Abstract
Potential human and environmental hazards resulting from the exposure of living organisms to silver nanoparticles (Ag NPs) have been the subject of intensive discussion in the last decade. Despite the growing use of Ag NPs in biomedical applications, a quantification of the toxic effects as a function of the total silver mass reaching cells (namely, target cell dose) is still needed. To provide a more accurate dose-response analysis, we propose a novel integrated approach combining well-established computational and experimental methodologies. We first used a particokinetic model (ISD3) for providing experimental validation of computed Ag NP sedimentation in static-cuvette experiments. After validation, ISD3 was employed to predict the total mass of silver reaching human endothelial cells and hepatocytes cultured in 96 well plates. Cell viability measured after 24 h of culture was then related to this target cell dose. Our results show that the dose perceived by the cell monolayer after 24 h of exposure is around 85% lower than the administered nominal media concentration. Therefore, accurate dosimetry considering particle characteristics and experimental conditions (e.g., time, size and shape of wells) should be employed for better interpreting effects induced by the amount of silver reaching cells.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Engineered nanomaterials (ENMs) are very successful in the bio-technology industry because of their exceptionally small size and unique physical and chemical properties.11,14 One of the most widely used ENMs is silver, popularly known for its antimicrobial properties. It can be coated on biomedical devices,10 used in medical contexts for personal health care17 or biological applications,36 and adapted for food products such as kitchen tools, storage containers and cutting boards.31 However, despite the growing use of silver nanoparticles (Ag NPs) in the last decade,1,8,26,44 potential human and environmental hazards resulting from exposure to Ag NPs continue to be the subject of attention38,54 owing to their well-documented toxicity both in vivo and in vitro. In fact, well-established in vivo7,13,46 and in vitro systems33,45,48 have been proposed for toxicological studies. Mouse and zebrafish models exposed to ENMs, for example, showed nanotoxicity effects on female reproductive and fetal development.27,40In vitro, dose-dependent Ag NPs induced cellular necrosis, inflammation and oxidative stress in living organisms in a size-specific manner.6,16,24,30,34,50 The oxidative stress was further related to the anti-microbial activity of Ag NPs affecting different types of pathogens.9,15,39 Le et al. also discussed short- and long-term toxicity induced by the interaction of Ag NPs with cellular interfaces (e.g., decreased cell viability and apoptosis).26 Genetic damage (e.g., DNA breakage) was additionally found within cells interacting with Ag NPs19 and associated with the production of reactive oxygen species.35,43 The interaction of nanoparticles with cellular interfaces can induce other effects such as biophysical changes in the cellular membrane and cytoskeleton, giving rise to changes in cell elasticity, morphology, motility and adhesion.51 All these alterations and biophysical changes are often misinterpreted since they are generally reported as a function of the initial silver nanoparticle concentration in liquid media (here referred to as nominal media concentration) and not as a function of that actually coming into contact with cells (i.e. target cell dose, here defined as the silver concentration reaching cells, including ions and NPs, divided by the total volume of the media). At a given experimental time, this target cell dose is lower than the nominal media concentration because of NP transport (e.g. particle settling in static experiments, Brownian motion) and dissolution in the culture media before reaching cells. Only under ideal conditions (t → ∞) are the two concentrations equal. Therefore, on the basis that the target cell dose is the cause of the decrease in cell viability, our aim was to provide a more accurate dose-response analysis by reporting the cellular viability (i.e. effect) as a function of the predicted dose coming into contact with cultures.4 Specifically, we propose a new approach integrating computational12,28,32 and experimental models.33,45,48 First, we experimentally validated Ag NP sedimentation by comparing the concentrations in static-cuvette experiments with theoretical values computed by an established particokinetic model (ISD3), published by Thomas et al.42 After verifying that ISD3 correctly predicts the sedimentation process, we proceeded to use this model for estimating the target cell dose (i.e. Ag NPs and dissolved Ag+ in cells) perceived by cells in culture. Cell viability was finally related to the computed target cell doses and compared to the nominal media concentrations of Ag NPs initially administered to cells.
Materials and Methods
In Vitro–In Silico Pipeline
Figure 1 schematises the integrated in vitro–in silico pipeline used in this study. We first adapted ISD342 to our experimental configuration (Fig. 1a).
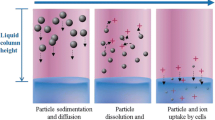
Particokinetics and experimental validation. (a) Graphical representation of Ag NP sedimentation, diffusion and dissolution in time. (b) Nanoparticle sedimentation in static-cuvette experiments for model validation. (c) 96-well micro-plates experiments for evaluating cell toxicity induced by computed effective Ag NP doses. (d) Cell viability vs. computed target cell doses at time t. Notably, at a finite experimental time t, target cell doses (here denoted as X′, Y′ and Z′) are lower than their respective nominal media concentrations (i.e. X, Y and Z).
