Abstract
Medical instrument detection is essential for computer-assisted interventions, since it facilitates clinicians to find instruments efficiently with a better interpretation, thereby improving clinical outcomes. This article reviews image-based medical instrument detection methods for ultrasound-guided (US-guided) operations. Literature is selected based on an exhaustive search in different sources, including Google Scholar, PubMed, and Scopus. We first discuss the key clinical applications of medical instrument detection in the US, including delivering regional anesthesia, biopsy taking, prostate brachytherapy, and catheterization. Then, we present a comprehensive review of instrument detection methodologies, including non-machine-learning and machine-learning methods. The conventional non-machine-learning methods were extensively studied before the era of machine learning methods. The principal issues and potential research directions for future studies are summarized for the computer-assisted intervention community. In conclusion, although promising results have been obtained by the current (non-) machine learning methods for different clinical applications, thorough clinical validations are still required.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
With the increasing financial pressure on the healthcare system, a general trend exists toward efficient workflow, shortening procedure time, and higher clinical outcomes in the treatment, resulting in fewer repeats for any given intervention or surgery. To guide interventional operations, e.g., cardiac intervention and needle biopsy, advanced medical imaging systems such as ultrasound (US) and fluoroscopy are utilized. The imaging system can display the interventional activities inside the patient’s body. It not only offers clinicians the visualization and measurement of anatomical structures, but also enables the guidance of medical instruments inside the patient’s body without an open incision. This approach is commonly known as image-guided minimally invasive intervention (Douglas et al. 2001; Germano 2002; Peters 2006; Cleary and Peters 2010), and is being increasingly adopted in many treatments and surgical applications because of its lower risk of complications, shorter patient recovery time, and therefore lower cost.
Among the key imaging modalities used in image-guided minimally invasive surgery, US imaging has received significant attention in recent years because of its advantages of being widely available, non-ionizing, and having real-time performance (Douglas et al. 2001). Furthermore, the US offers the unique benefit of having a wide range of transducers that can be used in different application scenarios from the operation room to emergency medical units (Scanlan et al. 2001). As a consequence, US-guided interventional procedures have been investigated and utilized in different clinical fields, such as biopsy (Hatada et al. 2000; Coplen et al. 1991), regional anesthesia (Barrington and Kluger 2013), ablation therapy (Sheafor et al. 1998; Machi et al. 2001), prenatal diagnosis and therapy (Oepkes et al. 2007) and cardiac interventions (structural and congenital heart disease) (Spencer et al. 2013; Jan et al. 2020).
There are commonly two US formats being used, i.e., 2D images or 3D volumes, which face different challenges for instrument guidance in clinical practice. As for 2D images, the alignment of the 2D US plane and the instrument is the most challenging part for the clinician. For example, in needle-based anesthesia, as shown in Fig. 1, the US plane needs to be placed parallel to the needle to visualize the instrument in the US image. Thus the clinical staff has to carefully align the needle and the US plane while looking at the screen for the operation. Besides the above alignment difficulty, it is still challenging for clinicians to distinguish the instrument from the background tissue in the B-mode images, which requires extra training to achieve the instrument interpretation, such as Fig. 1c. Regarding the 3D US, the 3D volumetric data is rendered and projected on the 2D display with compromised spatial information, which is hard to be interpreted directly. As a consequence, a common practice is to manually extract the slice containing the instrument from the volumetric data, which is time-consuming and unfavorable in clinical practice. An example is shown in Fig. 2, which demonstrates a rendered 3D volume and a sliced plane containing the instrument for cardiac intervention. Even if the slice is obtained, localizing the tiny instrument in the US image is still difficult. Therefore, automatic instrument detection and tracking could reduce procedure time for US-guided interventions. It would simplify the manipulations of the US transducer and reduce the total complexity, which would benefit to both patients and interventionalist. When comparing 2D US and 3D US for instrument guidance, it is easier to achieve real-time performance with the 2D US. This benefit is because the 2D US has a much lower amount of data to be processed. Many algorithms have been designed during past decades for the 2D US. From a financial perspective, 2D US imaging is much cheaper and accessible in most hospitals. In addition, it is not easy to navigate the US volumes, since sonographers are applying multi-planar slices to visualize an arbitrary slice in the volume, which requires time-consuming user manipulation. Therefore, the advantages of efficient intervention guidance are compromised. For these reasons, the 2D US is more widely utilized for procedure guidance than the 3D US. However, in contrast to 2D, 3D US provides better spatial information of the instrument position in the global view, and can better help the clinicians plan and make the decision in the procedures (St-Amant et al. 2016). With the recently developed algorithms in the 3D US, the slices containing the instrument can be automatically extracted from the complex 3D volumetric data, which drastically reduces the inefficiency of user manipulation (Pourtaherian et al. 2017a; Arif et al. 2019; Yang et al. 2020b).
US-guided needle therapy (Pourtaherian 2018): a the clinical staff has to manage the multi-fold coordination of \(\textcircled {1}\) needle, \(\textcircled {2}\) US transducer, while \(\textcircled {3}\) looking at the US screen (Courtesy of Philips Ultrasound). b Schematic representation of guiding a needle using US imaging, depicting an example situation for regional anesthesia, where the needle tip is outside the imaging plane and is approaching an erroneous target area. c B-mode US slice contains the needle, pointed by the green arrow
Existing approaches for instrument detection can be classified into two types: (1) instrument detection based on external or internal sensing devices (hardware-based), such as optical fiber sensing (Xia et al. 2015), electromagnetic tracking (Krücker et al. 2007), and robot-guided detection (Nadeau et al. 2014); (2) image-based approaches, without using any additional sensors or devices. Although sensing-based methods have achieved promising results, they have relatively high equipment costs, and the involved sensors complicate the system setup in the operation room. Therefore, a broad acceptance of the sensing-based approaches is hampered in clinical practice. In contrast, image-based approaches have been proposed to detect medical instrument in US images. Several relevant computer vision tasks are studied in the literature for image-based instrument detection. First, classification is commonly defined as the prediction of the type of object in the given images, such as whether the instrument exists or not. In addition, localization and detection are used to describe the task of finding the instrument’s position in the US images, such as finding the instrument tip point or its skeleton in the images. Finally, the most challenging task is image semantic (instance) segmentation, assigning the semantic class to each image pixel. Based on the segmentation results, instrument detection and localization can be easily achieved.
Various approaches have been introduced since the first literature on instrument modeling by Draper et al. (2000), including the latest deep learning-based methods. As illustrated in Fig. 3a, the first work on medical instrument detection appeared in 2000, but the related papers only grew rapidly after 2012. With these fast developments, it is necessary to have a dedicated review and summarize the current trends and directions in the research field. This paper presents a comprehensive review of image-based instrument detection in the US for minimally invasive interventions. To the best of our knowledge, this is the first review paper on medical instrument detection in US imaging. Recently Beigi et al. (2020) published a review on needle localization and visualization in US imaging. In contrast, our paper has a much broader scope, covering medical instrument detection in general, including the non-needle tool, such as a catheter. The non-needle tools (e.g., catheters) have a deformable shape compared to the rigid needle. Due to the different reflection patterns between metal and plastic, the catheter has a distorted thicker appearance than the needle. Moreover, the cardiac US image usually has a lower resolution than commonly used US images. All these challenges make instrument detection in the US more difficult.
The object of this paper is to review the state-of-the-art of medical instrument detection in the US. The main methods, from the conventional image processing methods to the latest deep learning methods, are discussed and compared. Key clinical applications are also introduced. This review article is organized as follows. Section 2 describes the literature selection method for this review article; an exhaustive literature search is performed to include all related papers. Section 3 discusses the main clinical applications before introducing the detailed technical methods, including needle anesthesia, biopsy, prostate brachytherapy, and cardiac interventions with catheters. A literature overview is summarized in Fig. 3b. Section 4 reviews the instrument detection methodologies, which clusters the related literature into non-machine-learning approaches and machine-learning approaches based on their key contributions. More specifically, non-machine-learning methods are grouped into physical space-based methods and projection space-based methods, while machine learning methods can be categorized into handcrafted feature-based methods and recent deep learning-based methods. The published papers, categorized based on the adopted taxonomy, are summarized in Fig. 3c. Section 5 systematically introduces the evaluation metrics, datasets, and experimental results of these papers. Section 6 summarizes the best-performing methods and discusses the remaining key issues and future research directions. Conclusions are given in Sect. 7.
2 Literature selection
We performed the literature search in the key search engines and indexing databases, including Google Scholar, PubMed, and Scopus. We utilized the search criteria ’(Needle OR Catheter OR Instrument) AND (Detection OR Segmentation OR Localization) AND Ultrasound’. We reviewed all the searched papers by title and abstract to ensure the content was relevant. If there is uncertainty, we read the paper to see if it is related. We also checked these papers’ references to ensure the related literature is not missing. The associated articles are mainly published in the journals of IEEE TMI, IEEE TBME, IEEE JBHI, IEEE TUFFC, Medical Physics, MedIA, IJCARS, Ultrasound in Medicine and Biology, etc. and the conference proceedings of MICCAI, SPIE Medical Imaging, IEEE ISBI, IPCAI, IEEE IUS, IEEE ICIP, and IEEE EMBC.
3 Clinical applications
This section discusses the main clinical applications related to medical instrument detection. The four clinical applications receiving the most attention in the literature are reviewed, as shown in Fig. 3b.
3.1 US-guided regional anesthesia
Needle-based regional anesthesia or imposing blockade is essential in current clinical practice, which provides a safer and more accurate intervention for further procedures. Conventional regional anesthesia requires experienced clinicians to deliver the medicine to the correct region, commonly guided by US imaging since it provides clinical staff with a fast and convenient visualization solution. However, as shown in Fig. 1, multi-fold coordination of the US screen, needle, and US probe complicates the procedure and hampers the operation outcomes with higher risks. As a result, extensive training is required to achieve a successful therapy under the guidance of the US.
To visualize the needle during the US-guided regional anesthesia or blockade, an essential condition should be achieved in conventional 2D US imaging: the needle should be positioned in-plane in 2D images, where the needle is visualized as a bright line, requiring the alignment between the instrument and the US plane (Mwikirize et al. 2016). However, this 2D US-guided therapy faces the challenges of the instrument being invisible (Beigi et al. 2017b) or the instrument being out of the plane (Pourtaherian et al. 2017a). Consequently, the 3D volumetric US has been gradually adopted in clinical practice because it can provide richer spatial information about the needle. However, the complicated 3D image visualization hampers the efficiency of the clinicians when they are searching for the needle and guiding it to the target region. Consequently, automatic needle detection in the US has been investigated to facilitate clinical interventions and improve operational outcomes.
Automated needle detection for anesthesia or blockade has been studied on clinical datasets (Mwikirize et al. 2016; Pourtaherian et al. 2017a). A needle tip localization method for the 2D US image is validated on bovine, porcine, kidney, and liver datasets (Mwikirize et al. 2016), which achieved a mean localization error of \(0.3\pm 0.06\) mm. Similarly, the Gabor-based needle detection method in the 3D US was validated on the patient dataset, which achieved a detection error of 0.68 mm (Pourtaherian et al. 2017a). These studies demonstrated good results from the clinical aspect, and needle detection in the US is proven to be a promising solution to facilitate this type of regional intervention. Nevertheless, these limited offline validations with small clinical datasets are insufficient; further extensive clinical validation is needed. In particular, clinical trials for online validation are necessary to validate the algorithm’s effectiveness for operation guidance.
