to combine Transformer features at different levels with the local features extracted by the CNN blocks. Furthermore, a Multi-Branch Skip Connection (MBSC) mechanism is proposed to enhance the traditional skip connection or Att-Unet architecture. MBSC extracts higher-level features from different branches and concatenates their outputs to pass them to the second filtering stage through the attention gates.
2.2 DL approaches for respiratory diseases analysis
Over the last decade, deep learning approaches have been widely utilized for the analysis of respiratory diseases, including chronic obstructive pulmonary disease, pneumonia, asthma, tuberculosis, fibrosis, lung cancer, etc Bharati et al. (2020); Jasmine Pemeena Priyadarsini et al. (2023). Despite the wide variety of deep learning methods available, most of the research on respiratory diseases has predominantly employed Convolutional Neural Networks (CNNs) as the primary technique. These applications encompass numerous tasks, such as the recognition of multiple respiratory diseases from X-ray images (Bharati et al. 2020; Jasmine Pemeena Priyadarsini et al. 2023), the detection of tuberculosis from X-ray images (Duong et al. 2021), the identification of Covid-19 from X-ray images (Duong et al. 2023; Vantaggiato et al. 2021), and Covid-19 segmentation from CT scans (Müller et al. 2021; Ilhan et al. 2023; Saood and Hatem 2021; Fung et al. 2021). In Bharati et al. (2020), S. Bharati et al. introduced a hybrid deep learning framework that combines CNNs, data augmentation, and a spatial transformer network (STN) for the detection of lung diseases in X-ray images. This framework includes 15 different classes, one of which is“No findings,”while the others represent various diseases. In Fung et al. (2021), the authors proposed a two-stage approach with a self-supervised learning strategy for Covid-19 segmentation. They utilized the InfNet architecture as the backbone and integrated generative adversarial image inpainting to enhance the segmentation of lung lesions.
In addition to the CNN architectures, transformers have been investigated for respiratory diseases tasks (Duong et al. 2021), especially for Covid-19 analysis (Liang et al. 2021; Chaudhary et al. 2022). In Duong et al. (2021), L. D. Tuan et al. proposed a hybrid CNN-Transformer architecture for detecting tuberculosis from X-rays. First, they used a CNN backbone (EfficientNet) to extract 2D feature maps from the input X-ray, then they used an Encoder-Decoder Transformers. The first Transformer aims to contextualize features from the CNN features, and the second Transformer aims to auto-regressively generate one channel for each input feature at every time step. In Liang et al. (2021), S. Liang et al. proposed a hybrid CTNet framework that exploits CNN and Transformer approaches. In this approach, they used a CNN feature extractor with an SE attention Block and then passed the obtained results to a ViT module. Their approach demonstrated a good capability to learn discriminative features from 3D CT scans for Covid-19 infection recognition. S. Chaudhary et al. proposed Chaudhary et al. (2022) exploited the strength of the Swin Transformer to extract the feature from the CT slice and pass it through MLP layers for Covid-19 infection percentage estimation. To utilize the recent developments in Vision Transformers, we proposed a Hybrid Transformer-CNN approach for Covid-19 infection segmentation with multiple classes from limited training data.
2.3 DL approaches for Covid-19 infection segmentation
Since the spread of the Covid-19 pandemic in 2019, Covid-19 infection segmentation has been extensively studied by the machine learning and computer vision community. Most of the existing works have been concentrating on segmenting Covid-19 infections as a binary task (infection and background) Bougourzi et al. (2023); Pezzano et al. (2021); Yao et al. (2021); Ding et al. (2021); Paluru et al. (2021); Cong et al. (2022a, 2022b). Fewer works, however, have attempted to segment Covid-19 infection as Multi-classes (Jin et al. 2022; Yu et al. 2022). In Wu et al. (2021), G. Pezzano et al. proposed a multi-stages approach which consists of the following steps: (i) lung delineation, (ii) Covid-19 detection, and (iii) Covid-19 infection segmentation. First hand, they proposed a Unet variant with Multiple Convolutional Layers structure, that performs lung segmentation within a novel pipeline for direct Covid-19 detection and segmentation. They also proposed a new customized loss function for optimizing the performance.
Mu et al. (2021) proposed a progressive global perception and local polishing (PCPLP) approach for Covid-19 infection segmentation. In detail, the PCPLP encoder follows the VGG-19 (Simonyan and Zisserman 2014) architecture to extract multi-level low and middle features. On the other hand, the decoder of PCPLP integrates the multi-scale to learn high-level features for Covid-19 segmentation. To this end, the following blocks were proposed GPM, LPM, and BMS, which aim to localise the global feature of the infected regions, extract the local features of the infected regions contours and preserve the edge-related features, respectively. In Wang et al. (2022), X. Wang et al. proposed a new architecture called Spatial Self-Attention Network (SSA-Net), which exploits both the self-attention mechanism and spatial convolution to expand the receptive field, strengthen the learning process, and accelerate the training convergence. In addition to the proposed SSA-Net architecture, a semi-supervised few-shot iterative approach is proposed to cope with the limited labelled data available for training (Wang et al. 2022).
Since there is a paucity of work addressing the segmentation of Covid-19 infections as Multi-classes, this work focuses on this task, which is very important for identifying the stage of infection and predicting its progression and severity (Hefeda 2020; Salehi et al. 2020). This allows following-up the patient’s state and taking the right actions to save the patient’s life (Hefeda 2020; Salehi et al. 2020). Unlike the state of the art, our proposed approach efficiently combines CNN and Transformer encoders to segment Covid-19 infection into multiple classes. Moreover, a new skip-connection block is proposed to interpret and extract higher-level features from the encoder features and pass them to the decoder layers to preserve the finer details associated with the Multi-classes Covid-19 infection. Finally, transfer learning and freezing strategies are exploited to segment complicated task from very limited data.
3 The proposed approach
Our proposed approach consists of two main components. First, we propose a CNN-Transformer based architecture, which is described in detail in Sect. 3.1. Second, a Multi-classes Boundary Aware Cross-Entropy (MBA-CE) Loss function is proposed. This is described in Sect. 3.2. Finally, the evaluation metrics are described in Sect. 3.3.
3.1 MB-TrAttUnet architecture
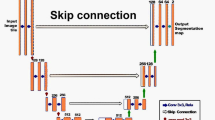
The proposed MB-TrAttUnet architecture is summarized in Fig. 1. Our proposed MB-TrAttUnet is an Encoder-Decoder architecture where the encoder consists of two components: Transformer path and Encoders Fusion Block (EFB). On the other hand, the decoder is an Att-Unet-like decoder consisting of Attention Gates and Deconvolution blocks. To connect the encoder and the decoder, an MBSC layer is proposed. Figure 2 shows the detailed structure of our proposed MB-TrAttUnet architecture.