Then we measured Ag NP sedimentation in static-cuvette experiments and compared the results with those computed by the model (Fig. 1b). In the next step, the model was used to estimate the target cell dose on cell monolayers cultured in 96-microwells for up to 24 h (Fig. 1c). The computed target cell doses as well as the nominal media concentrations of Ag NPs administered to cells were related to cell viability. (Fig. 1d).
Cell Cultures
Human umbilical vein endothelial cells (HUVECs) and the human hepatoma-derived hepatocyte C3A cell line (ATCC Culture, USA) were used for evaluating nanotoxicity.45 Specifically, HUVECs were obtained as described in our previous work45 and seeded at a concentration of 20,000 cells cm−2 on 1% w/v gelatin coated 96-well plates. Cells were allowed to reach confluence (typically 24 h) before exposure to Ag NPs. C3A hepatocytes were seeded at a density of 200,000 cells cm−2 on collagen coated plates and incubated for 24 h before experiments. In view of conducting future co-culture experiments in a flow through system, the same medium was used for Ag NP exposure experiments for both HUVEC and hepatocytes. The cells were cultured in Eagle’s Minimum Essential Medium (EMEM, Lonza Bioscience, Basel, Switzerland), supplemented with 10% Fetal Bovine Serum (FBS, PAA, Pasching, Austria), 1% Penicillin/Streptomycin/Amphotericin B, 2 mM l-Glutamine, 1% non-essential amino acids, 1% MEM vitamins solution (all from Lonza Bioscience, Basel, Switzerland), 10 µg mL−1 Endothelial Cell Growth Supplement (ECGS), 10 ng mL−1 Human Epidermal Growth Factor (hEGF), 3 ng mL−1 basic Fibroblast Growth Factor (bFGF), 1 µg mL−1 Hydrocortisone, and 10 µg mL−1 Heparin Sodium Salt (all from Sigma-Aldrich, St. Louis, USA).
Silver Nanoparticles
Ag NM300 from Ras GmbH, an OECD referenced nanomaterial (NM) with a nominal diameter of 20 nm, was purchased as 10% w/w suspension in a aqueous solution containing 7% v/v ammonium nitrate as stabilizing agent and 4% v/v Tween 20 and 4% v/v polyoxyethylene glycerol trioleate, as dispersants. The protocol developed by Klein et al.25 at the European Commission’s Joint Research Centre (JRC) and employed in several publications was used for a stock solution preparation.20,21 Ag NPs suspended in the same medium used in our experiments were characterized, applying the methods reported in Refs. 23 and 45, as discussed by JRC.25
Static Sedimentation Experiments
A 2 mL quartz cuvette (Fig. 1b) was filled with the Ag NP suspension and corked to avoid evaporation. Sample absorbance was measured at 414 nm for up to 24 h using a Varian Cary UV spectrophotometer equipped with a 1 mm hole positioned at half height of the cuvette. Nanoparticles were monitored at different nominal media concentrations (i.e. 5, 10, 15, 40, 50, 80 µg mL−1). For each Ag NP concentration used, the absorbance at the beginning of the experiment (time 0 h) was used to generate a calibration curve.
NP Exposure and Toxicity Assay
The medium of both cultures was replaced with 0.2 mL of medium containing homogeneously suspended Ag NPs at different concentrations (from 0 to 100 µg mL−1) at experimental time t = 0. Cells were then cultured for up to 24 h. To assess cell toxicity, medium containing Ag NPs or dispersant was removed after each incubation time and fresh medium and Alamar reagent (CellTiter-Blue® Promega, Madison, USA) were added. The assay reagent quantitatively measures mitochondrial activity through a cell-permeant fluorescent indicator. Cell viability was obtained from the slope in fluorescence emission within a 2 h time frame measured with a plate-reader (Omega-Fluostar Inc) and expressed as a percentage with respect to cells exposed to 0 µg mL−1 Ag NP. For each particle concentration was tested in the absence of cells in order to check for any interaction with the assay.