3.2 US-guided biopsy
Biopsy, i.e., taking a small sample of body tissue, is essential for diagnosis, such as liver, breast, or prostate biopsy, especially for finding the malignant tissue, as shown in Fig. 4. To obtain the tissue samples, needle biopsy or open surgical biopsy is considered, based on the suspected pathology, patient health condition, and procedure complexity. Although conventional open surgery biopsy provides good diagnostic results, the less invasive needle-based biopsy becomes attractive. Historically, a needle-based biopsy was performed by clinicians in special procedure rooms with interventional radiology suites, which is, however, gradually replaced by a US imaging system because of its lower cost, higher healthcare efficiency, and better tissue characterization. But the drawbacks of US imaging need to be addressed, like difficult interpretation, lower image contrast than traditional X-Ray imaging, and extra training required for clinicians (Patel et al. 2019).
Similar to the above regional anesthesia, multi-fold coordination has complicated the procedure of US-based needle guidance. As a result, automatic instrument detection is desired to help clinicians perform their tasks. Moreover, US imaging is also increasingly used for tissue characterization or abnormality detection, facilitating biopsy procedures and instrument detection. By providing richer 3D spatial information, 3D US can better support the clinicians to perform the operations and reduce the risk for patients.
Arif et al. (2019) have validated the automated needle detection on the liver biopsy dataset, where the needle can be detected with the mean error of 1 mm (position) and 2\(^\circ\) (orientation), respectively. This experimental result indicates that robust needle detection is possible for 3D US-guided liver biopsy taking. Nevertheless, their dataset is relatively small, including only 8–9 patient images. Because deep learning-based methods can easily overfit the background anatomical tissues when they are trained with limited data, further extensive validation on a large patient dataset is needed for clinical evaluation and practice.
3.3 US-guided prostate brachytherapy
Prostate brachytherapy is one of the popular treatments for patients at the early stage of cancer development. It delivers high-dose-rate (HDR) radiation or low-dose-rate (LDR) radiation to the tumor without affecting the normal tissue around abnormal areas. The needles or catheters are utilized to place radioactive sources in the tumor regions, as shown in Fig. 5. In practice, radioactive particles (i.e., small-size seeds) are only placed for LDR procedures. For HDR, catheters are inserted and a single radioactive source is brought to planned positions with those catheters. Transrectal Ultrasound (TRUS) has been utilized since the 1980s (Banerjee et al. 2017) to perform the operation, which is the key imaging modality for the radiologist to visualize the region-of-interested for the operation.
Example of US-guided prostate brachytherapy. Needles are guided by US images to place radiation seeds for prostate cancer (source: Understanding Brachytherapy for Prostate Cancer from https://www.prostate.org.au/)
In contrast to other applications, prostate brachytherapy requires that multiple instruments are inserted into the prostate, so that multi-instrument detection in TRUS is essential for successful operation planning (commonly 12–20 instruments inserted into the prostate (Zhang et al. 2020d)). However, because of this requirement and instrument placement condition, i.e., needles are close to each other, the detection algorithms need to be stable and accurate enough to detect multiple objects, which is rarely studied in state-of-the-art solutions. Moreover, besides this multi-detection challenge, an efficient detection algorithm is required because the typical operation time is around 90 minutes for prostate brachytherapy (Banerjee et al. 2017). Zhang et al. (2020c) have validated the needle detection on the patient dataset, which demonstrates the instrument detection procedures can be accelerated to a half minute with a tip location error of around 1 mm. In contrast, conventional needle digitization takes around 15-20 minutes by an experienced physician. Therefore, automated instrument detection can facilitate clinicians to find the instrument during the procedures.
3.4 US-guided catheterization
US-guided catheterizations have been utilized in clinical practice, especially for cardiac applications such as RF-ablation and Transcatheter Aortic Valve Implantation (TAVI) procedures. Similar to US-guided needle therapy, US-guided catheterization faces the challenge of multi-fold coordination. Moreover, due to complex anatomical structures in the heart chambers, clinicians have to localize the less obvious catheter (than the metal needle) in 3D volumetric data using a slice-by-slice tuning procedure, which is time-consuming and complicates the operation. Nevertheless, 3D US is attractive for catheterizations because it is radiation-free and easy-to-use, and offers more spatial information for tissues (Yang et al. 2019e). As a result, it has been investigated as a good choice to replace or support the current X-Ray imaging for cardiac interventions. An example of cardiac catheter detection in 3D volumetric data is shown in Fig. 6.
Several solutions have been proposed to detect the catheter in 3D US images (Cao et al. 2013; Yang et al. 2019e, 2020a), which employed different methods as reviewed in Sect. 4. However, several challenges still need to be addressed in future work: (1) US-guided catheterization is not widely accepted for clinical practice, so expert knowledge is limited by experience. Furthermore, it is unclear how accurate and fast instrument detection should be. (2) Due to the hardware limitations of the US probe, the 3D volumetric data only focuses on a small field-of-view of the heart chamber. So the 3D US cannot provide the guidance before the instrument is inserted into the target region, thus X-ray imaging is still required to perform catheter guidance in the vein, hampering the flexibility of US-guided interventions. (3) Only limited studies have been performed on US-guided catheterizations compared to needle-based interventions, which makes US-guided catheterizations less mature in the field of computer-assisted interventions.
4 Instrument detection methods
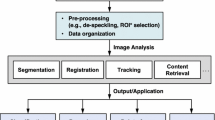
The systematic pipeline of the image-based instrument detection is shown in Fig. 7. As can be seen, the input US image is usually first pre-processed, such as region-of-interest (ROI) selection, image enhancement, and other transformations to prepare the image for the next steps. The instrument detection or segmentation algorithm is then applied to localize the instrument. Specifically, the detection is defined as localizing the instrument in the US image with, e.g., a bounding box, axis, or skeleton. Alternatively, the segmentation algorithm derives a more precise result by assigning each image pixel (voxel) a category, i.e., the instrument or not. Therefore, instrument segmentation is more complicated than instrument detection in most cases. In this article, detection is a more generalized term than segmentation since it is easy to obtain detection results based on segmented images. Finally, the post-processing steps can be applied to the detection or segmentation results to refine the output of the algorithm, such as model-fitting and bounding box refinement, which improves the accuracy of instrument detection.
Systematic pipeline of instrument detection in US image. First, the input US image is pre-processed by ROI selection and image enhancement methods. Then, as the key step of the pipeline, a detection or segmentation algorithm is applied to localize the instrument in the image, where the methods range from the conventional image processing methods to the state-of-the-art deep learning methods. Finally, based on the result of the detection or segmentation, the instrument is localized in the image after the post-processing step, such as model-fitting and bounding box refinement. Note that in some methods, pre-processing and/or post-processing may not be required
In this section, the detection step is considered as the key component of the instrument detection pipeline, and the existing detection methods are divided into two classes: non-machine-learning methods and machine learning methods. Most of the non-machine-learning publications originate from the period before the era of machine learning. Later on, driven by the rapid development of machine learning methods, high-performance computing, and advanced US imaging systems, instrument detection methods have shifted toward machine-learning-based methods.
4.1 Non-machine-learning methods
As for non-machine-learning approaches for instrument detection, there are two major classes based on how a priori knowledge is used: physical space methods and projection space methods. The physical space methods perform mathematical or geometrical modeling in the physical space with the standard spatial coordinate system in a straightforward manner. In contrast, the projection space methods first apply the specific spatial transformation or projection on the US image (after thresholding with the prior knowledge of the instrument), i.e., from the physical space to the projection space, and then detects the instrument in the projection space.
4.1.1 Physical space methods
The preliminary study for instrument modeling can be traced back to Draper et al. (2000) (for 2D US), Smith et al. (2001), Novotny et al. (2003) and Wei et al. (2005) (all for 3D US) with coarse segmentation by using simple thresholding or difference mapping. They modeled the instrument as a regional pixel or voxel cluster with the longest and straightest connectivity groups, which was implemented based on group connectivity analysis or Principal Component Analysis (PCA). After these initial studies, a texture-based instrument segmentation method (Linguraru and Howe 2006; Linguraru et al. 2007), which was described by statistical features, was proposed for 3D US images by applying Expectation-maximization, local texture analysis, and PCA, which iteratively segments the instrument from an in-vitro dataset. Zhao et al. (2009) and Qiu et al. (2014) introduced 3D gradient orientation to calculate the instrument phase information, which segments the needle by applying Line-Support-Region analysis for grouped regions. Similar to this local gradient analysis, a histogram analysis method was designed by McSweeney et al. (2014) to threshold the 3D US image, which localizes the needle by morphological operation and line fitting. The Frangi vesselness filter (line filter) (Frangi et al. 1998) was considered to describe the instrument better and filter out instrument-related points in 3D US images, based on assumptions on the high contrast of instrument edges compared to the background and a tubular structure (Ren and Dupont 2011; Mohareri et al. 2013; Zhao et al. 2013a; Malekian et al. 2014). Similarly, Yan et al. (2012) proposed segmenting the needle in the 3D US by a strong assumption of the instrument tube shape, obtained by shape-based level set segmentation. A complex decision-making chain is applied to track the needle with the obtained instruments set. Although these methods may include different pre-/post-processing steps, the core idea is to extract tubular-like structures by Hessian matrix analysis for local intensity distributions. Further processing steps range from simple thresholding (Ren and Dupont 2011; Mohareri et al. 2013; Agarwal et al. 2019) to Random sample consensus (RANSAC) model-fitting (Malekian et al. 2014) with Kalman filtering in time sequence-based 3D US datasets (Zhao et al. 2013a, b). Moreover, the local Hessian matrix is also applied to detect the shadow of steep needles in 3D US (Pourtaherian et al. 2016), which automatically extracts the 2D slice containing the needle for in-plane visualization.