3.1.1 Transformer path
As shown in Fig. 2, the input image \(x \in \mathbb {R}^{H\times W\times C}\), where H, W and C are the height, width, and input channels, respectively, is fed into two paths: Unet-like path and Transformer path. In the Transformer path, x is divided into uniform non-overlapping 2D patches \(x_v = [x_v^1;x_v^2; \ldots ;x_v^N]\), where each patch is represented by a row vector \(x_v^i \in \mathbb {R}^{S^2 \times C}\), where (\(S\times S\)) is the spatial size of the patch and N is the number of the patches \(N = (H\times W) /S^2\). These patches are projected into Embedding space \(z_0\) using a linear transformation \(E\in \mathbb {R}^{ (S^2. C)\times K}\), where K is the dimension of the embedding space, which is fixed for all of the transformer layers. \(z_0\) is defined by:
The embedded features \(z_0\in \mathbb {R}^{N\times K}\) are fed into Transformer layers similar to Dosovitskiy et al. (2020); Vaswani et al. (2017). As shown in Fig. 4c, the Transformer layer consists of two Layernorm (LN) blocks, a Multi-Head Self-Attention (MSA) block, a multilayer perceptron (MLP) block and residual connections. For the Transformer layer (l), the embedded input features \(z_{l-1}\) are fed into Layernorm (LN), followed by a Multi-Head Self-Attention block, which is then summed with \(z_{l-1}\) by a residual connection, as shown in equation (2):
The embedded features of \(z_{l-1}\) passed by the first LN are denoted by \(s = LN(z_{l-1})\).
These features are processed by h heads. Multi-head attention allows the model to jointly attend to information from different representational subspaces. Each head generates attended features with a dimensionof K/h. MSA is defined by:
where \(SA_1, SA_2,..., SA_h\) are self-Attention results provided by the individual heads, and \(U_{msa} \in \mathbb {R}^{ K\times K}\) is the global projection matrix for the SA features.
The \(z_l^\prime\) is fed into Layernorm block followed by MLP block, and then summed by \(z_{l-1}\) through the residual connection, as depicted in equation (4):
where MLP consists of two linear layers with a GELU nonlinearity. The first linear layer (\(MLP_1 \in \mathbb {R}^{ K\times K_{MLP}}\)) projects \(LN(z_l^\prime )\) into \(K_{MLP}\), then the second linear layer (\(MLP_2 \in \mathbb {R}^{ K_{MLP}\times K}\)) projects it back onto K features.
In our approach, the number of layers of the Transformer, L, is set to 12, h to 12, \(K = 786\) and \(K_{MLP} = 3072\). The input image x size is \(224\times 224 \times 3\) (\(W = H = 224\)) and the transformer patch size is \(S^2=16\times 16\) pixels. Consequently, the number of patches is 196. To obtain diversity of learned features from different Transformer layers (levels), the embedded features of layers 4, 7, 10 and 12 are selected. These layers are denoted as \(Tr_{1}\), \(Tr_{2}\), \(Tr_{3}\) and \(Tr_{4}\), respectively. Consequently, four layers from the Transformer path were injected into the Encoders Fusion Block (EFB), all of which are of shape \(196\times 786\). To obtain a 3D tensors, \(z_l\) is reshaped to \(14\times 14\times 786\), since \(14\times 14= 196\). The reshaped features of the embedded features of \(Tr_{1}\), \(Tr_{2}\), \(Tr_{3}\) and \(Tr_{4}\) are denoted by \(z_{1r}\), \(z_{2r}\), \(z_{3r}\) and \(z_{4r}\), respectively.
3.1.2 Encoders dusion block (EFB)
Encoders Fusion Block combines the classical CNN features and Transformer features through ResBlocks in a hierarchical manner. Its main goal is to enrich the representations of the encoder at several stages. To inject the transformer features into different layers of the EFB and combine them with the CNN features, UpResBlock is introduced as depicted in Fig. 4-b. UpResblock consists of linear upsampling followed by ResBlock, which is depicted in Fig. 4-a. ResBlock consists of two consecutive 3 by 3 convolutional blocks, each followed by Batch Normalization and ReLU activation function. In addition, the input is summed with the output of the two convolutional layers using the residual connection, which consists of a 1 by 1 convolutional block, followed by Batch Normalization and ReLU activation function, as shown in equations (5) and (6):
where \(x_{out_1}\) and \(x_{out}\) are the outputs of the first 3 by 3 convolutional block and the whole ResBlock, respectively. \(Conv 3\times 3_1\), \(Conv 3\times 3_2\) and \(Conv 1\times 1\) are convolutional kernels that transform the input number of channels into \(C_{out}\).
The injected transformer features into the EFB are denoted by \(z_{up_1}\), \(z_{up_2}\), \(z_{up_3}\) and \(z_{up_4}\), respectively, which are defined by:
Equations (7), (8) and (9) illustrate the number of UpResBlocks required to match the output of the transformer layers to the corresponding EFB level, using three, two, and one UpResBlock for \(Tr_{1}\), \(Tr_{2}\) and \(Tr_{3}\), respectively. For the \(Tr_{4}\) layer, ResBlock is used instead of UpResBlock since no upsampling is required here to match the last EFB level, as depicted in equation (10).
On the other hand, the Encoders Fusion Block has five layers which will be denoted by \(Un_{1}\), \(Un_{2}\), \(Un_{3}\), \(Un_{4}\) and \(Un_{5}\), respectively. The first layer uses ResBlock on the input image \(x \in \mathbb {R}^{H\times W\times C}\) to obtain the first EFB feature maps as shown in equation (11).
The second, third, fourth and fifth EFB layers combine the transformer features with the max-pooled features of the previous EFB layer as shown in equations (12), (13), (14) and (15):
where Max-Pooling layers (MP) reduces the spatial demensionality into half.
3.1.3 Multi-branches skip connection (MBSC)
Since the encoder of the proposed MB-TrAttUnet architecture generates composite and richer features by combining CNN and Transformer features, a more efficient skip connection is required to extract higher features and pass them to the decoder. As shown in Fig. 2, the encoder and decoder are connected thought Multi-Branches Skip Connections (MBSCs), where MBSC is depicted in Fig. 5. In detail, the proposed MBSC consists of four parallel paths, where each path performs different convolutional operations to learn the salient parts features from variety of feature sources (CNN and Transformer) \(x_i\). The first path consists of a 1 by 1 convolutional block followed by a BN layer. The second path consists of an Average Pooling (AP) block, followed of a 1 by 1 convolutional Block. The third path consists of three consecutive convolutional kernels, which are 1 by 3, 3 by 1 and 3 by 3, each followed by BN layer. The fourth path consists of five consecutive convolutional kernels, which are 1 by 3, 3 by 1, 1 by 5, 5 by 1 and 5 by 5, each followed by BN layer. It should be noted that each path reduces the number of input channels into half in the last convolutional layer. The four paths are joined by concatenation, resulting a doubled number of feature maps of the input features (\(x_i\)). The concatenated features are passed into 3 by 3 convoutional kernel followed by BN layer, which reduces the number of feature maps into half (match the \(x_i\) number of feature maps). Finally, a skip residual is used as depicted in Fig. 5 and followed by a ReLU activation function. For an input \(x_i\) and four MBSC paths output \(Skip_{i_1}\), \(Skip_{i_2}\), \(Skip_{i_3}\), and \(Skip_{i_4}\), respectively. The skip connection on the \(i^{th}\) level \(Skip_{i}\) is defined by:
where \(Skip_{c_i}\) and \(Skip_{i}\) are the concatenation of the four Skip paths output and the Skip features passed to the decoder, respectively. In the proposed MBSC the following properties are considered:
-
The \(n \times n\) convolution block is split into two consecutive convolution block of \(1 \times n\) and \(n \times 1\), to reduce the number of parameters.