The ISD3 Model
ISD3 simulates the sedimentation, diffusion and dissolution of spherical nanoparticles in liquid media.42 This particokinetic model was used for predicting silver concentrations (including Ag NPs and Ag+ ions) perceived by cultured cells (Fig. 1a). All the equations are reported in the Supplementary materials. In ISD3, the particle number density (N) is defined at any time (t) and space (x) as a function of the NP diameter (Dp) and depends on diffusion (coefficient Ddiff), sedimentation (characteristic velocity \(V_{t}\)), and dissolution rate:
The dissolution kinetics of Ag NPs are depicted in Fig. 2. Silver NPs are assumed to be immediately surrounded by a protein corona when exposed to cell culture media. Free Ag ions dissolve from the surface of the NPs and may be bound to proteins \(\left( {C^{{{\text{diss}},p}} } \right)\) or free \(\left( {C^{{{\text{diss}},f}} } \right)\). The ions are taken up by the cells through a membrane diffusion process (eqn. 4 in the Supplementary materials), while Ag NPs are instantaneously taken up by cells. Following the notation used in Ref. 42, \(k_{f}\) is the rate constant for the transfer of ions from the particle surface to the free ion state, kp the rate constant for the slow transfer of ions from the particle surface to the proteins, kp2 the rate constant for the initial fast transfer of silver ions from the particle surface to the protein-bound state, kf2p the rate constant for the transfer of free ions from solution to the protein-bound state, and kp2f the rate constant for the transfer of ions from the protein-bound state to the free ion state.
Schematic of Ag NP dissolution process. The dissolution rate constants from Ag NPs to free ions (kf), protein (kp) and protein-bound NPs (kp2) are described, as well as the rate constants for the free ions in solution to protein-bound NPs (kf2p) and the transfer of ions from protein-bound NPs to free ions (kp2f).
A sensitivity analysis conducted by Thomas et al. demonstrated that ISD3 can describe the dissolution behavior of silver nanoparticles within a 10% sensitivity range of the kinetic parameters. Because the concentration of serum in the media (10% FBS) and the nominal diameter of Ag NP (20 nm) used in this work are the same as those employed by the authors, all the parameters used here are from Ref. 42 except those in bold in Table 1. The latter were modified for modelling the dissolution process in our static-cuvette and 96-well micro-plates. Should other particles be used, they must first be characterized using the methods listed in Table S1 and S2 in the Supplementary Materials. Further information on the techniques used to obtain the rate constants and the use of the ISD3 Matlab code are also provided therein.
Data Processing
Static-cuvette experiments were performed in duplicate, while 96-well plate in vitro experiments were performed in triplicate. Data were processed and the dose-response curves plotted using MATLAB R2019b (Mathworks). Results are expressed as means ± standard deviation.
Results
The particles have an average size of 17.5 nm with euhedral morphology as measured using transmission electron microscopy (TEM). Finally, the mean effective diameter observed using Nanosight system is 120 nm with monomodal size distribution. Data are summarized in Table 2.
We first tracked the Ag NP dissolution and sedimentation process in static-cuvette experiments as a function of the initial administered concentrations (nominal media concentrations). The rationale was to validate ISD3 by quantifying how the predicted nano-particle concentrations fit with those experimentally measured at half height of the cuvette (Fig. 3; grey bars). A high Pearson coefficient (r = 0.9991) was observed, indicating a strong correlation between predicted and detected concentrations (inset), confirming the ISD3’s reliability in modelling particle sedimentation.
Validation of particle sedimentation in static-cuvette experiments. Ag NP concentrations measured at half height of the cuvette (grey bars) at 24 h fit well with those predicted by ISD3 (black dashed line). The scatter plot (inset) shows a strong correlation (r = 0.9991) between the experimental and computed Ag NP concentrations.
Subsequently, we compared predicted target doses reaching the cells (Fig. 4a, solid lines) with different Ag NP nominal media concentrations (dashed lines) over 24 h. The rationale was to provide a more accurate dose-response analysis since the amount of silver ions and Ag NPs coming into contact with cells at a given time is lower than the nominal media concentration administered because particles dissolve and settle. In fact, as represented by the vertical distance between black and grey lines in Fig. 4b, the target cell doses were much lower than their respective nominal media concentrations at the end of our experiment (24 h). These differences, here expressed as Δ = (nominal media concentration − target cell dose)/(nominal media concentration), are reported in Table 3. As expected, the target cell doses were proportional to their respective nominal media concentrations.
Target cell doses computed over 24 h. (a) Target cell doses (solid lines) and nominal media concentrations (dashed lines) as a function of time (red: 0.5 µg mL−1; blue 1 µg mL−1; green 5 µg mL−1; grey 10 µg mL−1; cyan 50 µg mL−1; black 100 µg mL−1). Target cell doses are equal to zero at t = 0 (not apparent in the semi-log plot). (b) Target cell doses computed at t = 24 h (black solid line) vs. nominal media concentrations. Notably, only under ideal conditions (t → ∞) are these two concentrations equal (grey solid line). The black dotted line denotes target cell doses at t = 0 h, which are all null regardless of the nominal media concentration.