Besides the above vesselness filter-based methods, a template matching with a pre-defined catheter filter was proposed by Cao et al. (2013) for coarse segmentation, which is considered a 3D catheter template for candidate voxel selection. Similarly, Novotny et al. (2007) proposed to detect the catheter in cardiac 3D US images by GPU-based template matching, which achieves fast execution based on the multiprocessing methods. A likelihood map with shape measurement optimized the resulting images. Buzurovic et al. (2012) proposed calculating the image’s vertical and horizontal gradients to enhance the needle-related information thresholded and fitted by transformation methods to detect the needle in the prostate brachytherapy 3D US images. Similarly, automatically optimized Gabor filter methods (Kaya and Bebek 2014a, b; Kaya et al. 2015; Hacihaliloglu et al. 2015; Mwikirize et al. 2016; Kaya et al. 2020) were used with different image processing steps for needle segmentation in the 2D US. Specifically, Kaya and Bebek (2014a, 2014b), Kaya et al. (2015, 2020) proposed to employ a two-stage method for needle localization based on Gabor filtering with an optimized insertion angle estimation. First, the Otsu’s method is applied to obtain the binary image. Then, the needle in the binarized image is localized by RANSAC model-fitting, which generates the region-of-interest (ROI) for needle-tip probability mapping and localizes the tip. Their methods were validated with static images (Kaya and Bebek 2014a) and real-time video (Kaya and Bebek 2014b). They further implemented a simulation platform for needle tracking (Kaya et al. 2015) for real-time localization. In contrast to Kaya et al. methods with complex post-processing, Hacihaliloglu et al. (2015) employed log-Gabor filters to extract phase-symmetry information, automatically selecting the scale, bandwidth, and orientation parameters to enhance the contrast of the needle. The needle is then detected by a modified Maximum Likelihood Estimation SAmple Consensus (MLESAC) method (Torr and Zisserman 2000). Furthermore, Mwikirize et al. (2016) proposed to localize the needle by introducing signal transmission maps for the 2D US, which firstly enhances the visibility of the needle in noisy US images. The needle is then localized by applying the algorithm from Hacihaliloglu et al. (2015). Like Gabor-based filtering, Qiu et al. (2014) proposed considering phase domain by applying Sobel operations, which are then grouped for needle segmentation by combining with model-fitting or image projection, such as least-squares fitting or Hough transform for the 3D US. For another recent method of needle detection in 3D imaging, Gillies et al. (2019) proposed to employ user-based supervision to guide the algorithm to find the instrument with an interactive approach, which shows promising results by considering human observation constraints.
In contrast to the above methods for static US images, some papers have focused on exploiting temporal information. Kaya et al. (2016) proposed to track the needle tip by applying a dynamically updated template to a 2D US video, which measures the similarity between the template and US images to identify the target. This method avoids needle localization in each 2D US video frame but requires a defined template. Beigi et al. (2015, 2016a, b) intensively studied needle detection by applying spectral analysis, using spatiotemporal information from natural hand tremors. This periodic pattern is hardly observed by human eyes but can be captured by spectral analysis of 2D B-mode images, which leads to a better result than static images. However, these image modalities were only applied to 2D+T format due to hardware constraints and complex filtering steps or real-time requirements for 3D imaging. Although a recent study focuses on 3D volumetric data with temporal information (Daoud et al. 2018a), they considered an extra camera for supporting information.
The above methods follow an algorithm pipeline, denoted as segmentation-modeling pipeline. First, carefully designed filters or instrument templates are applied to extract or enhance the instrument-related information in the image. The optimized thresholding is then applied to binarize the images to coarse segment the instrument from the data. Second, post-processing methods, such as model-fitting in the 2D/3D images can are applied to localize the target. Although the processing steps can be different, most of the above methods indicated that a successful segmentation method is a key step in detecting the instrument in challenging US images, which heavily depends on the first steps. However, these segmentation methods are limited by prior knowledge of the instrument and are sensitive to image modality or appearance. Moreover, simple thresholding with prior or empirical knowledge also limits the segmentation performances in different application cases. To better describe the instrument-related information and obtain more accurate segmentation results, machine learning methods have been exploited to better describe the instrument with knowledge learned from the data. As shown by Yang et al. (2020a), the physical space methods cannot handle the complex cardiac US images with noisy backgrounds and deformed instruments in the 3D US images, leading to much worse detection accuracy. It is mainly due to the limited discriminative information representation ability. However, Yang et al. (2019d) indicate that these conventional methods can be adopted in the pre-processing step to find the interest regions for further fine detection. Finally, it is worth mentioning that the above methods are general approaches, which can be applied to both 2D and 3D US images, even the initial proposals were 2D images for some methods, because of the advanced hardware in the current research, these 2D methods can easily be extended to 3D formate with proper software implementations.
4.1.2 Projection space methods
Besides the straightforwards studies in a common physical space, other explorations in space transformation were studied, denoted as projection methods. The methods in this category apply a spatial projection on US images with prior shape knowledge of the instruments, assuming, e.g., a straight or curved line in the 2D or 3D space. With the prior shape information, the instrument has a strong response after some projection-based spatial transformations, which accumulates the pixel or voxel intensities along the instrument propagation with respect to spatial location and direction. Here, it is assumed that the instruments yield a higher intensity value than the background in the B-mode or thresholded US images. Ding et al. (2002, 2003), Ding and Fenster (2004) proposed to segment a needle in the 3D US by applying spatial projection from 3D to 2D thresholded volumetric data, which iteratively adjusts the projection direction in 3D space to minimize the projected needle area in the 2D plane.
Ding and Fenster (2003) introduced the 2D Hough Transformation with prior knowledge of the needle insertion angle. The pixel values along the estimated direction are accumulated to generate a histogram of the projected voxels, which produces the corrected needle direction with post-processing, including iteratively direction rotation, points rejections, etc. Zhou et al. (2007, 2008), Qiu et al. (2013), and Hrinivich et al. (2017) presented the 3D Hough Transformation-based methods on a thresholded volumetric image, which select the highest accumulated values in the transformed space as the spatial parameters of the needle, which are mainly based thresholded images (with or without filtering). Similarly, Radon Transformation-based Parallel Integral Projection (Barva et al. 2008) on the voxel intensity was introduced by Barva et al. to detect the straight electrode in 3D US images. The instrument is detected as the maximized response point in the Radon projection space, accumulating the voxel intensity values along with the propagation of the instrument. However, the essential insight of their methods is similar to the Hough Transformation-based approaches, except for the case of thresholding. Later on, the Hough Transformation method was also applied to the 2D images by projecting 3D images using a ray-casting approach (Aboofazeli et al. 2009). Some recent literature also proposed transform-based methods with software-based optimizations. For example, Rodgers et al. (2020b) proposed combining filter-based edge enhancement with randomized 3D Hough transform, implemented based on CPU with GPU accelerations. The results show comparable performance to experience clinicians on brachytherapy operations. Similarly, Neshat and Patel (2008) also proposed implementing the 3D Radon/Hough transformation by GPU acceleration by employing the CUDA library, which is now commonly used for deep learning accelerations. Recently, Beigi and Rohling have employed temporal information to enhance the ability of the Hough Transformation (Beigi and Rohling 2014), which detects the needle in the 2D+T(time) US images. Daoud et al. (2018b) also applied the 2D Hough Transformation technique to needle localization in 2D B-mode images, where they introduced Power Doppler as a supporting modality to improve performance.
The above methods have some prior knowledge or requirement for instrument detection: (1) instruments are straight or a little bit curved in the images, which can be modeled by accumulating the values along the instrument axis through a spatial transformation; (2) the instrument has higher intensity values than the background such that a simple threshold or intensity-based transformation can be directly applied to detect the target. With the previous assumptions, most methods were validated based on computer simulations (Barva et al. 2008) or phantom environment (in-vitro) (Ding et al. 2002; Okazawa et al. 2006; Zhou et al. 2007, 2008; Qiu et al. 2013); the challenges and difficulties were underestimated for instrument detection in noisy B-mode images. Although there are some studies (Ding et al. 2002; Zhou et al. 2007; Qiu et al. 2013; Beigi and Rohling 2014) consider more challenging datasets from isolated tissue or even patient data, i.e., ex-vivo and in-vivo datasets, the simplified voxel thresholding without considering sufficient local or contextual information hampers the capacity of the detection algorithms, leading to many outliers or an under-segmented instrument. In contrast, instrument model-fitting methods with proper pre-processing, such as better segmentation, could be more suitable to model the instrument in B-mode images and improve the detection performance (Zhao et al. 2017). With the above considerations and conclusions from the previous physical space methods, the conventional non-machine-learning methods should be re-designed with more complex mathematical modeling in order to better handle more complex images. Possible improvement can be the combination of projection and physical spaces for a stable instrument estimation.
4.2 Machine learning methods
In recent years, machine learning methods have been intensively exploited in computer vision and medical image analysis areas. The main idea of machine learning methods is to model task-related information by designing a proper mathematical representation from the training dataset, e.g., feature vector and pre-trained classifier, which is then used to make a prediction or decision. There are two kinds of approaches in the literature. The first is a handcrafted feature design with a machine learning classifier, depicted in Fig. 8. This method employs feature vector extraction and task classification, commonly applied at the pixel(voxel) level for instrument segmentation. Specifically, the segmentation-modeling pipeline is often utilized, where the model fitting is applied on the pixel(voxel)-level classification results to obtain the final instrument segmentation result. However, the design of handcrafted features requires task-related knowledge and experience, which hampers the classification performance. The traditional machine learning approach is gradually replaced by the recently developed deep learning technology. Deep learning is a kind of machine learning method that combines feature extraction and classifier training in a fully automated information learning style. Deep learning methods can automatically learn task-related information from the provided data, and with well-designed network architecture and sufficient training data, they can learn more powerful representations than handcrafted feature design methods. However, as argued in the literature, the ability of model explanation is one key limitation of the deep learning methods, as the learned features are less understandable than the handcrafted features.
4.2.1 Handcrafted feature-based methods
Block diagram of the CRF-based needle detection (Zanjani et al. 2018). The feature vector for each voxel is extracted for classification. At the same time, the feature selection on the vector elements is applied for fully-connected 3D CRF processing, which is calculated based on the input image and classified volumetric output. With the analysis of spatial correlations for each voxel point, the 3D CRF refines the segmentation outputs
Because machine learning methods provide better instrument segmentation performance (Krefting et al. 2007; Uherčík et al. 2013) than conventional image processing methods, e.g., Hough transformation methods, they have been intensively studied in recent years. Uherčík et al. (2013) proposed the use of voxel intensity, Frangi vesselness response and axis descriptors as the discriminating features, categorized by a cascade classifier for needle detection. Compared to the non-machine-learning methods, such as Parallel Integral Projection and Random Hough Transformation, the voxel-level classification provides a higher detection accuracy because a better segmentation result can be obtained. Similar to Uherčík, Hatt et al. (2015) proposed to describe the pixels in 2D US images with the second-order Gaussian derivative filters, which are classified by AdaBoost for segmenting the needle. Their results demonstrated their method achieved better performance than the straightforward thresholding methods like intensity thresholding, filtered thresholding, or Frangi vesselness thresholding.
Pourtaherian et al. extensively studied needle segmentation by applying 3D orientation-invariant Gabor features with Linear Support Vector Machine (LSVM) (Suykens and Vandewalle 1999), where the segmentation results are further processed by a RANSAC algorithm to localize the instrument (Pourtaherian et al. 2015a, b, 2017a). Specifically, the Gabor filter bank with a pre-defined scale and multiple orientations are applied to the 3D volumes. The voxel is classified by the SVM classier to determine whether it is a needle or not. With classification on all voxels of the volume, the 3D US is segmented for further post-processing, e.g., RANSAC model-fitting. Their studies show that the Gabor filter with SVM could adequately capture the spatial information for a long but thin instrument in complex 3D US images. Later on, Zanjani et al. (2018) demonstrated that the Gabor features could be simplified by feature selection, and the segmentation performance can be boosted by conditional random field (CRF) (Lafferty et al. 2001) compared to a simple SVM, which segments the images based on contextual-level information correlation (as shown in Fig. 9).