-
The last branch is implemented by increasing the kernel size to capture progressively higher features.
-
Since the higher number of parameters block in the third and fourth branches (\(n \times n\) convolutional block) are the last convolutional blocks, the number of channels of this block is reduced to the half to decrease the number of parameters.
-
All branches features are concatenated, and then passed to a 3 by 3 convolutional block to summarize and capture the most important features from all of the four branches. Finally, the residual connection is added, and then passed to the ReLU activation function. As can be seen in Fig 5, the MBSC blocks do not change the dimesion of the 3D input tensors.
3.1.4 The decoder of MB-TrAttUnet architecture
The decoder of MB-TrAttUnet is an Att-Unet-like decoder, as shown in Fig. 2. In detail, the bottleneck feature maps \(x_{5}\) of the encoder are fed into the first expansion layer of the decoder. First, \(x_{5}\) is up-sampled using a linear transformation to obtain \(d_{5}\), and then passed to the decoder as shown in equation (18). On the other hand, the encoder feature maps \(x_{1}, x_{2}, x_{3}\) and \(x_{4}\) are forwarded to the decoder layers of MB-TrAttUnet via MBSC connections, as shown in Fig. 2. Following the Att-Unet architecture Oktay et al. (2018), three linear upsampling layers (US), four decoder layers, four Attention Gates (AG), and four ResBlocks are used, as shown in the following equations:
where ’;’ denotes features concatenation.
Finally, a 1 by 1 convolutional block is used to match the feature map dimension of \(d_{1}\) to the infection mask prediction, which consists of three channels for the Multi-classes segmentation (Background, GGO, and Consolidation).
The Attention Gate (AG) is depicted in Fig. 3 (Oktay et al. 2018), and is defined as follows:
where \(W_{x}\) and \(W_{g}\) are two 1 by 1 convolution kernels performing linear transformations of the number of channels (\(c_{x}\) and \(c_{g}\) of \(x_{i}\) and \(g_{i}\), respectively) to \(c_{int}\). \(\psi _{i}\) consists of \(W_{\psi _{i}} \in \mathbb {R}^{1\times 1}\) followed by Batch Normalization (BN) and sigmoid activation function to learn the spatial attention coefficient \(M_{att_{i_p}}\) for each pixel. The obtained spatial coefficients \(M_{att_i}\) are applied to the skip feature maps of the encoder \(x_{i}\).
where \(\otimes\) is element-wise multiplication operator.
3.2 Multi-classes boundary aware cross-entropy (MBA-CE) loss function
In the field of medical imaging, objects can exhibit high variability in terms of their shapes, intensities, and positions (Bougourzi et al. 2023). This can make it challenging for traditional loss functions, such as Cross-Entropy, to accurately segment the target boundaries, resulting in fuzzy segmented object boundaries (Qin et al. 2019; Bougourzi et al. 2023). To address this issue, we propose a new Edge Aware Loss function for multi-class segmentation that specifically concentrates on segmenting infection boundaries, improving the separation between different classes, and giving greater attention to minority classes. Our proposed Multi-classes Boundary Aware Cross-Entropy (MBA-CE) Loss Functionis defined as following:
Here, \(\mathcal {L}_{Inf}\) and \(\mathcal {L}_{Edges}\) are the Multi-classes infection and edges losses, respectively. Moreover, the value of the weight \(\alpha\) is obtained experimentally and equals 2. By emphasizing the edges between infection types, the MBA-CE Loss Function encourages the model to focus on correctly predicting the boundaries between them, thereby improving its ability to distinguish between different types of infections. For the ground-truth mask width (W) and height (H), \(\mathcal {L}_{Inf}\) loss is defined by:
where \(G_{i}\) and \(p_{i}\) are the infection types ground-truth probability distribution and the model prediction probability distribution of pixel i, respectively. M is the total number of classes.
The \(\mathcal {L}_{Edges}\) is defined by:
where, \(E_{i}\) and \(q_{i}\) are the boundary ground-truth probability distribution and boundary prediction probability distribution of pixel i, respectively. As depicted in Fig. 6, the edge ground-truth E is obtained by multiplying the ground-truth mask G with the union of the morphological gradient applied to the ground-truth masks of GGO and Consolidation. In other words:
Here, Grad() denotes the morphological gradient operation.
Similarly, the edge prediction maps q is obtained by multiplying the model output map p with the union of the morphological gradient applied to the ground-truth masks of GGO and Consolidation. In other words:
3.3 Evaluation measurements
The following evaluation metrics are used: F1-score (F1-S), Dice-score (D-S), Intersection over Union (IoU), and Hausdorff Distance (HD95) in pixels, to compare between different architectures and items. It should be noted that each infection type is evaluated using the four metrics.
It should be noted that F1-score and IoU (Jaccard Index) are micro metrics, where they are calculated for all images at one time using True Positives (TP), True Negatives (TN), False Positives (FP) and False Negatives (FN). However, the Dice-score is the macro version of the \(F1-score\). For N training or test images, it is defined by:
where \(TP_i\), \(TN_i\), \(FP_i\) and \(FN_i\) are True Positives, True Negatives, False Positives and False Negative for the ith image, respectively.
4 Experiments and results
4.1 Datasets and splitting configurations
Table 1 displays datasets labeled for multi-class COVID-19 infection segmentation. To replicate the urgency of pandemic situations, our approach is assessed in two scenarios: Scenario 1, depicting standard data availability, and Scenario 2, concentrating on the influence of limited training data on the segmentation approaches performance. Crafting an effective approach in the latter scenario is especially vital in pandemic situations.
In total, we performed four splitting configurations, in the first two, Dataset_1 RADIOLOGISTS (2019) and Dataset_2 RADIOLOGISTS (2019) are used. In the first splitting configuration, Dataset_1 and 50% of Dataset_2 are used as training data and the remaining 50% of Dataset_2 is used as test data. In the second splitting configuration, the training data are reduced to half to study the efficiency of the proposed approach in a more challenging few-shot learning strategy (less training data are available). More specifically, for the second splitting configuration experiment only 50% of CT-scans from Dataset_1 and 25% of slices from Dataset_2 are used as training data. In the third and fourth configuration splittings, Dataset_3 is used. Where in the third one, 60% of Dataset_3 is used as training data and the remaining 40% is used as testing data. The fourth configuration corresponds to the second evaluation scenario where only 15% of Dataset_3 is used as training data.
Table 2 provides an overview of the four evaluation configurations. It’s important to note that not all slices necessarily exhibit infection, mirroring the real-world scenario where the prevalence of infected slices can vary across cases. Additionally, an infected slice may contain either a single infection type (GGO or Consolidation) or both, a critical aspect for tracking disease progression and severity. In the first two splitting configurations (as shown in Table 2), it’s evident that the number of infected slices constitutes approximately half of the total slices in the training dataset. However, when considering all four splitting configurations, it becomes apparent that the number of slices infected with Consolidation is notably lower, especially in the second evaluation scenario. This imbalance in the training data poses a significant challenge for multi-class Covid-19 infection segmentation.