Having predicted target cell doses during the entire exposure period (i.e., from 0 to 24 h) and quantified differences with increased nominal media concentrations (Δ values), we investigated time-dependent nanotoxicity effects induced by the effective silver concentrations coming into contact with cells reporting HUVEC and C3A viability at 6 h (Fig. 5a), 16 h (Fig. 5b) and 24 h (Fig. 5c) of culture as a function of the target cell doses. Both HUVEC (gray line) and C3A (black line) viability decreased with increasing target (and nominal) cell dose. From the graphs it is also evident that the Δ has implications on the estimation of the IC50 values. In fact, referring to the nominal media concentration, the IC50 value at 24 h is estimated as 41.04 µg mL−1 for HUVEC and 54.78 µg mL−1 for C3A. However, if one considers the target cell dose (actually experienced by cells), the IC50 drops significantly (~ 80%): 8.13 µg mL−1 for HUVEC and 10.95 µg mL−1 for C3A.
Discussion
An integrated pipeline based on computational (ISD3) and experimental approaches to provide more accurate dose-response analyses and to minimize time-consuming, expensive and ethically sensitive in vivo tests52 is described. Having verified the accuracy of ISD3 through sedimentation experiments, we evaluated the viability of HUVEC and C3A cells as a function of the total silver mass reaching cells (namely, target cell dose). The results show that target cell doses computed at 24 h are much lower (85%) than the nominal media concentrations initially administered to the cells (t = 0 h), thus the nanotoxic effects induced by Ag NP exposure may be underestimated if they are related to the initial administered concentrations.
Much uncertainty still remains regarding which constituent (i.e., NPs, free and protein-bound ions) contributes to cellular toxicity.2,37 Bouwmeester et al.5 as well as Wang et al.49 suggest that exposure to silver ions formed extracellularly is responsible for observed toxic effects. Conversely, other groups assume that the internalized nanoparticles (i.e. NPs taken up by cells) undergo rapid dissolution resulting in silver ions inducing toxicity.18,41 Therefore, to take into account both NP and ion related toxic effects, the target cell dose in this work includes both dissolved ions and particles reaching cells (i.e. Ag NPs + dissolved Ag+ in cells).
Some other fundamental concerns in in vitro nanotoxicology still need to be addressed before data from cell culture experiments can be considered reliable for predicting NP toxicity. The ISD3 model, for example, assumes Ag NPs are immediately absorbed once the cell surface is reached. Furthermore, ISD3 does not simulate the intercellular dissolution of NPs once they have been taken up by the cells and it is limited to a 2 dimensional cell configuration, which is not considered “physiologically relevant”. By modelling the kinetics involved within the three-dimensional geometries of cellular constructs, we should be able to predict the Ag NP uptake through multiple layers in a more realistic in vivo-like multilayer configuration. Moreover, the computed target cell doses sequentially absorbed “layer by layer” would allow an even more accurate analysis of the nanotoxicity effects. In this regard, the three-dimensional and scaffold-less cellular aggregates in the form of “organoids”, “microtissues” or “spheroids” which resemble the cytoarchitectural arrangement of human tissues (e.g., liver, lung and specific brain regions) are currently being used as more physiologically relevant in vitro systems for experimental validation.29 For example, Kermanizadeh et al.22 have investigated three-dimensional human liver microtissue models, exposing them to different types of nanomaterials (including Ag NPs). They used Ag NP concentrations within the range employed here, demonstrating that repeated exposure is more damaging to the liver tissue in comparison to a single exposure. In this context, as there is some interest in the effects of chronic exposure, it would be useful to incorporate dosing regimens into ISD3. The boundary conditions, as well as all variables regarding nanoparticles and dissolved ions in liquid medium, could be updated taking into account multiple doses which are sequentially exposed to the cells and change over every grid point in space at any simulation time. Under the assumption that the chemical equilibrium in the system depends principally on the concentration of dissolved silver ions in liquid \(\left( {C^{{{\text{diss}},f}} } \right)\) and in cells \(\left( {C^{{{\text{diss}},c}} } \right)\),3 we could iteratively add \(C^{{{\text{diss}},f}}\) and subtract \(C^{{{\text{diss}},c}}\) at any given time in space (Eqs. 3 and 4, respectively, in the Supplementary Materials) and then recalculate the cumulative particle number density. The application of the model to chronic exposure regimens is currently being validated using the same framework as described herein.
It should be noted that for the model configuration described in our work, we would need to compute and experimentally validate not only the NPs diffusing towards the cells cultured on the bottom of the wells, but also those particles coming into contact with the cells adjacent to the walls. To the best of our knowledge, this second computational aspect has been exclusively provided by Bohemert et al.4 They indeed modelled the dosimetry and the exposure to nanoparticles of Caco-2 cells forming a confluent monolayer on the bottom of the cell culture dish, but also growing up its encircling walls.