Mwikirize et al. (2017) also considered log-Gabor features as the local phase extractor, which is processed by a locally normalized histogram of orientated gradients (HOG) features, to describe the needle in 2D US slices. The constructed HOGs are then classified by SVM to segment and enhance the needle in 2D US slices. In the meantime, Younes et al. (2018) proposed using a Gaussian mixture model-based classifier to segment the needle in 3D prostate brachytherapy US. The needle is finally localized by a standard RANSAC-based model fitting. To further exploit complex anatomical information in the 3D US for catheter segmentation in catheterization, Yang et al. proposed multi-scale and multi-definition features for supervised learning classifiers, which demonstrated a better discriminating information extraction than techniques solely based on Gabor features (Yang et al. 2018a, 2019e). The segmented instrument was fitted by a more complex Sparse-Plus-Dense RANSAC algorithm to fit the curvature instrument in the cardiac chambers.
The above methods are applied to static images because the voxel-level feature extraction and classification in 3D+T US images are computationally expensive for current hardware. Nevertheless, several papers were proposed to focus on the 2D US with temporal information. Beigi et al. (2017b) proposed to detect a hardly visible needle in 2D+T US by applying local phase extraction with temporal sequence analysis. More specifically, the phase information for each frame is extracted to formulate the element from a time-sequence-based phase video, which is then processed by the Auto-Regressive Moving-Average (ARMA) model to extract the feature vector. With classification from a modified SVM, the small motion of the needle can be characterized for needle detection. Furthermore, Beigi et al. employed spectral feature analysis on spatiotemporal features derived from optical-flow analysis (Beigi et al. 2017a), which allows detecting and tracking the needle in a 2D US video. In contrast to these off-line learning methods, Mathiassen et al. (2016) applied online learning, i.e., learning and updating the needle-related information during the video progress. They applied the statistical filtering methods, i.e., Kalman filter and particle filter, to learn the appearance of the instrument in the video with real-time performance.
Besides the above machine learning methods, Zhang et al. (2020b, c) proposed to detect multi-needle in 3D brachytherapy by unsupervised sparse dictionary learning. Specifically, the needles and tissue information in the 3D images are encoded into latent space, distinguished by the sparse dictionary model. Based on the sparse dictionary learning, the needle in the 3D space can be captured and reconstructed in the volume so that multiple needles can be localized by region-of-interest-based RANSAC.
Even though the above methods achieved promising detection results for the given tasks, it is challenging to design the optimal feature representation, which is one of the key factors that hamper the segmentation performance. Because of this, complex post-processing is needed to avoid outliers or false positives. Moreover, the designed features can only focus on local information while ignoring contextual and semantic information (Zanjani et al. 2018). To handle these limitations, many deep learning methods have been studied recently (Litjens et al. 2017).
4.2.2 Deep learning-based methods
At the beginning of the deep learning era, Geraldes and Rocha proposed using neural networks, i.e., the Multilayer Perceptron network (MLP), to segment the needle in 2D US images (Geraldes and Rocha 2014; Rocha and Geraldes 2014). The segmented results guide the Kalman filter to track the needle tip in the video sequences (Geraldes and Rocha 2014). These papers demonstrated the feasibility of deep learning to detect a medical instrument in challenging US images.
The conventional idea of voxel-based classification was extended into deep learning methods, which employed convolutional neural networks (CNN, a famous type of deep neural network, DNN) to replace the feature extraction and classification steps in Fig. 8. The CNN model determines the voxels’ category by a classification strategy on the whole 3D image. A typical approach is the tri-planar CNN method (Pourtaherian et al. 2017b, 2018b; Yang et al. 2018b; Min et al. 2020), which segments the instrument in 3D US volume by voxel-level classification with decomposed 3D local patches. The volume is segmented by classifying all the voxels. Compared to conventional handcrafted feature-based methods, this deep learning method automatically learns the discriminative representation based on the training data, which better exploits the information for classification. Nevertheless, the exhaustive strategy is time-consuming for the segmentation of each image. Later on, this exhaustive strategy was overcome by applying the Frangi vesselness filter as a voxel-of-interest (VOI) pre-selection step (Yang et al. 2019d), which is shown in Fig. 10. A fast region-based CNN (Fast R-CNN) is combined with a region proposal network (RPN) to efficiently detect the needle in 2D US images (Mwikirize et al. 2018). However, this method cannot accurately segment and localize the instrument skeleton at the pixel or voxel level. To overcome this limited performance and leverage the powerful fully convolutional network (FCN), which assigns the class categories to all the points of the input image by using semantic information, semantic segmentation was introduced and studied for instrument segmentation.
Block diagram of VOI-based CNN for catheter detection (Yang et al. 2019d). The input volume is first processed by a Frangi filter to select the VOI voxels, which are then classified by a tri-planar-based CNN for voxel-based classification. The RANSAC model-fitting is applied to localize the catheter in 3D B-mode images
A systematic example of U-Net structure. The U-Net includes an encoder and a decoder. The input image is encoded by several convolutional and max pooling operations. The intermediate feature maps are processed by the decoder via several convolutional and up-convolutional operations. The encoder feature maps are skipping connected with the decoder to preserve the low-level information
To semantically segment the instrument in the US, an FCN with a U-Net structure (Ronneberger et al. 2015) (shown in Fig. 11) was considered since it exploits the semantic information at different image scales with skipping connections for data flow. This approach leads to state-of-the-art performance in most applications in the medical imaging area. A 2D FCN is applied to segment the needle/catheter in 2D US (Lee et al. 2020; Rodgers et al. 2020a; Gillies et al. 2020; Wijata et al. 2021; Liu et al. 2022) and in 3D US where the volume is decomposed into stacks of slices (Pourtaherian et al. 2018a; Yang et al. 2019b) for segmentation with the U-Net. However, for 3D volumetric data, the decomposition approach may limit semantic information usage due to the compromised 3D information after slicing. To address this, patch-based 2.5D or 3D U-Net were proposed to segment the cardiac catheter (Yang et al. 2019a, c, f, 2020a, c) or prostate needles (Zhang et al. 2020d; Andersén et al. 2020) in 3D volumetric data by dividing the image into smaller patches or reducing the image size. In this way, the 3D contextual information is preserved and the requirement on GPU memory is reduced for 3D deep learning. Nevertheless, this patch-based strategy limits the whole image contextual information usage. To overcome this limitation, Arif et al. applied an extremely simplified 3D U-Net on a complete 3D image to segment the needle in US imaging for the liver (Arif et al. 2019). Although their method showed promising results, its generalization and segmentation abilities are constrained by the simplified network design (Yang et al. 2019f). Alternatively, a multi-dimensional FCN was proposed to decompose the high-level 3D feature maps into 2D space, which simplifies the decoder and reduces GPU usage (Yang et al. 2020b).
Due to the requirements of a large number of training images and high GPU memory usage for deep learning, temporal information is not widely investigated in deep learning-based medical instrument detection. Mwikirize et al. (2019a, b) proposed time-difference-based regression and classification CNNs to detect the needle in 2D US sequences. The differences between two adjacent frames are obtained by applying pixel-wise logical operation, which captures the subtle motion of the needle and feeds it into CNNs for detection. Nevertheless, these methods process the temporal information outside the CNN such that the deep learning approach may not properly handle the spatial-temporal information. Later on, Mwikirize et al. (2021) further exploited temporal information by employing long short-term memory (LSTM) module with CNN, which achieved a 30% performance improvement compared to their previous work while achieving the detection rate of about 15 frames per second (FPS). In contrast to the aforementioned works, Chen et al. (2021) made use of temporal information in the CNN’s input channel by employing the adjacent frames, which achieved promising results for robot-assisted needle insertion processes.
In addition to commonly used CNNs, convolutional dictionary learning was proposed by Zhang et al. (2020a) as an extension of their work (Zhang et al. 2020b). Instead of sparse dictionary learning, they considered a convolutional sparse coding model to replace the sparse dictionary learning method (Zhang et al. 2020a), which used CT images as the supervisory signal to create the dictionary for reconstructing the detected needles in the 3D volumetric data.
Although the deep learning methods provide superior segmentation performance and better information description than the conventional image filtering and/or transformation methods, these machine learning methods require a large amount of training data with annotations. A semi-supervised learning method has been proposed to address this challenge by jointly considering uncertainty estimation and contextual constraints (Yang et al. 2020d, 2021). Later on, Yang et al. (2022) also exploited the weakly-supervised catheter segmentation in 3D Frustum US imaging, which achieves much lower computational cost than commonly considered Cartesian imaging, while the state-of-the-art performances are obtained in their study. With the proposed methods, the deep learning network requires much fewer annotations than the conventional supervised learning method, while the segmentation results are comparable. Nevertheless, this study still has some limitations, which include but are not limited to (1) semi-supervised learning methods that have not been validated on the large-scale dataset, (2) these methods have not been extensively compared to state-of-the-art methods. Moreover, most state-of-the-art instrument detection or tracking methods are far from real-time performance, so optimizations are still required to improve computational efficiency. More specifically, real-time performance is more crucial for instrument tracking in clinical practice, while non-real-time performance is accepted for instrument detection in some operations (Zhang et al. 2020a, b). These limitations also form key issues for employing deep learning for real clinical applications.
To give an overview, the existing papers using non-machine-learning techniques and machine learning techniques are summarized in Tables 1, 2, respectively.
5 Evaluation datasets, metrics and results
5.1 Evaluation datasets
Various datasets have been utilized in the existing studies on instrument detection. Specifically, there are four types of data: simulation dataset, in-vitro dataset, ex-vivo dataset, and in-vivo dataset.
Simulation: Computer software-based simulation was used to generate US images with well-defined instrument information with image content. Field II (Jensen 1996) and kWave (Treeby and Cox 2010) are the commonly used simulation platform for US image generation. Because they can provide satisfying image results without extra cost for US equipment and experiment labs, such as a biological lab for tissue experiments and target instrument operations, which require extra clinical doctors to guide the experiments. As a result, a simulation dataset can be used to validate the scientific idea and perform a feasibility study, e.g., mimicking the needle or catheter insertion in 3D US images (Barva et al. 2008; Zhao et al. 2013a). Although kWave has ability to model nonlinearities in tissue, which achieves better simulation results than Field II, this simulation approach lacks complex anatomical structures of the tissue and is far from the real images from the operation, which limit the clinical value and stability of the methods, especially for the learning methods. This is because the learning algorithm learns all available information from the images, while the simulated tissue or background may not be the correct information for the learning procedure, which will hamper the generalization of the algorithms. Therefore, a thorough validation of the methods, which were validated on simulated datasets only, should be performed on clinical datasets.
In-vitro: In contrast to simulation datasets, in-vitro datasets provide a more realistic case, which employs polyvinyl alcohol (PVA) or agar phantoms to mimic the human tissue (Draper et al. 2000; Okazawa et al. 2006; Zhao et al. 2009). Moreover, these datasets were recorded from real US imaging equipment with proper post-processing, which mimics real clinical scenarios. However, this approach has a significant limitation: the phantom cannot include complex and detailed anatomical structures in clinical applications, such as vessels or muscles. Moreover, due to the different physical properties between a phantom and natural tissue, the US imaging results are also different, limiting the algorithms’ clinical value. For instance, a line-filtering-based needle detection algorithm was validated on a phantom dataset (Zhao et al. 2009), proving to be unstable for real tissue data (Pourtaherian et al. 2017a).