4.2 Experimental setup
Paszke et al. (2019) Library with NVIDIA RTX A5000 GPU with 24 GB of memory is used to train and test our approach and the comparison ones. The used machine has 11th Gen Intel(R) Core(TM) i9-11900KF (3.50GHz) CPU and 64 of RAM. All models are trained for 60 epochs with batch size of 6 images and a learning rate schedule where the initial \(lr = 0.01\) and then it decays by 0.1 twice after 30 and 50 epochs, respectively. In the training, two augmentation techniques are used: (i) Active Rotation using random angle between \([-35^\circ , 35 ^\circ ]\), and (ii) random horizontal and vertical flipping. Multi-classes CE loss is used for all experiments as the loss function. To investigate the stability of the each architecture performance, each experiment is repeated five times and then the mean and standard deviation are declared for each evaluation metric.
4.3 Experimental results of the first splitting configuration
Table 3 summarizes the experimental results of the first splitting configuration. In order to compare our approach performance with state-of-the-art architectures, three CNN-Based Segmentation architectures (Unet Ronneberger et al. (2015), Attention-Unet Oktay et al. (2018)and Nested-Unet Zhou et al. (2018)), three recent approaches that were designed to segment Covid-19 from the CT-scans (CopleNet Wang et al. (2020), AnamNet Paluru et al. (2021), and SCOATNet Zhao et al. (2021)), and four Transformer-Based architectures (SwinUnet Cao et al. (2021), MTUnet Wang et al. (2022), MISSFormer Huang et al. (2022), and UCTransNet Wang et al. (2022)) are evaluated as shown in Table 3. These results show that our proposed approach outperforms the comparison approaches by considerable margin for both GGO and Consolidation segmentation. For the GGO segmentation results, most of the comparison architectures achieved close performance (except for CopleNet and MTUnet, which achieved the lowest performance). The best comparison approach is MISSFormer for both GGO and Consolidation segmentation. Our approach outperformed the performance of MISSFormer by 1.41%, 6.75%, 1.6% and 3.97 for F-S, D-S, IoU and HD95 for GGO segmentation, respectively. By looking on the standard deviation values of the best comparison architecture, we find that our approach not only outperforms the best competing architecture, but also exhibits more stable performance at different running.
On the other hand, MISSFormer architecture performed the best in all metrics compared to the other state-of-the-art approaches for Consolidation segmentation. However, our approach outperformed its performance by 7%, 7.15%, 6.81% and 0.9 for F-S, D-S, IoU, and HD95 respectively. Compared with the results of GGO, it is noticed that the performance margin between our approach and the state-of-the-art architectures is larger. This proves that our approach can perform accurately even in unbalanced class segmentation scenarios, as is the case in most of Multi-class segmentation tasks in medical imaging domain. The results of GGO and Consolidation show that our approach has a high ability to segment different types of Covid-19 infections.
4.4 Experimental results of the second splitting configuration
Table 4 summarizes the experimental results of the second evaluation scenario, where half of the training data of first splitting configuration is used to investigate the efficiency of the proposed approach in more critical few shot-shot learning scenario. To compare the performance of our approach with state-of-the-art architectures, three CNN-based segmentation architectures (Unet Ronneberger et al. (2015), Att-Unet Oktay et al. (2018), Unet++ Zhou et al. (2018)), three recent approaches that were designed to segment Covid-19 from the CT-scans (CopleNet Wang et al. (2020), AnamNet Paluru et al. (2021), and SCOATNet Zhao et al. (2021)) and four and four Transformer-Based architectures (SwinUnet Cao et al. (2021), MTUnet Wang et al. (2022), MISSFormer Huang et al. (2022), and UCTransNet Wang et al. (2022)), are evaluated as shown in Table 4. Similar to what noticed in the first evaluation scenario, our approach outperforms all of the comparison architectures on all of the four evaluation metrics for both GGO and Consolidation segmentation.
In more details, the three CNN baseline architectures (Unet Ronneberger et al. (2015), Att-Unet Oktay et al. (2018), Unet++ Zhou et al. (2018)) performed similar to the first scenario for GGO segmentation. In contrast, the three CNN baseline architectures failed to segment the Consolidation infection type. Similar behavior is observed for the CopleNet and AnamNet architectures. Table 4 shows that the SCOATNet architecture outperforms the other comparison architectures in segmenting GGO for F1-S, D-S and IoU metrics. Comparing the performance of SCOATNet in the first and second evaluation scenarios (Tables 3 and 4), it is noticed that SCOATNet achieves similar results for GGO segmentation. However, the segmentation results for Consolidation dropped to one-third by reducing the training data to half. For GGO segmentation in the second evaluation scenario, our approach outperforms SCOATNet architecture by 1.91%, 5.67%, and 1.65% for F-S, D-S, and IoU, respectively and outperforms MISSFormer by 7.12 for the HD95 metric. For the transformer based architecture, it is also noticed that their performance in segmentation GGO is decreased. From Consolidation results, it is noticed that the proposed approach surpasses MISSFormer architecture (the best comparison architecture) by a large margin: 8.68%, 3.7%, 7.26% and 1.8 for F-S, D-S, IoU and HD95, respectively. On the other hand, the comparison between the standard deviation values of our approach and the MISSFormer architecture in the second evaluation scenario shows that the performance of our approach is consistent across different runs.
The comparison between the results of the first and the second evaluation scenarios (from the first and second splitting configurations) shows that our approach performs consistently, although the training data in the second evaluation scenario are reduced to half. This proves the efficiency of the proposed components, which will be discussed in detail in the ablation study section.
4.5 Experimental results of the third splitting configuration
Table 5 summarizes the results of the third splitting configuration, where our proposed approach is compared with three baseline CNN architectures, three state-of-the-art approaches for Covid-19 segmentation, and four Transformer-based architectures similar to the first and second splitting configurations. From these results, it is noticed that SCOATNet and UCTransNet achieve similar performance in segmenting both classes (GGO and Consolidation), outperforming the other comparison approaches. However, our approach demonstrates its efficiency by outperforming the best competitor for each metric. For GGO segmentation results, our approach achieves better performance than the top competitor by 0.11%, 1.66%, 0.12%, and 0.87 for F-S, D-S, IoU, and HD95, respectively. Similarly, for Consolidation segmentation, our approach outperforms the best competitor by 2.14%, 1.66%, 0.12%, and 0.87 for F-S, D-S, IoU, and HD95, respectively. In this splitting configuration, with a substantial training dataset from nearly 100 CT scans, it is noticeable that some state-of-the-art approaches come close to our approach, but our approach still exhibits superiority. Furthermore, when compared to the previous two splitting configurations, UCTransNet emerges as one of the top competitors instead of the MISSFormer architecture, which showed lower performance than the best competitors. On the other hand, SCOATNET continues to perform as one of the best competitors. This demonstrates that the performance of certain architectures can vary depending on the scenario and the dataset used. In contrast, the proposed EMB-TrAttUnet consistently delivers strong performance across different scenarios.
4.6 Experimental results of the fourth splitting configuration
Table 6 summarizes the results obtained from the fourth splitting configuration. These results represent the second evaluation scenario of the third splitting configuration, in which the training dataset is reduced to a fourth. The comparison with state-of-the-art approaches reveals the superiority of our approach. When compared with the results from the first evaluation scenario in Table 5, it is noticed that the performance of all approaches has significantly decreased. For example, the F1-score of the top two competitors (SCOATNET and UCTransNet) has decreased by 12.34% and 11.11% for GGO segmentation, respectively. Similarly, their F1-score for Consolidation has decreased by 15.32% and 17.99%, respectively. In contrast, our approach demonstrates consistent performance, with the F1-score decreasing by only 4.2% and 3.86% for GGO and Consolidation, respectively.