Another aspect to consider is that Ag NP sedimentation and cell toxicity were modelled and experimentally evaluated under static conditions. It would be interesting to extend the combined computational–experimental approach to dynamic experiments, implemented with a modular bioreactor similar to that described in our previous work47 or a microfluidic system.53 A multiphysics computational approach could be used to model the combined effects of particle sedimentation, diffusion, dissolution and convection as a function of the involved fluxes characterizing the dynamics of this type of more “physiologically relevant” in vitro model. It would also be interesting to quantify the effects of absorbed nanoparticles of different materials and diameter sizes. These features, including the protein corona on the surface of the nanoparticles (here represented by a 51.25 nm thick protein layer; see Table 1), are known to modulate nanotoxicity.4,42
In conclusion, combining computational and experimental approaches, we provide more reliable dose–response analyses and hazard assessment. This study paves the way for the development of a methodological framework which combines the expertise of modellers, toxicologists and scientists studying nanotechnology and nanoscience to enable good experimental design and more accurate in vitro to in vivo data extrapolation.
Change history
30 January 2020
The article An Integrated In Vitro–In Silico Approach for Silver Nanoparticle Dosimetry in Cell Cultures, written by Ahluwalia et al, was originally published electronically on the publisher’s internet portal (currently SpringerLink) on 13 January 2020 without open access.
References
AshaRani, P. V., G. Low Kah Mun, M. P. Hande, and S. Valiyaveettil. Cytotoxicity and genotoxicity of silver nanoparticles in human cells. ACS Nano 3:279–290, 2009.
Beer, C., R. Foldbjerg, Y. Hayashi, D. S. Sutherland, and H. Autrup. Toxicity of silver nanoparticles—nanoparticle or silver ion? Toxicol. Lett. 208:286–292, 2012.
Behra, R., L. Sigg, M. J. D. Clift, F. Herzog, M. Minghetti, B. Johnston, A. Petri-Fink, and B. Rothen-Rutishauser. Bioavailability of silver nanoparticles and ions: from a chemical and biochemical perspective. J. R. Soc. Interface 10:20130396, 2013.
Böhmert, L., L. König, H. Sieg, D. Lichtenstein, N. Paul, A. Braeuning, A. Voigt, and A. Lampen. In vitro nanoparticle dosimetry for adherent growing cell monolayers covering bottom and lateral walls. Part. Fibre Toxicol. 15:42, 2018.
Bouwmeester, H., J. Poortman, R. J. Peters, E. Wijma, E. Kramer, S. Makama, K. Puspitaninganindita, H. J. P. Marvin, A. A. C. M. Peijnenburg, and P. J. M. Hendriksen. Characterization of translocation of silver nanoparticles and effects on whole-genome gene expression using an in vitro intestinal epithelium coculture model. ACS Nano 5:4091–4103, 2011.
Carlson, C., S. M. Hussain, A. M. Schrand, L. K. Braydich-Stolle, K. L. Hess, R. L. Jones, and J. J. Schlager. Unique cellular interaction of silver nanoparticles: size-dependent generation of reactive oxygen species. J. Phys. Chem. B 112:13608–13619, 2008.
Chakraborty, C., A. R. Sharma, G. Sharma, and S.-S. Lee. Zebrafish: a complete animal model to enumerate the nanoparticle toxicity. J. Nanobiotechnology 14:65, 2016.
Chen, N., Z.-M. Song, H. Tang, W.-S. Xi, A. Cao, Y. Liu, and H. Wang. Toxicological effects of Caco-2 cells following short-term and long-term exposure to Ag nanoparticles. Int. J. Mol. Sci. 17:974, 2016.
Comfort, K. K., E. I. Maurer, L. K. Braydich-Stolle, and S. M. Hussain. Interference of silver, gold, and iron oxide nanoparticles on epidermal growth factor signal transduction in epithelial cells. ACS Nano 5:10000–10008, 2011.
Cronholm, P., H. L. Karlsson, J. Hedberg, T. A. Lowe, L. Winnberg, K. Elihn, I. O. Wallinder, and L. Möller. Intracellular uptake and toxicity of Ag and CuO nanoparticles: a comparison between nanoparticles and their corresponding metal ions. Small 9:970–982, 2013.
de Faria, A. F., D. S. T. Martinez, S. M. M. Meira, A. C. M. de Moraes, A. Brandelli, A. G. S. Filho, and O. L. Alves. Anti-adhesion and antibacterial activity of silver nanoparticles supported on graphene oxide sheets. Colloids Surf. B Biointerfaces 113:115–124, 2014.