Ex-vivo: Similar to in-vitro data, ex-vivo data is another commonly used dataset type, which replaces PVA or agar phantoms with isolated real animal tissue, such as chicken breast for needle detection (Pourtaherian et al. 2017a) or porcine heart for catheter detection (Yang et al. 2018a). These tissues provide a more complex image appearance due to anatomical structures, which are more similar to clinical applications. Nevertheless, this dataset type still has limitations in recording conditions, such as less complex muscle and vessel structure of chicken breast when considering a needle detection for anesthesia or a water-filled (rather than real blood) heart chamber for cardiac catheterization. Although these limitations hamper the clinical value, they provide a more stable and promising comparison than the above non-tissue-based datasets. Because of the data recording difficulties, such as biological experiment certification and support from clinical experts, ex-vivo datasets are still important and are considered in a majority of recent papers, which indicates their importance for algorithm validation. In addition, by comparing the performance stability, ex-vivo dataset has more challenges than the in-vitro images (Pourtaherian et al. 2017a; Yang et al. 2018a).
In-vivo: This article defines in-vivo as the off-line dataset from a real clinical operation, such as prostate biopsy or live animal dataset, e.g., live porcine for cardiac catheterization. Because the datasets were recorded from real clinical usage or mimicking it, these in-vivo datasets demonstrated important clinical value for the algorithm validation. However, recording this type of dataset is challenging compared to the above three dataset types. First, the research protocol on data collection needs to be approved by the ethical committee and the consent form needs to be signed by the patient. The data collection must avoid any risk for the patient, and the procedure should be fully accepted in the institution. Second, it is difficult to obtain the real patient dataset considering the complex anonymization steps taken to avoid privacy issue. Third, cooperation with clinical experts can be challenging because different clinicians may have different preferences for image appearance and configurations, making it difficult to agree on similar image quality and appearance for the algorithm.
As can be observed, the early studies on instrument detection were mostly validated on the simulation data and in-vitro images. Mainly non-learning methods were investigated in these studies. In recent years, with the development and increasing maturity of image-guided operations, instrument detection in ultrasound has attracted more attention, and more and more ex-vivo and in-vivo datasets have been collected for validation. At the same time, machine learning-based methods have eventually become the mainstream technique. Therefore, learning-based methods are mostly validated on ex-vivo and in-vivo datasets, which produce promising results for clinical applications. The learning-based method learns the anatomical structures from the input images, which leads to less generalization when it is trained on the simulation and in-vitro datasets. Therefore, a carefully prepared dataset, preferably collected in clinical scenarios, is more meaningful for algorithm validation since the domain gap between technical experiments and clinical practice should be as small as possible.
5.2 Evaluation metrics and performance
As for evaluation metrics, the used metrics and their definitions are summarized in Table 3. In the detection algorithm, the instrument detection is successful when the detected instrument belongs to the true target (Krefting et al. 2007). Based on this and specifically, metrics AE to PE are commonly considered to evaluate the performance of the detection. In contrast, the rest metrics are used to evaluate the segmentation performance. The performances of medical instrument detection algorithms are summarized in Tables 4, 5. To evaluate the detection performance, several validation methods are used: (1) non-machine-learning methods are commonly validated on the dataset as described, i.e., validating the method on the whole dataset; (2) learning-based methods are commonly validated on a testing dataset after using a different training dataset for model training. More specifically, cross-validation like leave-one-out cross-validation (LOOCV), k-fold cross-validation (k-CV), or a straightforward dataset split (S), i.e., dividing the dataset into training, validation, and testing subsets, are commonly used in learning-based methods. Some table cells are empty due to unclear descriptions from the considered papers.
Although there is no commonly used benchmarking dataset for a fair comparison, the detection performance from the literature shows a satisfactory accuracy for clinical usage as all the performances are consistent by considering similar evaluation metrics. However, in terms of real-time efficiency required in clinical applications for operation guidance, it is far from real-time in most papers. Specifically for papers validated on the in-vivo datasets, most are far from real clinical use. Most of the articles were validated on limited clinical data, e.g., around ten patients or even less, without reporting the time efficiency (especially for 3D imaging).
6 Discussion
Medical instrument detection in the US has attracted much attention in the last twenty years. Particularly much progress has been made in the last ten years, with various approaches being reported in the literature. Although promising results have been obtained in the literature, some challenges remain, which should be further investigated in the future.
6.1 Current status
As can be observed in Tables 4, 5, the state-of-the-art methods, mainly deep CNNs, have achieved good performance on medical instrument detection in US images. We summarize the current progress from the aspect of US imaging format, clinical applications, validation strength, and clinical applicability.
6.1.1 2D versus 3D US imaging
Regarding the US imaging format considered for instrument detection, 2D US imaging is much more studied because of its easy access and wide availability in clinical practice. At the same time, because the 2D US images have a smaller data size, it is easier to realize real-time guidance for the clinician, which is a key factor for real clinical applications. Moreover, as shown in the above tables, the datasets of the 2D US are commonly much larger than the 3D datasets, because of their easier access and lower annotation effort. In the current literature, the CNN-based instrument detection methods for the 2D US can have an execution speed of around 15–20 FPS (Mwikirize et al. 2021; Chen et al. 2021) and sub-millimeter accuracy. In contrast, the state-of-the-art CNN method for the 3D US achieves a speed of about 4 FPS with a detection accuracy of about 1–2 mm (Arif et al. 2019; Yang et al. 2020b). Therefore, medical instrument in the 2D US is more mature than 3D US-guided operations. Nevertheless, as discussed above, the 2D US has its limitations, and the 3D US-based instrument detection is still attractive and important for operations like catheterization, anesthesia, and brachytherapy, because it can provide richer spatial information and better anatomical structures.
6.1.2 Clinical applications
As for different clinical applications of medical instrument detection, needle-based operations (such as anesthesia, biopsy, and brachytherapies) have been widely studied, while other instrument-based procedures (e.g., catheter-based) have received less attention. For needle-based procedures, since the metal target has high contrast, it is easier to localize the needle by image processing and/or learning-based methods. In the current literature, the best performing methods for needle localization can achieve high detection accuracy of around sub-millimeter by employing CNNs (Zhang et al. 2020d; Mwikirize et al. 2021; Chen et al. 2021; Liu et al. 2022) . In contrast, the instrument with a plastic coating, such as catheters, instrument detection in the US is more challenging due to the subtle difference between the target and the background. The best state-of-the-art method of catheter detection achieves an accuracy of 2.3 mm with an execution time of about 0.12 sec per volume (Yang et al. 2020b). In clinical practice, needle-based operations are more mature with US guidance, which provides more possibility to acquire a large dataset for validation. In contrast, catheterization in cardiac intervention or other procedures is less guided by the US in practice, which leads to difficulties for data collection and clinical validation.
6.1.3 Validation strength and clinical applicability
When considering the validation strength and clinical applicability, instrument detection in the operations of anesthesia, biopsy taking, and brachytherapies have been extensively validated in large ex-vivo and/or in-vivo datasets consisting of more than thousands of images (Mwikirize et al. 2021; Chen et al. 2021; Liu et al. 2022). Moreover, the approaches for these applications are studied by different groups worldwide, and these studies and their performance can be reproduced more easily. Nevertheless, catheter detection, especially for US-guided cardiac catheterization, is less studied and not mature for clinical applications due to its low validation strength with limited studies (Yang et al. 2022), e.g., small datasets with less than 100 images from just 1–2 research groups.
To summarize, needle detection in both 2D and 3D US formats has been extensively studied for different clinical applications, which have shown great potential for clinical use. These methods can be adopted in operation to provide real-time guidance. However, the detection of other instruments such as a catheter (e.g., cardiac catheterization) is less studied with limited validations, and not mature for clinical use considering its non-real-time performance.
6.2 Challenges and future directions
6.2.1 Non-learning versus learning methods
When considering the commonly used segmentation-modeling in both non-machine-learning and machine learning methods, this is a straightforward method without complex pre- and post-processing compared to state-of-the-art methods in computer vision. A successful segmentation may lead to better model fitting and detection results, but this approach was only validated on limited datasets. Therefore, it cannot ensure generalization and robustness for real clinical applications. For example, US consoles from different vendors with different US probes have various US transmit and receive settings, leading to different US image appearances and qualities. The lack of standardization forms a challenge for algorithms’ desired robustness and generalization. A standardization in pre-processing, such as domain adaptation or image normalization, may be required for future improvement. Instead of the popular RANSAC modeling, standardization of model fitting should be applied to improve the post-processing efficiency. Meanwhile, this model-fitting method has performance limitations when the instrument has a geometry different from a curved tube. For example, this occurs when instruments with a ball or circular shape are used in cardiology.
As for machine learning methods and especially deep-learning-based methods, algorithms are trained with information from a large amount of annotated data. This may be difficult to realize for clinical applications due to the limited access to clinical data. Privacy regulations are in place to protect against patient data misuse, and patient informed consent must be present to use clinical data and facilitate the training of supervised learning algorithms. To obtain the reliability of learning, sufficient data should be available. In addition, interpretation of medical data is complex, and only experts can do this, which makes annotated data even more desired, but difficult to collect because of the annotation cost. Therefore, it is a tedious and expensive solution to train a satisfying model for clinical practice, because of the complex network design and the required large data collection. Besides the above challenges, the generation of an annotated dataset with one user may have a bias and intrinsic variability, which would lead to evaluation bias. Therefore, a multi-annotator framework is required to reduce the ground truth bias, and consider different US machines. Nevertheless, this consideration would lead to higher data collected expenses.
Besides the above challenges for data collection, how to exploit the available datasets, especially for unlabeled images, is important for the deep-learning-based algorithm design. Several solutions could be considered to decrease annotated effort and exploit unlabeled images. (1) A task-specific CNN design should be employed to decrease overfitting and the total detection time to support a real-time application. (2) Domain adaptation may be a solution to address the dataset limitation, which can train a network based on extensive in-vitro/ex-vivo datasets and adapt the network to the in-vivo dataset, which has a limited clinical data size. This approach is more cost-effective than directly training from patient data since it requires less annotation by clinical experts. (3) Semi-supervised or weakly supervised learning approach should be considered for the algorithm design, which requires fewer annotation efforts and can easily extend the training dataset for a better and more stable detection algorithm.