4.7 Ablation study
In this section, the significance of each component of the proposed EMB-TrAttUnet approach is studied, which includes Transformer Encoder Path (TE), MBSC connection blocks (MBS), and Multi-classes Boundary Aware Loss Function (MBA). Tables 7 and 8 summarize the ablation study for the first and second evaluation scenarios correspond to the first and second splitting configurations, respectively. The Att-Unet architecture is considered as the baseline architecture for both evaluation scenarios. From Table 7, the comparison between the baseline architecture and the first ablation experiment shows that the Transformer path significantly improves the GGO and Consolidation segmentation results. This shows that the Transformer Path provides diversity of features in the encoding phase. In other words, the fusion of the global contextual and long-range dependencies features using the Transformer path and the local contextual features using the CNN Convolutional blocks provides finer details in the encoding phase, allowing to distinguish between infected, non-infected tissues and different infection types. In the second ablation experiment of Table 7, the results of the Att-Unet architecture with MBSC blocks are depicted. The results show that there is no improvement for GGO segmentation, but the results for Consolidation have dropped. The explanation of this result is that adding more Convolutional layers without rich and diverse features extraction tends to lead to more over-fitting. Experiment (3) shows that adding the Transformer path and MBSC connection blocks improves the results compared to both the baseline experiment and experiment (1) for both segmentation classes. Experiments (4) proves the effectiveness of the proposed Boundary Aware loss function in segmenting the two classes of Covid-19 infections. Compared with the results of Experiment (3), the GGO results are improved by 1%, 3.5%, and 1.19% for F-S, D-S, and IoU, respectively. Similarly, the Consolidation segmentation results are considerably improved by 2.51%, 5.46%, and 2.51% for F-S, D-S, and IoU, respectively.
Table 8 summarizes the ablation study results of the second evaluation scenario. Similar to what were noticed in the first evaluation scenario (Table 7), the Transformer Path provides richer features in the encoding phase, leading to better segmentation performance, especially for the minor class (Consolidation). On the other hand, it is noticed from Experiment (2) that adding the MBSC connection blocks to the baseline architecture (Att-Unet) significantly improves the Consolidation segmentation results, while the GGO results slightly decrease. This shows that MBSC connection blocks can help hard class segmentation when less labelled data is available. However, this can affect the performance of the other classes in the case of only CNN encoder is used. Similar to the first evaluation scenario, Experiment (4) prove the effectiveness of the proposed loss function. By comparing with the results of Experiment (3), the GGO results are improved by 0.8%, 1.7%, and 0.38% for F-S, D-S, and IoU, respectively. Significantly, the Consolidation segmentation results are considerably improved by 8%, 3.93%, and 6.69% for F-S, D-S, and IoU, respectively. These results shows the efficiency of the proposed MBA-CE loss function in few-shot learning scenario, with imbalanced classes.
Visual Comparison of COVID-19 Segmentation Results from the First Evaluation Scenario. The seven rows represents the input slice, ground truth, and the segmentation results of Att-Unet, SCOATNET, UCTransNet, MISSFormer and our proposed EMB-TrAttUnet, respectively. GGO is presented by the green color and Consolidation by the red color
Visual Comparison of COVID-19 Segmentation Results from the Second Evaluation Scenario. The seven rows represents the input slice, ground truth, and the segmentation results of Att-Unet, SCOATNET, UCTransNet, and MISSFormer and our proposed EMB-TrAttUnet, respectively. GGO is presented by the green color and Consolidation by the red color
5 Discussion
5.1 Segmentation visualization analysis
Figures 7 and 8 depict visual comparison examples from the first and second evaluation scenarios which correspond to the first and second splitting configurations, respectively. The approaches that demonstrated the best segmentation performance in comparison to our approach are Att-Unet, SCOATNET, UCTransNet, and MISSFormer. All six examples in Fig. 7 demonstrate that our approach achieves superior segmentation performance for both GGO and Consolidation when compared to the other methods.
In more detail, the first example in Fig. 7 illustrates cases where the infection has spread in both lungs, manifesting as multiple small regions of GGO and Consolidation. In contrast to the comparison methods, our approach effectively segments most of the infected regions, including the smaller ones. This highlights the efficiency of combining CNN and Transformer encoders, where the Transformer path treats the input image as a collection of 2D patches, enabling the extraction of finer infection details, including small regions. Additionally, our proposed loss function places more emphasis on smaller regions.
The second example depicts a case where the infection has spread throughout most of the lung field as GGO, with Consolidation appearing in the lower region of the right lung. The segmentation masks generated by the comparison methods reveal their difficulty in accurately matching the ground truth masks for the GGO regions, particularly near the boundaries. In contrast, our approach demonstrates a strong ability to match the infection boundaries, thanks to the use of our proposed boundary-aware loss function. While some of the comparison approaches have identified Consolidation in the lower regions, many of them misalign with the ground truth mask. Notably, Att-Unet, SCOATNET, and UCTransNet show Consolidation in both lungs, which is inconsistent with the ground truth. In contrast, MISSFormer and our approach accurately locate the region of Consolidation infection (the lower region of the right lung).
The third example presents a case where the infection appears in both lungs and is a combination of medium and small spots, with small Consolidation areas in the lower lung regions. The segmented masks demonstrate that most of the comparison methods struggle to segment the small regions and differentiate between the infection types. In contrast, our approach efficiently segments both medium and small infections and accurately distinguishes between the two classes (GGO and Consolidation), aligning with the ground truth. This underscores that our proposed approach has a better capability to segment small regions, as well as medium and large ones, compared to the state-of-the-art approaches.
The last three examples depict cases where the infection has a peripheral distribution with a mixture of GGO and Consolidation infection types, and Consolidation appears in the lower half of the lung regions. In the fourth example, the predicted masks of the segmentation methods struggle to distinguish between the infection classes and their sizes. For instance, the MISSFormer approach segments all infections as Consolidation, while other approaches consider most of the infected areas as Consolidation. Although our approach identifies most of the infected regions, it shows limited distinguishability between the two infection types in this case.
In the fifth example, the comparison approaches segment all infections as GGO. In contrast, our approach demonstrates a high capability to segment both classes accurately. Similarly, our approach performs well in the last example. However, some of the comparison approaches exhibit lower performance, such as UCTransNet and SCOATNet, as they struggle to segment the infection due to the low contrast between the infection and lung borders.
Figure 8 displays the segmented masks for the second evaluation scenario, where the training data was reduced to half compared to the first splitting configuration. From the visualized six examples, the following observations can be made: In the first example, almost all comparison approaches failed to segment any infection. However, our approach continues to demonstrate good performance in scenarios with more challenging data available. In the second example, all approaches are still capable of segmenting the widely spread infection and also successfully segment the Consolidation type in the lower part of the right lung. However, the accuracy varies from one approach to another. MISSFormer matches the Consolidation prediction, while our approach accurately captures the details of GGO infection in terms of shape and boundaries. This once again highlights the efficiency of the proposed methods, particularly the proposed loss function in this case. In the third example, our approach continues to perform well in segmenting small and medium infection regions with the correct infection type. The last three examples reveal that the comparison approaches miss a significant portion of the infection, especially UCTransNet and SCOATNet in examples 4 and 6. Additionally, it’s worth noting that, as in the previous evaluation scenario, some approaches entirely predict only one infection class. However, our approach still demonstrates a high segmentation capability to accurately segment the infection and efficiently identify its type in scenarios with more data availability limitation regime.