De Loid, G. M., J. M. Cohen, G. Pyrgiotakis, S. V. Pirela, A. Pal, J. Liu, J. Srebric, and P. Demokritou. Advanced computational modeling for in vitro nanomaterial dosimetry. Part. Fibre Toxicol. 12:32, 2015.
Dhakshinamoorthy, V., V. Manickam, and E. Perumal. Neurobehavioural toxicity of iron oxide nanoparticles in mice. Neurotox. Res. 32:187–203, 2017.
Durán, N., C. P. Silveira, M. Durán, and D. S. T. Martinez. Silver nanoparticle protein corona and toxicity: a mini-review. J. Nanobiotechnology 13:55, 2015.
Filipović, N., M. Stevanović, A. Radulović, V. Pavlović, and D. Uskoković. Facile synthesis of poly(ε-caprolactone) micro and nanospheres using different types of polyelectrolytes as stabilizers under ambient and elevated temperature. Compos. Part B Eng. 45:1471–1479, 2013.
Foldbjerg, R., P. Olesen, M. Hougaard, D. A. Dang, H. J. Hoffmann, and H. Autrup. PVP-coated silver nanoparticles and silver ions induce reactive oxygen species, apoptosis and necrosis in THP-1 monocytes. Toxicol. Lett. 190:156–162, 2009.
Ge, L., Q. Li, M. Wang, J. Ouyang, X. Li, and M. M. Q. Xing. Nanosilver particles in medical applications: synthesis, performance, and toxicity. Int. J. Nanomed. 9:2399–2407, 2014.
Hsiao, I.-L., Y.-K. Hsieh, C.-F. Wang, I.-C. Chen, and Y.-J. Huang. Trojan-horse mechanism in the cellular uptake of silver nanoparticles verified by direct intra- and extracellular silver speciation analysis. Environ. Sci. Technol. 49:3813–3821, 2015.
Kawata, K., M. Osawa, and S. Okabe. In vitro toxicity of silver nanoparticles at noncytotoxic doses to HepG2 human hepatoma cells. Environ. Sci. Technol. 43:6046–6051, 2009.
Kermanizadeh, A., B. K. Gaiser, G. R. Hutchison, and V. Stone. An in vitro liver model—assessing oxidative stress and genotoxicity following exposure of hepatocytes to a panel of engineered nanomaterials. Part. Fibre Toxicol. 9:28, 2012.
Kermanizadeh, A., I. Gosens, L. MacCalman, H. Johnston, P. H. Danielsen, N. R. Jacobsen, A.-G. Lenz, T. Fernandes, R. P. F. Schins, F. R. Cassee, H. Wallin, W. Kreyling, T. Stoeger, S. Loft, P. Møller, L. Tran, and V. Stone. A multilaboratory toxicological assessment of a panel of 10 engineered nanomaterials to human health—ENPRA Project—the highlights, limitations, and current and future challenges. J. Toxicol. Environ. Heal. Part B 19:1–28, 2016.
Kermanizadeh, A., M. Roursgaard, S. Messner, P. Gunness, J. M. Kelm, P. Møller, V. Stone, and S. Loft. Hepatic toxicology following single and multiple exposure of engineered nanomaterials utilising a novel primary human 3D liver microtissue model. Part. Fibre Toxicol. 11:56, 2014.
Kermanizadeh, A., S. Vranic, S. Boland, K. Moreau, A. Baeza-Squiban, B. K. Gaiser, L. A. Andrzejczuk, and V. Stone. An in vitroassessment of panel of engineered nanomaterials using a human renal cell line: cytotoxicity, pro-inflammatory response, oxidative stress and genotoxicity. BMC Nephrol. 14:96, 2013.
Kim, S., J. E. Choi, J. Choi, K.-H. Chung, K. Park, J. Yi, and D.-Y. Ryu. Oxidative stress-dependent toxicity of silver nanoparticles in human hepatoma cells. Toxicol. Vitr. 23:1076–1084, 2009.
Klein, C. L., S. Comero, B. Stahlmecke, J. Romazanov, T. A. J. Kuhlbusch, E. V Doren, P. J. D. Temmerman, J. Mast, P. Wick, and H. Krug. NM-series of representative manufactured nanomaterials. NM-300 silver. characterisation, stability, homogeneity. JRC Scientific and Technical Reports 2011. Google Sch. 2013.
Le, Q. H., and A. T. Le. Silver nanoparticles: synthesis, properties, toxicology, applications and perspectives. Adv. Nat. Sci. Nanosci. Nanotechnol. 4:33001, 2013.
Li, Y., Y. Zhang, and B. Yan. Nanotoxicity overview: nano-threat to susceptible populations. Int. J. Mol. Sci. 15:3671–3697, 2014.