Comparing the above two methods of non-machine-learning techniques and recent popular machine learning methods, they have different advantages and limitations. For the non-machine-learning techniques, which are mainly based on properly designed image filtering and instrument geometrical modeling. The key advantages can be summarized in two key points. First, these methods are designed based on the domain knowledge of the instruments, which are easier to be accepted by clinicians. Second, these methods are less data-hungry, requiring fewer annotation efforts and reducing implementation costs. With proper optimization on GPU, semi-real-time efficiency can be obtained, for example, by employing the post-training quantization on trained CNNs (Abadi et al. 2016). However, these methods face a limitation of parameter sensitivity, i.e., high requirement of the image recording settings. Once images have a different appearance, the algorithm can be failed with pre-defined parameters, such as instrument size, dynamic ranges or threshold, etc. As for machine learning methods, they are more stable for variation of the image quality, as the advanced feature designing and classifiers can better handle the image variation. In addition, as the parts of instrument knowledge are obtained from data-driven approaches, more instrument discriminative information can be obtained. However, as stated above, the limited training data hampers the performances and generalizations to real clinical applications. From the literature review in this article, the non-machine-learning methods are much more mature than machine-learning approaches. In addition, by comparing the performance and efficiency, they are comparable on the validated datasets w.r.t. detection time within seconds and accuracy around 1 mm. However, a fair comparison should be conducted in future studies, as some of the methods are specifically designed without publicly available sources.
6.2.2 Clinical requirements in different aspects
Considering the existing literature survey about the clinical applications, most technical researchers focus on the most manageable tasks for US-guided interventions, i.e., needle-based anesthesia delivery, biopsy taking, and prostate brachytherapy, since these datasets are more accessible to obtain than cardiac catheterizations. This trend also reflects that these interventions are more mature and widely accepted by hospitals. Nevertheless, researchers should cooperate with hospitals and industries to develop instrument detection algorithms for different clinical practices and better surgical outcomes. In addition, a more intensively survey should be conducted on both medical doctors and technical researchers, which can lead to a high agreement on the application scenarios with detailed requirements, e.g., make the algorithm easier to be implemented in the operating procedures with a minimized learning curve and design the algorithm pipeline in the way of following the clinicians’ preference. In addition, as stated at the beginning of the introduction section, the financial requirement is one of the reasons for investigating the automatic algorithm design. However, due to the expensive devices of the 3D US probe, conventional 2D probes are still widely employed in the common clinical practice for procedure guidance. In the future, as the cost of the device is reduced, the 3D image-based algorithm might be more applicable for clinicians.
As discussed in this paper, the current literature emphasizes detection accuracy rather than time efficiency. For US-guided intervention therapies, real-time execution for device detection or tracking is crucial, but the requirement for real-time detection during interventions is under discussion. Specifically, real-time performance is crucial for instrument tracking or guidance-based tasks, such as needle tracking. Alternatively, instrument detection might require less time efficiency, such as finding the instrument from a complex 3D imaging modality. Nevertheless, some tracking algorithms are based on a frame-by-frame detection algorithm, which is crucial for detection efficiency. Moreover, it is unclear for clinical applications how accurate the detection should be for broad acceptance by clinicians, e.g., whether 1 mm detection accuracy or 0.7 DSC segmentation accuracy is sufficient for specific clinical applications. In the future, a more comprehensive study should be performed in cooperation with interventionists, which should validate the importance and different value settings for detection accuracy and detection efficiency.
Recently proposed solutions for device segmentation are based on US images, which implies performing detection algorithms on a frame-by-frame basis for an eventual application. However, real-time US imaging is captured as a video sequence in both 2D and 3D US formats in clinical scenarios. 3D+time data should be investigated in future research to exploit better temporal information, which is currently limited by software and hardware implementations. Moreover, most detection algorithms are executed with more than one second to find the instrument, which is accepted for instrument localization. Nevertheless, this efficiency is far from the requirement of the operation guidance, which asks for a real-time segmentation or detection result for the radiologist during the operation.
6.2.3 Public dataset for benchmarking
Finally, it is important to collect public benchmarking datasets for different clinical applications, such as biopsy, anesthesia, and catheterization. This could enable a fair comparison of different methods on their efficiency, robustness, and accuracy. The benchmarking datasets can lead to a higher diversity of dedicated solutions and their acceptance in the community. Here are a few suggestions on designing and building such public a benchmarking dataset. First, the dataset should consist of all-aspect of the data distribution, including the challenging cases, which is to make the dataset representative and to contain different examples, based on the knowledge from clinical experts. Second, the dataset should include as wide as possible institution varieties when obtaining the images, including both equipment distributions and end-user preferences, since the domain gaps among different institutions are always challenging for state-of-the-art algorithms and this issue should be carefully covered. Third, the dataset should cover different regions of the world, as different countries have different image appearances due to different equipment, operators, and patients.
7 Conclusion
This article has provided a comprehensive survey of medical instrument detection methods in US images used to guide various operations. Different clinical applications are reviewed first, showing that most topics are related to needle-based interventions since they have been widely used in current clinical practice, ranging from anesthesia to biopsy. In contrast, catheter-based interventions are currently less guided by the US, because other image modalities can provide a better contrast-to-noise ratio. From the methodology perspective, although both machine learning and non-machine learning methods have been studied in recent years, the research attentions are more focused on the machine learning methods (especially deep learning methods), because of their better discriminative information descriptions. Nevertheless, these machine learning methods require a large amount of training data with annotations, which is expensive. The public clinical benchmarking datasets are necessary for a fair algorithm comparison. Finally, the advantages and limitations of US-guided interventions are discussed. Though much progress has been achieved in the past decades, there are remaining issues for US-guided instrument detection algorithms. This review could encourage other researchers to explore further US-guided intervention therapy and pave the way for more mature clinical applications.
Data availability
Data sharing not applicable to this article as no datasets were generated or analyzed during the current study, as this is a review article and all the information can be obtained from the referred papers.
References
Abadi M, Barham P, Chen J, Chen Z, Davis A, Dean J, Devin M, Ghemawat S, Irving G, Isard M, et al. (2016) {TensorFlow}: a system for {Large-Scale} machine learning. In: 12th USENIX symposium on operating systems design and implementation (OSDI 16). pp 265–283
Aboofazeli M, Abolmaesumi P, Mousavi P, Fichtinger G (2009) A new scheme for curved needle segmentation in three-dimensional ultrasound images. In: 2009 IEEE ISBI. IEEE, pp 1067–1070
Agarwal N, Yadav AK, Gupta A, Orlando MF (2019) Real-time needle tip localization in 2d ultrasound images using kalman filter. In: 2019 IEEE/ASME International conference on advanced intelligent mechatronics (AIM). IEEE, pp 1008–1012
Andersén C, Rydén T, Thunberg P, Lagerlöf JH (2020) Deep learning-based digitization of prostate brachytherapy needles in ultrasound images. Med Phys 47:6414.
Arif M, Moelker A, van Walsum T (2019) Automatic needle detection and real-time bi-planar needle visualization during 3d ultrasound scanning of the liver. MedIA 53:104–110
Banerjee S, Kataria T, Gupta D, Goyal S, Bisht SS, Basu T, Abhishek A (2017) Use of ultrasound in image-guided high-dose-rate brachytherapy: enumerations and arguments. J Contemp Brachytherapy 9(2):146
Barrington MJ, Kluger R (2013) Ultrasound guidance reduces the risk of local anesthetic systemic toxicity following peripheral nerve blockade. Reg Anesthesia Pain Med 38:289.
Barva M, Uhercik M, Mari JM, Kybic J, Duhamel JR, Liebgott H, Hlavác V, Cachard C (2008) Parallel integral projection transform for straight electrode localization in 3-d ultrasound images. IEEE TUFFC 55(7):1559–1569
Beigi P, Rohling R (2014) Needle localization using a moving stylet/catheter in ultrasound-guided regional anesthesia: a feasibility study. In: Medical imaging 2014: image-guided procedures, robotic interventions, and modeling, international society for optics and photonics, vol 9036, p 90362Q
Beigi P, Salcudean T, Rohling R, Lessoway VA, Ng GC (2015) Needle detection in ultrasound using the spectral properties of the displacement field: a feasibility study. In: Medical imaging 2014: image-guided procedures, robotic interventions, and modeling, international society for optics and photonics, vol 9415, p 94150U
Beigi P, Rohling R, Salcudean SE, Ng GC (2016a) Spectral analysis of the tremor motion for needle detection in curvilinear ultrasound via spatiotemporal linear sampling. IJCARS 11(6):1183–1192
Beigi P, Salcudean SE, Rohling R, Ng GC (2016b) Automatic detection of a hand-held needle in ultrasound via phased-based analysis of the tremor motion. In: Medical imaging 2014: image-guided procedures, robotic interventions, and modeling, international society for optics and photonics, vol 9786, p 97860I