5.2 Early stage infection localization and segmentation
One of the most important steps to stop the spreading of Covid-19 disease and save the infected person live is to detect the infection in the early stages. Where the golden standard RT-PCR test has a considerable false negative rate in this case (Jin et al. 2020). We propose to test the efficiency of our approach in detecting the early infection regions. To do so, we manually selected four CT-scans from Dataset_1 as the testing data, which shows infection in early stages. The remaining CT-scans of Dataset_1 and Dataset_2 are used as the training data. In order to compare with the state-of-the-art, the best performed approaches in the previous evaluated splitting configurations, which are Att-Unet (Oktay et al. 2018), and SCOATNet (Zhao et al. 2021), MISSFormer (Huang et al. 2022), and UCTransNet (Wang et al. 2022). Table 9 summarizes the obtained results, which show the superiority of our approach compared with the comparing ones for both infection types segmentation.
Figure 9 depicts three slices from two CT scans (each scan is represented by 3 slices) and the predicted masks of the four comparison approaches and our approach. The first three examples show that our approach correctly localizes the position and the number of tiny infected regions and classifies them as GGO. However, most of the comparison approaches failed to segment these infected regions. This once again demonstrates the efficiency of the proposed approach in dealing with even the tiniest infected regions in the early stage, allowing for the identification of infections in their very early stages. The second case (examples 4 to 6) depicts a situation where the infection is beginning to develop, and consolidation is starting to appear. In this case as well, it is evident that our approach has a high ability to detect small consolidation regions attached to the GGO infection, as well as small GGO infections spreading in both lungs. These two cases highlight our approach’s capability to localize and segment infections at different stages, including the early stage, which is crucial for detecting infected individuals, preventing the spread of the Covid-19 pandemic, and monitoring the patient’s condition.
Visual Comparison of COVID-19 Segmentation Results for Early Infection Segmentation Scenario. The seven rows represents the input slice, ground truth, and the segmentation results of The seven rows represents the input slice, ground truth, and the segmentation results of Att-Unet, SCOATNET, UCTransNet, and MISSFormer and our proposed EMB-TrAttUnet, respectively. GGO is presented by the green color and Consolidation by the red color
5.3 Model size and inference time comparison
In this section, we investigate the number of parameters and inference times of our approach and the compared ones. Table 10 summarizes these comparisons. It is worth noting that our approach features a higher number of parameters compared to most of the compared architectures. This is due to the proposed Hybrid Encoder and the MBSC block, both of which have proven their efficiency in handling the complex task with very limited training data. Despite the larger parameter count, our approach still delivers competitive inference times. In fact, the inference time for a batch size of 50 slices is less than half a second, which is suitable for real-time scenarios.
6 Conclusion
In this paper, we proposed a Hybrid CNN-Transformer (MB-TrAttUnet) architecture and a Multi-classes Boundary Aware Cross-Entropy (MBA-CE) Loss function for Multi-classes Covid-19 infection segmentation. The Encoder of the proposed MB-TrAttUnet architecture fuses CNN and Transformer blocks to extract richer features, including local, global, and long-range dependencies features. On the other hand, the Decoder of the MB-TrAttUnet is similar to the Att-Unet decoder, which consists of AG and deconvolutional blocks. In addition, we proposed a new skip connection (MBSC) to integrate the encoder features into the decoder layers, where the MBSC blocks extract and combine higher level features related to the finer features of different infection types. On the other hand, MBA-CE Loss function is designed to enhance boundaries segmentation, the separability between classes and minority classes decision.
The performance of the proposed approach is evaluated and compared with three CNN-Based Segmentation architectures (Unet, Nested-Unet and Attention Unet), three proposed segmentation approaches for Covid-19 infection segmentation (CopleNet, AnamNet, and SCOATNet) and four recent Transformer based segmentation architectures (SwinUnet, MTUnet, MISSFormer, and UCTransNet) in two evaluation scenarios using two splitting configurations. From which, our approach outperformed the comparison methods for both Ground-glass opacity (GGO) and Consolidation segmentation. On the other hand, our approach showed consistent performance in the more critical“few-shot”learning scenario (training data, in terms of scans, were reduced by half). In contrast, the performance of the comparison methods decreased in this scenario. Moreover, our approach showed a good ability to deal with imbalanced data and segmenting the infection in early stages. These advantages demonstrate the effectiveness and efficiency of the proposed EMB-TrAttUnet approach in a pandemic scenario where time is critical to save patient lives. As part of our future work, we intend to leverage recent advancements in vision transformer architectures to reduce the number of parameters in our model. Additionally, we plan to extend the application of our approach to various other medical imaging segmentation tasks.
References
Amyar A, Modzelewski R, Li H, Ruan S (2020) Multi-task deep learning based CT imaging analysis for covid-19 pneumonia: classification and segmentation. Comput Biol Med 126:104037
Bharati S, Podder P, Mondal MRH (2020) Hybrid deep learning for detecting lung diseases from x-ray images. Inf Med Unlocked 20:100391
Bougourzi F, Distante C, Ouafi A, Dornaika F, Hadid A, Taleb-Ahmed A (2021) Per-COVID-19: a benchmark dataset for COVID-19 percentage estimation from CT-scans. J Imaging 7:189
Bougourzi F, Contino R, Distante C, Taleb-Ahmed A (2021) Recognition of COVID-19 from CT scans using two-stage deep-learning-based approach: CNR-IEMN. Sensors 21:5878
Bougourzi F, Distante C, Dornaika F, Taleb-Ahmed A, Hadid A (2022) Ilc-unet++ for covid-19 infection segmentation. In: International conference on image analysis and processing, pp 461–472. Springer, Berlin
Bougourzi F, Dornaika F, Taleb-Ahmed A (2022) Deep learning based face beauty prediction via dynamic robust losses and ensemble regression. Knowl-Based Syst 242:108246