Liu, R., H. H. Liu, Z. Ji, C. H. Chang, T. Xia, A. E. Nel, and Y. Cohen. Evaluation of toxicity ranking for metal oxide nanoparticles via an in vitro dosimetry model. ACS Nano 9:9303–9313, 2015.
Mattei, G., C. Magliaro, S. Giusti, S. D. Ramachandran, S. Heinz, J. Braspenning, and A. Ahluwalia. On the adhesion-cohesion balance and oxygen consumption characteristics of liver organoids. PLoS ONE 12:e0173206, 2017.
McShan, D., P. C. Ray, and H. Yu. Molecular toxicity mechanism of nanosilver. J. Food Drug Anal. 22:116–127, 2014.
Morris, V. J. Emerging roles of engineered nanomaterials in the food industry. Trends Biotechnol. 29:509–516, 2011.
Mukherjee, D., B. F. Leo, S. G. Royce, A. E. Porter, M. P. Ryan, S. Schwander, K. F. Chung, T. D. Tetley, J. Zhang, and P. G. Georgopoulos. Modeling physicochemical interactions affecting in vitro cellular dosimetry of engineered nanomaterials: application to nanosilver. J. Nanoparticle Res. 16:2616, 2014.
Oomen, G. A., A. E. Bleeker, M. P. Bos, F. van Broekhuizen, S. Gottardo, M. Groenewold, D. Hristozov, K. Hund-Rinke, M.-A. Irfan, A. Marcomini, J. W. Peijnenburg, K. Rasmussen, S. A. Jiménez, J. J. Scott-Fordsmand, M. van Tongeren, K. Wiench, W. Wohlleben, and R. Landsiedel. Grouping and read-across approaches for risk assessment of nanomaterials. 12:13415–13434, 2015.
Park, M. V. D. Z., A. M. Neigh, J. P. Vermeulen, L. J. J. de la Fonteyne, H. W. Verharen, J. J. Briedé, H. van Loveren, and W. H. de Jong. The effect of particle size on the cytotoxicity, inflammation, developmental toxicity and genotoxicity of silver nanoparticles. Biomaterials 32:9810–9817, 2011.
Rahman, M. F., J. Wang, T. A. Patterson, U. T. Saini, B. L. Robinson, G. D. Newport, R. C. Murdock, J. J. Schlager, S. M. Hussain, and S. F. Ali. Expression of genes related to oxidative stress in the mouse brain after exposure to silver-25 nanoparticles. Toxicol. Lett. 187:15–21, 2009.
Rai, M., K. Kon, A. Ingle, N. Duran, S. Galdiero, and M. Galdiero. Broad-spectrum bioactivities of silver nanoparticles: the emerging trends and future prospects. Appl. Microbiol. Biotechnol. 98:1951–1961, 2014.
Smith, J. N., D. G. Thomas, H. Jolley, V. K. Kodali, M. H. Littke, P. Munusamy, D. R. Baer, M. J. Gaffrey, B. D. Thrall, and J. G. Teeguarden. All that is silver is not toxic: silver ion and particle kinetics reveals the role of silver ion aging and dosimetry on the toxicity of silver nanoparticles. Part. Fibre Toxicol. 15:47, 2018.
Sohal, I. S., K. S. O’Fallon, P. Gaines, P. Demokritou, and D. Bello. Ingested engineered nanomaterials: state of science in nanotoxicity testing and future research needs. Part. Fibre Toxicol. 15:29, 2018.
Stevanović, M., B. Kovačević, J. Petković, M. Filipič, and D. Uskoković. Effect of poly-α, γ, l-glutamic acid as a capping agent on morphology and oxidative stress-dependent toxicity of silver nanoparticles. Int. J. Nanomed. 6:2837–2847, 2011.
Sun, J., Q. Zhang, Z. Wang, and B. Yan. Effects of nanotoxicity on female reproductivity and fetal development in animal models. Int. J. Mol. Sci. 14:9319–9337, 2013.
Theodorou, I. G., K. H. Müller, S. Chen, A. E. Goode, V. Yufit, M. P. Ryan, and A. E. Porter. Silver nanowire particle reactivity with human monocyte-derived macrophage cells: intracellular availability of silver governs their cytotoxicity. ACS Biomater. Sci. Eng. 3:2336–2347, 2017.
Thomas, D. G., J. N. Smith, B. D. Thrall, D. R. Baer, H. Jolley, P. Munusamy, V. Kodali, P. Demokritou, J. Cohen, and J. G. Teeguarden. ISD3: a particokinetic model for predicting the combined effects of particle sedimentation, diffusion and dissolution on cellular dosimetry for in vitro systems. Part. Fibre Toxicol. 15:6, 2018.