Beigi P, Rohling R, Salcudean SE, Ng GC (2017a) Casper: computer-aided segmentation of imperceptible motion-a learning-based tracking of an invisible needle in ultrasound. IJCARS 12(11):1857–1866
Beigi P, Rohling R, Salcudean T, Lessoway VA, Ng GC (2017b) Detection of an invisible needle in ultrasound using a probabilistic svm and time-domain features. Ultrasonics 78:18–22
Beigi P, Salcudean SE, Ng GC, Rohling R (2020) Enhancement of needle visualization and localization in ultrasound. Int J Comput Assist Radiol Surg 2020:1–10
Buzurovic I, Misic V, Yu Y (2012) Needle identification in high-dose-rate prostate brachytherapy using ultrasound imaging modality. In: 2012 Annual international conference of the IEEE engineering in medicine and biology society. IEEE, pp 476–479
Cao K, Mills D, Patwardhan KA (2013) Automated catheter detection in volumetric ultrasound. In: 2013 IEEE ISBI. IEEE, pp 37–40
Chen S, Lin Y, Li Z, Wang F, Cao Q (2021) Automatic and accurate needle detection in 2d ultrasound during robot-assisted needle insertion process. Int J Comput Assist Radiol Surg 2021:1–9
Cleary K, Peters TM (2010) Image-guided interventions: technology review and clinical applications. Annu Rev Biomed Eng 12:119–142
Coplen DE, Andrile GL, Yuan JJ, Catalona WJ (1991) The ability of systematic transrectal ultrasound guided biopsy to detect prostate cancer in men with the clinical diagnosis of benign prostatic hyperplasia. J Urol 146(1):75–77
Daoud MI, Alshalalfah AL, Mohamed OA, Alazrai R (2018) A hybrid camera-and ultrasound-based approach for needle localization and tracking using a 3d motorized curvilinear ultrasound probe. MedIA 50:145–166
Daoud MI, Shtaiyat A, Zayadeen AR, Alazrai R (2018) Accurate needle localization using two-dimensional power doppler and b-mode ultrasound image analyses: a feasibility study. Sensors 18(10):3475
Ding M, Fenster A (2003) A real-time biopsy needle segmentation technique using hough transform. Med Phys 30(8):2222–2233
Ding M, Fenster A (2004) Projection-based needle segmentation in 3d ultrasound images. Comput Aided Surg 9(5):193–201
Ding M, Cardinal HN, Guan W, Fenster A (2002) Automatic needle segmentation in 3d ultrasound images. In:Medical imaging 2002: visualization, image-guided procedures, and display, international society for optics and photonics, vol 4681, pp 65–76
Ding M, Cardinal HN, Fenster A (2003) Automatic needle segmentation in three-dimensional ultrasound images using two orthogonal two-dimensional image projections. Med Phys 30(2):222–234
Douglas BR, Charboneau JW, Reading CC (2001) Ultrasound-guided intervention: expanding horizons. Radiol Clin North Am 39(3):415–428
Draper KJ, Blake CC, Gowman L, Downey DB, Fenster A (2000) An algorithm for automatic needle localization in ultrasound-guided breast biopsies. Med phys 27(8):1971–1979
Frangi AF, Niessen WJ, Vincken KL, Viergever MA (1998) Multiscale vessel enhancement filtering. In: MICCAI. Springer, pp 130–137
Geraldes AA, Rocha TS (2014) A neural network approach for flexible needle tracking in ultrasound images using kalman filter. In: 5th IEEE RAS/EMBS international conference on biomedical robotics and biomechatronics. IEEE, pp 70–75
Germano IM (2002) Advanced techniques in image-guided brain and spine surgery. Thieme Medical Publishers Incorporated
Gillies DJ, Awad J, Rodgers JR, Edirisinghe C, Cool DW, Kakani N, Fenster A (2019) Three-dimensional therapy needle applicator segmentation for ultrasound-guided focal liver ablation. Med Phys 46(6):2646–2658
Gillies DJ, Rodgers JR, Gyacskov I, Roy P, Kakani N, Cool DW, Fenster A (2020) Deep learning segmentation of general interventional tools in two-dimensional ultrasound images. Med Phys 47:4956
Hacihaliloglu I, Beigi P, Ng G, Rohling RN, Salcudean S, Abolmaesumi P (2015) Projection-based phase features for localization of a needle tip in 2d curvilinear ultrasound. In: MICCAI. Springer, pp 347–354
Hatada T, Ishii H, Ichii S, Okada K, Fujiwara Y, Yamamura T (2000) Diagnostic value of ultrasound-guided fine-needle aspiration biopsy, core-needle biopsy, and evaluation of combined use in the diagnosis of breast lesions. J Am Coll Surg 190(3):299–303
Hatt CR, Ng G, Parthasarathy V (2015) Enhanced needle localization in ultrasound using beam steering and learning-based segmentation. Comput Med Imaging Gr 41:46–54
Hrinivich WT, Hoover DA, Surry K, Edirisinghe C, Montreuil J, D’Souza D, Fenster A, Wong E (2017) Simultaneous automatic segmentation of multiple needles using 3d ultrasound for high-dose-rate prostate brachytherapy. Med Phys 44(4):1234–1245
Jan M, Kalinšek TP, Štublar J, Jelenc M, Pernat A, Žižek D, Lakič N (2020) Intra-cardiac ultrasound guided approach for catheter ablation of typical right free wall accessory pathways. BMC Cardiovasc Disord 20(1):1–8
Jensen JA (1996) Field: a program for simulating ultrasound systems. In: 10th Nordicbaltic conference on biomedical imaging, vol 4, part 1. Citeseer, pp 351–353
Kaya M, Bebek O (2014a) Gabor filter based localization of needles in ultrasound guided robotic interventions. In: 2014 IEEE IST. IEEE, pp 112–117
Kaya M, Bebek O (2014b) Needle localization using gabor filtering in 2d ultrasound images. In: 2014 IEEE ICRA. IEEE, pp 4881–4886
Kaya M, Senel E, Ahmad A, Orhan O, Bebek O (2015) Real-time needle tip localization in 2d ultrasound images for robotic biopsies. In: 2015 ICAR. IEEE, pp 47–52
Kaya M, Senel E, Ahmad A, Bebek O (2016) Visual tracking of biopsy needles in 2d ultrasound images. In: 2016 IEEE ICRA. IEEE, pp 4386–4391
Kaya M, Senel E, Bebek O (2020) Gabor filter-based localization of straight and curved needles in 2d ultrasound images. Turk J Electric Eng Comput Sci 28:2940-2955
Krefting D, Haupt B, Tolxdorff T, Kempkensteffen C, Miller K (2007) Segmentation of prostate biopsy needles in transrectal ultrasound images. In: Medical imaging 2007: image processing, international society for optics and photonics, vol 6512, p 65122Y
Krücker J, Xu S, Glossop N, Viswanathan A, Borgert J, Schulz H, Wood BJ (2007) Electromagnetic tracking for thermal ablation and biopsy guidance: clinical evaluation of spatial accuracy. J Vasc Intervent Radiol 18(9):1141–1150
Lafferty J, McCallum A, Pereira FC (2001) Conditional random fields: probabilistic models for segmenting and labeling sequence data
Lee JY, Islam M, Woh JR, Washeem TM, Ngoh LYC, Wong WK, Ren H (2020) Ultrasound needle segmentation and trajectory prediction using excitation network. IJCARS 15(3):437–443
Linguraru MG, Howe RD (2006) Texture-based instrument segmentation in 3d ultrasound images. In: Medical imaging 2006: image processing, international society for optics and photonics, vol 6144, p 61443J
Linguraru MG, Vasilyev NV, Del Nido PJ, Howe RD (2007) Statistical segmentation of surgical instruments in 3-d ultrasound images.Ultrasound Med Biol 33(9):1428–1437
Litjens G, Kooi T, Bejnordi BE, Setio AAA, Ciompi F, Ghafoorian M, Van Der Laak JA, Van Ginneken B, Sánchez CI (2017) A survey on deep learning in medical image analysis. MedIA 42:60–88
Liu D, Tupor S, Singh J, Chernoff T, Leong N, Sadikov E, Amjad A, Zilles S (2022) The challenges facing deep learning-based catheter localization for ultrasound guided high-dose-rate prostate brachytherapy. Med Phys 49(4):2442–2451
Machi J, Uchida S, Sumida K, Limm WM, Hundahl SA, Oishi AJ, Furumoto NL, Oishi RH (2001) Ultrasound-guided radiofrequency thermal ablation of liver tumors: percutaneous, laparoscopic, and open surgical approaches. J Gastrointest Surg 5(5):477–489
Malekian L, Talebi HA, Towhidkhah F (2014) A noise adaptive method for needle localization in 3d ultrasound images. In: 2014 Iranian conference on intelligent systems (ICIS), IEEE, pp 1–5
Mathiassen K, Dall’Alba D, Muradore R, Fiorini P, Elle OJ (2016) Robust real-time needle tracking in 2-d ultrasound images using statistical filtering. IEEE Trans Control Syst Technol 25(3):966–978
McSweeney I, Murphy B, Wright WM (2014) Estimation of needle tip location using ultrasound image processing and hypoechoic markers. In: 2014 IEEE IUS. IEEE, pp 1876–1879
Min L, Yang H, Shan C, Kolen AF, et al. (2020) Feasibility study of catheter segmentation in 3d frustum ultrasounds by dcnn. In: Medical imaging 2020: image-guided procedures, robotic interventions, and modeling, international society for optics and photonics, vol 11315, p 1131521
Mohareri O, Ramezani M, Adebar TK, Abolmaesumi P, Salcudean SE (2013) Automatic localization of the da vinci surgical instrument tips in 3-d transrectal ultrasound. IEEE TBME 60(9):2663–2672
Mwikirize C, Nosher JL, Hacihaliloglu I (2016) Enhancement of needle tip and shaft from 2d ultrasound using signal transmission maps. In: MICCAI. Springer, pp 362–369
Mwikirize C, Nosher JL, Hacihaliloglu I (2017) Local phase-based learning for needle detection and localization in 3d ultrasound. In: Computer assisted and robotic endoscopy and clinical image-based procedures. Springer, pp 108–115
Mwikirize C, Nosher JL, Hacihaliloglu I (2018) Convolution neural networks for real-time needle detection and localization in 2d ultrasound. IJCARS 13(5):647–657
Mwikirize C, Nosher JL, Hacihaliloglu I (2019) Learning needle tip localization from digital subtraction in 2d ultrasound. IJCARS 14(6):1017–1026
Mwikirize C, Nosher JL, Hacihaliloglu I (2019b) Single shot needle tip localization in 2d ultrasound. In: MICCAI. Springer, pp 637–645
Mwikirize C, Kimbowa AB, Imanirakiza S, Katumba A, Nosher JL, Hacihaliloglu I (2021) Time-aware deep neural networks for needle tip localization in 2d ultrasound. Int J Comput Assist Radiol Surg 16(5):819–827
Nadeau C et al (2014) Intensity-based visual servoing for instrument and tissue tracking in 3d ultrasound volumes. IEEE TASE 12(1):367–371
Neshat HRS, Patel RV (2008) Real-time parametric curved needle segmentation in 3d ultrasound images. In: 2008 2nd IEEE RAS & EMBS international conference on biomedical robotics and biomechatronics. IEEE, pp 670–675
Novotny PM, Cannon JW, Howe RD (2003) Tool localization in 3d ultrasound images. In: MICCAI. Springer, pp 969–970
Novotny PM, Stoll JA, Vasilyev NV, Pedro J, Dupont PE, Zickler TE, Howe RD (2007) Gpu based real-time instrument tracking with three-dimensional ultrasound. Med Image Anal 11(5):458–464
Oepkes D, Devlieger R, Lopriore E, Klumper F (2007) Successful ultrasound-guided laser treatment of fetal hydrops caused by pulmonary sequestration. Ultrasound Obstet Gynecol 29(4):457–459
Okazawa SH, Ebrahimi R, Chuang J, Rohling RN, Salcudean SE (2006) Methods for segmenting curved needles in ultrasound images. MedIA 10(3):330–342
Patel SA, Pierko K, Franco-Sadud R (2019) Ultrasound-guided bedside core needle biopsy: a hospitalist procedure team’s experience. Cureus 11(1):e3817
Peters TM (2006) Image-guidance for surgical procedures. Phys Med Biol 51(14):R505
Pourtaherian A (2018) Robust needle detection and visualization for 3d ultrasound image-guided interventions. PhD thesis, Department of Electrical Engineering, proefschrift
Pourtaherian A, Zinger S, Korsten HH, Mihajlovic N, et al. (2015a) Benchmarking of state-of-the-art needle detection algorithms in 3d ultrasound data volumes. In: Medical imaging 2015: image-guided procedures, robotic interventions, and modeling, international society for optics and photonics, vol 9415, p 94152B