Bougourzi F, Distante C, Dornaika F, Taleb-Ahmed A (2023) Cnr-iemn-cd and cnr-iemn-csd approaches for covid-19 detection and covid-19 severity detection from 3d ct-scans. In: Computer vision–ECCV 2022 workshops: Tel Aviv, Israel, October 23–27, 2022, Proceedings, Part VII, pp 593–604. Springer, Berlin
Bougourzi F, Distante C, Dornaika F, Taleb-Ahmed A (2023) Pdatt-unet: pyramid dual-decoder attention unet for covid-19 infection segmentation from ct-scans. Med Image Anal 886:102797
Cao H, Wang Y, Chen J, Jiang D, Zhang X, Tian Q, Wang M (2021) Swin-unet: unet-like pure transformer for medical image segmentation. arXiv preprint arXiv:2105.05537
Chaudhary S, Yang W, Qiang Y (2022) Swin transformer for covid-19 infection percentage estimation from ct-scans. In: International conference on image analysis and processing, pp 520–528. Springer, Berlin
Cong R, Yang H, Jiang Q, Gao W, Li H, Wang C, Zhao Y, Kwong S (2022) Bcs-net: boundary, context, and semantic for automatic covid-19 lung infection segmentation from CT images. IEEE Trans Instrum Meas 71:1–11
Cong R, Zhang Y, Yang N, Li H, Zhang X, Li R, Chen Z, Zhao Y, Kwong S (2022) Boundary guided semantic learning for real-time covid-19 lung infection segmentation system. IEEE Trans Consum Electron 68(4):376–386
Ding W, Abdel-Basset M, Hawash H, Elkomy OM (2021) Mt-ncov-net: a multitask deep-learning framework for efficient diagnosis of covid-19 using tomography scans. IEEE Trans Cybern 99:1
Dosovitskiy A, Beyer L, Kolesnikov A, Weissenborn D, Zhai X, Unterthiner T, Dehghani M, Minderer M, Heigold G, Gelly S (2020) An image is worth 16x16 words: transformers for image recognition at scale. arXiv preprint arXiv:2010.11929
Duong LT, Le NH, Tran TB, Ngo VM, Nguyen PT (2021) Detection of tuberculosis from chest x-ray images: boosting the performance with vision transformer and transfer learning. Expert Syst Appl 184:115519
Duong LT, Nguyen PT, Iovino L, Flammini M (2023) Automatic detection of covid-19 from chest x-ray and lung computed tomography images using deep neural networks and transfer learning. Appl Soft Comput 132:109851
Fung DL, Liu Q, Zammit J, Leung CK-S, Hu P (2021) Self-supervised deep learning model for covid-19 lung CT image segmentation highlighting putative causal relationship among age, underlying disease and covid-19. J Transl Med 19:1–18
Ghosh S, Das N, Das I, Maulik U (2019) Understanding deep learning techniques for image segmentation. ACM Comput Surv 52(4):1–35
Goceri E (2023) Medical image data augmentation: techniques, comparisons and interpretations. Artif Intell Rev 20:1–45
Hatamizadeh A, Tang Y, Nath V, Yang D, Myronenko A, Landman B, Roth H. R, Xu D (2022) Unetr: transformers for 3d medical image segmentation. In: Proceedings of the IEEE/CVF winter conference on applications of computer vision, pp 574–584
Hefeda MM (2020) CT chest findings in patients infected with covid-19: review of literature. Egypt J Radiol Nucl Med 51(1):1–15
Huang X, Deng Z, Li D, Yuan X, Fu Y (2022) Missformer: an effective transformer for 2d medical image segmentation. IEEE Trans Med Imaging
Ilhan A, Alpan K, Sekeroglu B, Abiyev R (2023) Covid-19 lung CT image segmentation using localization and enhancement methods with u-net. Proc Comput Sci 218:1660–1667
Jasmine Pemeena Priyadarsini M, Rajini G, Hariharan K, Utkarsh Raj K, Bhargav Ram K, Indragandhi V, Subramaniyaswamy V, Pandya S et al (2023) Lung diseases detection using various deep learning algorithms. J Healthcare Eng 2023:1
Jin Y-H, Cai L, Cheng Z-S (2020) A rapid advice guideline for the diagnosis and treatment of 2019 novel coronavirus (2019-nCoV) infected pneumonia (standard version), Mil Med Res 7:4
Jin G, Liu C, Chen X (2022) An efficient deep neural network framework for covid-19 lung infection segmentation. Inf Sci 612:745–758
Khan S, Naseer M, Hayat M, Zamir S. W, Khan F. S, Shah M (2021) Transformers in vision: a survey, ACM Computing Surveys (CSUR). ACM, New York
Li Z, Zhao S, Chen Y, Luo F, Kang Z, Cai S, Zhao W, Liu J, Zhao D, Li Y (2021) A deep-learning-based framework for severity assessment of covid-19 with CT images. Expert Syst Appl 185:115616
Li Y, Wang S, Wang J, Zeng G, Liu W, Zhang Q, Jin Q, Wang Y (2021) Gt u-net: a u-net like group transformer network for tooth root segmentation. In: International workshop on machine learning in medical imaging, pp 386–395. Springer, Berlin
Li J, Chen J, Tang Y, Wang C, Landman B. A, Zhou S. K (2023) Transforming medical imaging with transformers? a comparative review of key properties, current progresses, and future perspectives. Med Image Anal p 102762
Liang S, Zhang W, Gu Y (2021) A hybrid and fast deep learning framework for covid-19 detection via 3d chest CT images. In: Proceedings of the IEEE/CVF international conference on computer vision, pp 508–512
Litjens G, Kooi T, Bejnordi BE, Setio AAA, Ciompi F, Ghafoorian M, Van Der Laak JA, Van Ginneken B, Sánchez CI (2017) A survey on deep learning in medical image analysis. Med Image Anal 42:60–88
Liu Z, Lin Y, Cao Y, Hu H, Wei Y, Zhang Z, Lin S, Guo B (2021) Swin transformer: hierarchical vision transformer using shifted windows. In: Proceedings of the IEEE/CVF international conference on computer vision, pp 10012–10022
Liu X, Wang K, Wang K, Chen T, Zhang K, Wang G (2020) Kiseg: a three-stage segmentation framework for multi-level acceleration of chest CT scans from covid-19 patients. In: International conference on medical image computing and computer-assisted intervention, pp 25–34. Springer, Berlin
Maji D, Sigedar P, Singh M (2022) Attention res-unet with guided decoder for semantic segmentation of brain tumors. Biomed Signal Process Control 71:103077
Minaee S, Boykov YY, Porikli F, Plaza AJ, Kehtarnavaz N, Terzopoulos D (2021) Image segmentation using deep learning: a survey. IEEE transactions on pattern analysis and machine intelligence
Mu N, Wang H, Zhang Y, Jiang J, Tang J (2021) Progressive global perception and local polishing network for lung infection segmentation of covid-19 CT images. Pattern Recogn 120:108168
Müller D, Soto-Rey I, Kramer F (2021) Robust chest CT image segmentation of covid-19 lung infection based on limited data. Inf Med Unlocked 25:100681
Oktay O, Schlemper J, Folgoc L. L, Lee M, Heinrich M, Misawa K, Mori K, McDonagh S, Hammerla N. Y, Kainz B, et al.(2018) Attention u-net: learning where to look for the pancreas. arXiv preprint arXiv:1804.03999
Paladini E, Vantaggiato E, Bougourzi F, Distante C, Hadid A, Taleb-Ahmed A (2021) Two ensemble-cnn approaches for colorectal cancer tissue type classification. J Imaging 7(3):51
Paluru N, Dayal A, Jenssen HB, Sakinis T, Cenkeramaddi LR, Prakash J, Yalavarthy PK (2021) Anam-net: anamorphic depth embedding-based lightweight cnn for segmentation of anomalies in covid-19 chest CT images. IEEE Trans Neural Netw Learn Syst 32(3):932–946