Tiwari, D. K., T. Jin, and J. Behari. Dose-dependent in-vivo toxicity assessment of silver nanoparticle in Wistar rats. Toxicol. Mech. Methods 21:13–24, 2011.
Tlili, A., J. Jabiol, R. Behra, C. Gil-Allué, and M. O. Gessner. Chronic exposure effects of silver nanoparticles on stream microbial decomposer communities and ecosystem functions. Environ. Sci. Technol. 51:2447–2455, 2017.
Ucciferri, N., E.-M. Collnot, B. K. Gaiser, A. Tirella, V. Stone, C. Domenici, C.-M. Lehr, and A. Ahluwalia. In vitro toxicological screening of nanoparticles on primary human endothelial cells and the role of flow in modulating cell response. Nanotoxicology 8:697–708, 2014.
Vibe, C. B., F. Fenaroli, D. Pires, S. R. Wilson, V. Bogoeva, R. Kalluru, M. Speth, E. Anes, G. Griffiths, and J. Hildahl. Thioridazine in PLGA nanoparticles reduces toxicity and improves rifampicin therapy against mycobacterial infection in zebrafish. Nanotoxicology 10:680–688, 2016.
Vinci, B., D. Cavallone, G. Vozzi, D. Mazzei, C. Domenici, M. Brunetto, and A. Ahluwalia. In vitro liver model using microfabricated scaffolds in a modular bioreactor. Biotechnol. J. 5:232–241, 2010.
Wang, Y., W. G. Aker, H. Hwang, C. G. Yedjou, H. Yu, and P. B. Tchounwou. A study of the mechanism of in vitro cytotoxicity of metal oxide nanoparticles using catfish primary hepatocytes and human HepG2 cells. Sci. Total Environ. 409:4753–4762, 2011.
Wang, X., Z. Ji, C. H. Chang, H. Zhang, M. Wang, Y.-P. Liao, S. Lin, H. Meng, R. Li, B. Sun, L. Van Winkle, K. E. Pinkerton, J. I. Zink, T. Xia, and A. E. Nel. Use of coated silver nanoparticles to understand the relationship of particle dissolution and bioavailability to cell and lung toxicological potential. Small 10:385–398, 2014.
Williams, K. M., K. Gokulan, C. E. Cerniglia, and S. Khare. Size and dose dependent effects of silver nanoparticle exposure on intestinal permeability in an in vitro model of the human gut epithelium. J. Nanobiotechnol. 14:62, 2016.
Wu, Y.-L., N. Putcha, K. W. Ng, D. T. Leong, C. T. Lim, S. C. J. Loo, and X. Chen. Biophysical responses upon the interaction of nanomaterials with cellular interfaces. Acc. Chem. Res. 46:782–791, 2013.
Wu, T., and M. Tang. Review of the effects of manufactured nanoparticles on mammalian target organs. J. Appl. Toxicol. 38:25–40, 2018.
Yang, C.-H., L.-S. Wang, S.-Y. Chen, M.-C. Huang, Y.-H. Li, Y.-C. Lin, P.-F. Chen, J.-F. Shaw, and K.-S. Huang. Microfluidic assisted synthesis of silver nanoparticle–chitosan composite microparticles for antibacterial applications. Int. J. Pharm. 510:493–500, 2016.
Zhang, Y. Cell toxicity mechanism and biomarker. Clin. Transl. Med. 7:34, 2018.
Acknowledgments
The work leading to this paper has received funding from the European Union’s H2020 research and innovation programme under Grant Agreement No. 760813 (PATROLS).
Author Contributions
DP analyzed the data. NU performed experiments. DP, GM and AA wrote the paper and interpreted the data. DP and AA edited and prepared the final layout. All authors gave final approval of the paper.
Disclosure
The authors have no relevant interests to disclose.
Data Availability
Data are available from the corresponding author upon reasonable request.
Author information
Authors and Affiliations
Corresponding author
Additional information
Associate Editor Merryn Tawhai oversaw the review of this article.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
The original version of this article was revised due to a retrospective Open Access order.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
This article is published under an open access license. Please check the 'Copyright Information' section either on this page or in the PDF for details of this license and what re-use is permitted. If your intended use exceeds what is permitted by the license or if you are unable to locate the licence and re-use information, please contact the Rights and Permissions team.
About this article
Cite this article
Poli, D., Mattei, G., Ucciferri, N. et al. An Integrated In Vitro–In Silico Approach for Silver Nanoparticle Dosimetry in Cell Cultures. Ann Biomed Eng 48, 1271–1280 (2020). https://doi.org/10.1007/s10439-020-02449-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10439-020-02449-5