Pourtaherian A, Zinger S, Mihajlovic N, Huang J, Ng GC, Korsten HH, et al. (2015b) Multi-resolution gabor wavelet feature extraction for needle detection in 3d ultrasound. In: Eighth International conference on machine vision (ICMV 2015), International society for optics and photonics, vol 9875, p 987513
Pourtaherian A, Mihajlovic N, Zinger S, Korsten HH, de With PH, Huang J, Ng GC (2016) Automated in-plane visualization of steep needles from 3d ultrasound data volumes. In: 2016 IEEE IUS. IEEE, pp 1–4
Pourtaherian A, Scholten HJ, Kusters L, Zinger S, Mihajlovic N, Kolen AF, Zuo F, Ng GC, Korsten HH, de With PH (2017) Medical instrument detection in 3-dimensional ultrasound data volumes. IEEE TMI 36(8):1664–1675
Pourtaherian A, Zanjani FG, Zinger S, Mihajlovic N, Ng G, Korsten H, et al. (2017b) Improving needle detection in 3d ultrasound using orthogonal-plane convolutional networks. In: MICCAI. Springer, pp 610–618
Pourtaherian A, Mihajlovic N, GhazvinianZanjani F, Zinger S, Ng GC, Korstcn HH, De With PH (2018a) Localization of partially visible needles in 3d ultrasound using dilated cnns. In: 2018 IEEE IUS. IEEE, pp 1–4
Pourtaherian A, Zanjani FG, Zinger S, Mihajlovic N, Ng GC, Korsten HH et al (2018) Robust and semantic needle detection in 3d ultrasound using orthogonal-plane convolutional neural networks. IJCARS 13(9):1321–1333
Qiu W, Yuchi M, Ding M, Tessier D, Fenster A (2013) Needle segmentation using 3d hough transform in 3d trus guided prostate transperineal therapy. Med Phys 40(4):042902
Qiu W, Yuchi M, Ding M (2014) Phase grouping-based needle segmentation in 3-d trans-rectal ultrasound-guided prostate trans-perineal therapy. Ultrasound Med Biol 40(4):804–816
Ren H, Dupont PE (2011) Tubular structure enhancement for surgical instrument detection in 3d ultrasound. In: 2011 Annual international conference of the IEEE engineering in medicine and biology society, IEEE, pp 7203–7206
Rocha TS, Geraldes AA (2014) Flexible needles detection in ultrasound images using a multi-layer perceptron network. In: 5th ISSNIP-IEEE biosignals and biorobotics conference (2014): biosignals and robotics for better and safer living (BRC), IEEE, pp 1–5
Rodgers JR, Gillies DJ, Hrinivich WT, Gyackov I, Fenster A (2020a) Automatic needle localization in intraoperative 3d transvaginal ultrasound images for high-dose-rate interstitial gynecologic brachytherapy. In: Medical imaging 2020: image-guided procedures, robotic interventions, and modeling, international society for optics and photonics, vol 11315, p 113150K
Rodgers JR, Hrinivich WT, Surry K, Velker V, D’Souza D, Fenster A (2020) A semiautomatic segmentation method for interstitial needles in intraoperative 3d transvaginal ultrasound images for high-dose-rate gynecologic brachytherapy of vaginal tumors. Brachytherapy 19(5):659–668
Ronneberger O, Fischer P, Brox T (2015) U-net: Convolutional networks for biomedical image segmentation. In: MICCAI. Springer, pp 234–241
Scanlan KA, Propeck PA, Lee FT Jr (2001) Invasive procedures in the female pelvis: value of transabdominal, endovaginal, and endorectal us guidance. Radiographics 21(2):491–506
Sheafor DH, Paulson EK, Simmons CM, DeLong DM, Nelson RC (1998) Abdominal percutaneous interventional procedures: comparison of ct and us guidance. Radiology 207(3):705–710
Smith WL, Surry K, Mills G, Downey DB, Fenster A (2001) Three-dimensional ultrasound-guided core needle breast biopsy. Ultrasound Med Biol 27(8):1025–1034
Spencer KT, Kimura BJ, Korcarz CE, Pellikka PA, Rahko PS, Siegel RJ (2013) Focused cardiac ultrasound: recommendations from the american society of echocardiography. J Am Soc Echocardiogr 26(6):567–581
St-Amant P, Foster W, Forment MA, Aubin S, Beaulieu L (2016) Use of 3d-ultrasound for cervical cancer brachytherapy: an imaging technique to improve treatment planning. Brachytherapy 15:S92–S93
Suykens JA, Vandewalle J (1999) Least squares support vector machine classifiers. Neural Process Lett 9(3):293–300
Torr PH, Zisserman A (2000) Mlesac: A new robust estimator with application to estimating image geometry. Comput Vision Image Underst 78(1):138–156
Treeby BE, Cox BT (2010) k-wave: Matlab toolbox for the simulation and reconstruction of photoacoustic wave fields. J Biomed Optics 15(2):021314
Uherčík M, Kybic J, Zhao Y, Cachard C, Liebgott H (2013) Line filtering for surgical tool localization in 3d ultrasound images. Comput Biol Med 43(12):2036–2045
Wei Z, Gardi L, Downey DB, Fenster A (2005) Oblique needle segmentation and tracking for 3d trus guided prostate brachytherapy. Med Phys 32(9):2928–2941
Wijata A, Andrzejewski J, Pyciński B (2021) An automatic biopsy needle detection and segmentation on ultrasound images using a convolutional neural network. Ultrason Imaging 43(5):262–272
Xia W et al (2015) In-plane ultrasonic needle tracking using a fiber-optic hydrophone. Med Phys 42(10):5983–5991
Yan P, Cheeseborough JC III, Chao KC (2012) Automatic shape-based level set segmentation for needle tracking in 3-d trus-guided prostate brachytherapy. Ultrasound Med Biol 38(9):1626–1636
Yang H, Pourtaherian A, Shan C, Kolen AF, et al. (2018a) Feature study on catheter detection in three-dimensional ultrasound. In: Medical imaging 2018: image-guided procedures, robotic interventions, and modeling, international society for optics and photonics, vol 10576, p 105760V
Yang H, Shan C, Kolen AF, de With PH (2018b) Catheter detection in 3d ultrasound using triplanar-based convolutional neural networks. In: 2018 25th IEEE ICIP. IEEE, pp 371–375
Yang H, Shan C, Kolen AF, de With PH (2019a) Automated catheter localization in volumetric ultrasound using 3d patch-wise u-net with focal loss. In: 2019 IEEE ICIP. IEEE, pp 1346–1350
Yang H, Shan C, Kolen AF, de With PH (2019b) Efficient catheter segmentation in 3d cardiac ultrasound using slice-based fcn with deep supervision and f-score loss. In: 2019 IEEE ICIP. IEEE, pp 260–264
Yang H, Shan C, Kolen AF, de With Peter H (2019c) Improving catheter segmentation & localization in 3d cardiac ultrasound using direction-fused fcn. In: 2019 IEEE ISBI. IEEE, pp 1122–1126
Yang H, Shan C, Kolen AF et al (2019) Catheter localization in 3d ultrasound using voxel-of-interest-based convnets for cardiac intervention. IJCARS 14(6):1069–1077
Yang H, Shan C, Pourtaherian A, Kolen AF et al (2019) Catheter segmentation in three-dimensional ultrasound images by feature fusion and model fitting. JMI 6(1):015001
Yang H, Shan C, Tan T, Kolen AF, et al. (2019f) Transferring from ex-vivo to in-vivo: Instrument localization in 3d cardiac ultrasound using pyramid-unet with hybrid loss. In: MICCAI. Springer, pp 263–271
Yang H, Shan C, Bouwman A, Kolen AF, de With PH (2020) Efficient and robust instrument segmentation in 3d ultrasound using patch-of-interest-fusenet with hybrid loss. Med Image Anal 67:101842
Yang H, Shan C, Kolen A, de With PH (2020b) Efficient medical instrument detection in 3d volumetric ultrasound data. IEEE TBME
Yang H, Shan C, Kolen AF, de With PH (2020c) Deep q-network-driven catheter segmentation in 3d us by hybrid constrained semi-supervised learning and dual-unet. arXiv preprint arXiv:2006.14702
Yang H, Shan C, Kolen AF, et al. (2020d) Deep q-network-driven catheter segmentation in 3d us by hybrid constrained semi-supervised learning and dual-unet. In: International conference on medical image computing and computer-assisted intervention. Springer, pp 646–655
Yang H, Shan C, Bouwman RA, Dekker L, Kolen A, De With PH (2021) Medical instrument segmentation in 3d us by hybrid constrained semi-supervised learning. IEEE J Biomed Health Info 26:762
Yang H, Shan C, Kolen AF, de With PH (2022) Weakly-supervised learning for catheter segmentation in 3d frustum ultrasound. Comput Med Imaging Gr 96:102037
Younes H, Voros S, Troccaz J (2018) Automatic needle localization in 3d ultrasound images for brachytherapy. In: 2018 IEEE ISBI. IEEE, pp 1203–1207
Zanjani FG, Pourtaherian A, Tang X, Zinger S, Mihajlovic N, Ng GC, Korsten HH, et al. (2018) Coherent needle detection in ultrasound volumes using 3d conditional random fields. In: Medical imaging 2018: image-guided procedures, robotic interventions, and modeling, international society for optics and photonics, vol 10576, p 105760W
Zhang Y, Harms J, Lei Y, Wang T, Liu T, Jani AB, Curran WJ, Patel P, Yang X (2020a) Weakly supervised multi-needle detection in 3d ultrasound images with bidirectional convolutional sparse coding. In: Medical imaging 2020: ultrasonic imaging and tomography, international society for optics and photonics, vol 11319, p 1131914
Zhang Y, He X, Tian Z, Jeong J, Lei Y, Wang T, Zeng Q, Jani AB, Curran W, Patel P, et al. (2020b) Multi-needle detection in 3d ultrasound images with sparse dictionary learning. In: Medical imaging 2020: ultrasonic imaging and tomography, international society for optics and photonics, vol 11319, p 113190I
Zhang Y, He X, Tian Z, Jeong JJ, Lei Y, Wang T, Zeng Q, Jani AB, Curran WJ, Patel P, et al. (2020c) Multi-needle detection in 3d ultrasound images using unsupervised order-graph regularized sparse dictionary learning. IEEE TMI
Zhang Y, Lei Y, Qiu RL, Wang T, Wang H, Jani AB, Curran WJ, Patel P, Liu T, Yang X (2020d) Multi-needle localization with attention u-net in us-guided hdr prostate brachytherapy. Med Phys 47:2735
Zhao S, Qiu W, Ming Y, Ding M (2009) Needle segmentation in 3d ultrasound images based on phase grouping. In: MIPPR 2009: medical imaging, parallel processing of images, and optimization techniques, international society for optics and photonics, vol 7497, p 74971L
Zhao Y, Cachard C, Liebgott H (2013) Automatic needle detection and tracking in 3d ultrasound using an roi-based ransac and kalman method. Ultrasonc Imaging 35(4):283–306
Zhao Y, Cachard C, Liebgott H (2013b) A new automatically biopsy needle tracking method using 3d ultrasound. In: 2013 IEEE IUS. IEEE, pp 844–847
Zhao Y, Shen Y, Bernard A, Cachard C, Liebgott H (2017) Evaluation and comparison of current biopsy needle localization and tracking methods using 3d ultrasound. Ultrasonics 73:206–220
Zhou H, Qiu W, Ding M, Zhang S (2007) Automatic needle segmentation in 3d ultrasound images using 3d hough transform. In: MIPPR 2007: Medical imaging, parallel processing of images, and optimization techniques, international society for optics and photonics, vol 6789, p 67890R
Zhou H, Qiu W, Ding M, Zhang S (2008) Automatic needle segmentation in 3d ultrasound images using 3d improved hough transform. In: Medical imaging 2008: visualization, image-guided procedures, and modeling, international society for optics and photonics, vol 6918, p 691821
Author information
Authors and Affiliations
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Yang, H., Shan, C., Kolen, A.F. et al. Medical instrument detection in ultrasound: a review. Artif Intell Rev 56, 4363–4402 (2023). https://doi.org/10.1007/s10462-022-10287-1
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10462-022-10287-1