Parasher A (2021) COVID-19: current understanding of its pathophysiology. Clin Present Treat Postgrad Med J 97:312–320
Paszke A, Gross S, Massa F, Lerer A, Bradbury J, Chanan G, Killeen T, Lin Z, Gimelshein N, Antiga L et al (2019) Pytorch: an imperative style, high-performance deep learning library. Adv Neural Inf Process Syst 32
Pezzano G, Díaz O, Ripoll VR, Radeva P (2021) Cole-cnn+: context learning-convolutional neural network for covid-19-ground-glass-opacities detection and segmentation. Comput Biol Med 136:104689
Qin X, Zhang Z, Huang C, Gao C, Dehghan M, Jagersand M (2019) Basnet: boundary-aware salient object detection. In: Proceedings of the IEEE/CVF conference on computer vision and pattern recognition, pp 7479–7489
RADIOLOGISTS (2019) COVID-19 CT-scans segmentation datasets, http://medicalsegmentation.com/covid19/. Accessed 18 Aug 2021
Rehman A, Khan A, Fatima G, Naz S, Razzak I (2023) Review on chest pathogies detection systems using deep learning techniques. Artif Intell Rev 1–47
Ronneberger O, Fischer P, Brox T(2015) U-Net: convolutional networks for biomedical image segmentation. In: Medical image computing and computer-assisted intervention - MICCAI 2015, Lecture Notes in Computer Science, pp 234–241. Springer, Cham
Roy S, Menapace W, Oei S, Luijten B, Fini E, Saltori C, Huijben I, Chennakeshava N, Mento F, Sentelli A et al (2020) Deep learning for classification and localization of covid-19 markers in point-of-care lung ultrasound. IEEE Trans Med Imaging 39(8):2676–2687
Salehi S, Abedi A, Balakrishnan S, Gholamrezanezhad A et al (2020) Coronavirus disease 2019 (covid-19): a systematic review of imaging findings in 919 patients. AJR Am J Roentgenol 215(1):87–93
Saood A, Hatem I (2021) Covid-19 lung CT image segmentation using deep learning methods: U-net versus segnet. BMC Med Imaging 21(1):1–10
Shi H, Han X, Jiang N, Cao Y, Alwalid O, Gu J, Fan Y, Zheng C (2020) Radiological findings from 81 patients with covid-19 pneumonia in Wuhan, China: a descriptive study. Lancet Infect Dis 20(4):425–434
Simonyan K, Zisserman A (2014) Very deep convolutional networks for large-scale image recognition. arXiv preprint arXiv:1409.1556
Soomro TA, Zheng L, Afifi AJ, Ali A, Yin M, Gao J (2022) Artificial intelligence (ai) for medical imaging to combat coronavirus disease (covid-19): a detailed review with direction for future research. Artif Intell Rev 1–31
Tomar N. K, Jha D, Riegler M. A, Johansen H. D, Johansen D, Rittscher J, Halvorsen P, Ali S (2022) Fanet: a feedback attention network for improved biomedical image segmentation. IEEE Trans Neural Netw Learn Syst 34:1
Touvron H, Cord M, Douze M, Massa F, Sablayrolles A, Jégou H (2021) Training data-efficient image transformers & distillation through attention, in International conference on machine learning, pp 10347–10357, PMLR
Valanarasu JMJ, Sindagi VA, Hacihaliloglu I, Patel VM (2021) Kiu-net: overcomplete convolutional architectures for biomedical image and volumetric segmentation. IEEE Trans Med Imaging 41(4):965–976
Van der Velden BH, Kuijf HJ, Gilhuijs KG, Viergever MA (2022) Explainable artificial intelligence (xai) in deep learning-based medical image analysis. Med Image Anal 79:102470
Vantaggiato E, Paladini E, Bougourzi F, Distante C, Hadid A, Taleb-Ahmed A (2021) Covid-19 recognition using ensemble-cnns in two new chest x-ray databases. Sensors 21(5):1742
Vaswani A, Shazeer N, Parmar N, Uszkoreit J, Jones L, Gomez A. N, Kaiser Ł, Polosukhin I (2017) Attention is all you need. Adv Neural Inf Process Syst 30:5998
Vaswani A, Shazeer N, Parmar N, Uszkoreit J, Jones L, Gomez A.N, kaiser, Polosukhin I (2017) Attention is all you need. Adv Neural Inf Process Syst 30
WHO (2023) World Health Organization. https://covid19.who.int/. Accessed 08 Nov 2023
Wang G, Liu X, Li C, Xu Z, Ruan J, Zhu H, Meng T, Li K, Huang N, Zhang S (2020) A noise-robust framework for automatic segmentation of covid-19 pneumonia lesions from CT images. IEEE Trans Med Imaging 39(8):2653–2663
Wang X, Yuan Y, Guo D, Huang X, Cui Y, Xia M, Wang Z, Bai C, Chen S (2022) Ssa-net: spatial self-attention network for covid-19 pneumonia infection segmentation with semi-supervised few-shot learning. Med Image Anal 79:102459
Wang H, Cao P, Wang J, Zaiane OR (2022) Uctransnet: rethinking the skip connections in u-net from a channel-wise perspective with transformer. Proc AAAI Conf Artif Intell 36:2441–2449
Wang H, Xie S, Lin L, Iwamoto Y, Han X.-H, Chen Y.-W, Tong R (2022) Mixed transformer u-net for medical image segmentation. In: ICASSP 2022–2022 IEEE international conference on acoustics, speech and signal processing (ICASSP), pp 2390–2394, IEEE
Wu Y-H, Gao S-H, Mei J, Xu J, Fan D-P, Zhang R-G, Cheng M-M (2021) Jcs: an explainable covid-19 diagnosis system by joint classification and segmentation. IEEE Trans Image Process 30:3113–3126
Xie L, Huang J, Yu J, Zeng Q, Hu Q, Chen Z, Xie G, Feng Y (2023) Cntseg: a multimodal deep-learning-based network for cranial nerves tract segmentation. Med Image Anal 86:102766
Yao Q, Xiao L, Liu P, Zhou SK (2021) Label-free segmentation of covid-19 lesions in lung ct. IEEE Trans Med Imaging 40(10):2808–2819
Yu Q, Qi L, Gao Y, Wang W, Shi Y (2022) Crosslink-net: double-branch encoder network via fusing vertical and horizontal convolutions for medical image segmentation. IEEE Trans Image Process 31:5893–5908
Zhang K, Liu X, Shen J, Li Z, Sang Y, Wu X, Zha Y, Liang W, Wang C, Wang K et al (2020) Clinically applicable ai system for accurate diagnosis, quantitative measurements, and prognosis of covid-19 pneumonia using computed tomography. Cell 181(6):1423–1433
Zhao S, Li Z, Chen Y, Zhao W, Xie X, Liu J, Zhao D, Li Y (2021) Scoat-net: a novel network for segmenting covid-19 lung opacification from CT images. Pattern Recogn 119:108109
Zhou Z, Rahman Siddiquee M. M, Tajbakhsh N, Liang J (2018) UNet++: a nested U-net architecture for medical image segmentation. In: Stoyanov D, Taylor Z, Carneiro G, eds, Deep learning in medical image analysis and multimodal learning for clinical decision support, Lecture Notes in Computer Science, pp 3–11. Springer, Cham
Acknowledgements
This work is partially supported by Grant PID2021-126701OB-I00 funded by MCIN/AEI/10.13039/501100011033 and by“ERDF A way of making Europe”.
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Bougourzi, F., Dornaika, F., Nakib, A. et al. Emb-trattunet: a novel edge loss function and transformer-CNN architecture for multi-classes pneumonia infection segmentation in low annotation regimes. Artif Intell Rev 57, 90 (2024). https://doi.org/10.1007/s10462-024-10717-2
Accepted:
Published:
DOI: https://doi.org/10.1007/s10462-024-10717-2