Abstract
Since 2007, an ongoing African swine fever (ASF) pandemic has significantly impacted Eurasia. Extensive field evidence and modeling confirm the central role of wild boar in ASF epidemiology. To effectively control and eradicate the infection, rapid detection of the ASF virus (ASFV) is crucial for prompt intervention in areas of recent viral introduction or ongoing outbreaks. Environmental DNA (eDNA) is a cost-effective and non-invasive technique that has shown promising results in monitoring animal species and their pathogens and has the potential to be used for wildlife disease surveillance. In this study, we designed and evaluated an eDNA sampling method for highly turbid water and soil samples to detect ASFV and wild boar (Sus scrofa) DNA as a control using qPCR while ensuring biosafety measures and evaluating ASF epidemiology. To validate our method, we obtained samples from La Mandria Regional Park (LMRP) in northwestern Italy, an area free of ASFV, and spiked them in a laboratory setting with an ASFV’s synthetic DNA template. Our findings highlight the potential of eDNA monitoring as a reliable, rapid, and safe method for early detection of ASFV from soil and turbid water samples.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Since 2007, an ongoing African swine fever (ASF) pandemic has been affecting vast parts of Eurasia (Morelle et al. 2020; Sánchez-Cordón et al. 2018; Sauter-Louis et al. 2021), and the Eurasian wild boar (Sus scrofa) has been shown to be a relevant wildlife host for ASF virus (ASFV), contributing to infection maintenance and spread and representing a challenge for disease control (Iacolina et al. 2021).
Links between ASF in domestic pigs and in wild boar are strongly supported by both field evidence and modeling (EFSA (European Food Safety Authority) et al. 2023; EFSA (European Food Safety Authority) et al. 2022) even if the importance of wild boar in ASF maintenance and spread can vary among epidemiological situations and geographical settings (Dixon et al. 2020). In order to control and eradicate the infection, rapid detection of ASFV in an area is crucial for prompt intervention in both long-distance introduction points such as the outbreaks reported in Czech Republic and Belgium (Cukor et al. 2020); Linden et al. 2019) or in front-like situations as those of several other European countries or South Korea (Jo and Gortázar 2021; Lim et al. 2023; Sauter-Louis et al. 2021). Moreover, the evaluation of the persistence of ASFV in an area is crucial in evaluating ASF epidemiology and developing an eradication campaign for its control.
Environmental DNA (eDNA) is a cost-effective non-invasive technique that has been widely employed to monitor biodiversity in wild flora and fauna populations using water, soil, and air samples (Thomsen and Willerslev 2015). Although most eDNA studies are focused on aquatic populations, recent eDNA surveys have shown remarkably promising results in terrestrial mammals monitoring as well as their pathogens (Alfano et al. 2021; ENETWILD-consortium et al. 2022; Leempoel et al. 2020; Williams et al. 2017). Previously, this method has been applied to the detect feral pig in captivity from filtered water (Williams et al. 2017) and also ASFV in pig breeding facilities from eDNA isolated from dry sponges (3M) pre-hydrated with a specific surfactant liquid applied to animals’ skin and contaminated surfaces (Kosowska et al. 2021). While these studies demonstrate promising results in detecting both wild boar and ASFV, further adaptation of eDNA sampling and extraction methods is necessary for its application in the context of wildlife surveillance.
In order to evaluate the possibility of using eDNA-based methods for the detection of ASFV in the natural environment, we developed a simple eDNA method compatible with highly turbid water and soil samples that is in compliance with all the necessary biosafety issues.
Methods
Study area and sample collection
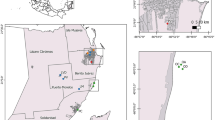
The study was conducted in La Mandria Regional Park (LMRP) near Turin in northwestern Italy, which encompasses a fenced area of approximately 2700 ha housing four ungulate species, including wild boar, red deer, roe deer, and fallow deer. The wild boar population density in this area was estimated at 15.26 individuals per square kilometer (Enetwild Consortium et al. 2023,). LMRP is ASF-free, and it is located in an area exempt from any restrictions for pathogen management and control.
Four separate mudholes (designated 1–4, respectively) were identified randomly and monitored using camera traps to confirm the local presence of wild boars and their use of the identified mudholes (Fig. 1). Field sampling activities were carried out in a single day in accordance with the park authority for both water and soil collection at each site. Specifically, seven L of turbid water were collected from each mudhole site using a diaphragm pump (connected to a 12V battery) and separately stored in water tanks. To prevent cross-contamination between sampling events, a closed circuit of one L bleach solution (20%) was run between each collection using the same tubes. Additionally, 5 mL volume of soil was collected from each site, using 50 mL falcon tubes. Buffer AVL™ (Qiagen GmbH, Hilden, Germany) (25 mL) was added to each tube. Buffer AVL is a viral lysis buffer that effectively deactivates ASFV even at high titer stock when mixed at a 1:5 v/v ratio for 10 min (McCleary et al. 2021). It has also been shown to preserve viral RNA for up to 35 days at 4 °C (Blow et al. 2008).
a Collection sites for water and soil samples at LMRP’s fenced-off area. b Sample collection and preparation workflow. c qPCR results for samples collected at LMRP and spiked in laboratory — results (dots represent every qPCR replicate value and horizontal red line depicts the replicate values’ mean) are reported as genome copy number/µL of eDNA for ASF virus (ASFV) and wild boar (WB). Line types demonstrate limit of quantification (LOQ) thresholds for both assays
Laboratory procedures and sample preparation
To validate the eDNA extraction method for detecting ASFV, a synthetic DNA fragment was designed (Supplementary materials A). The synthetic DNA fragment was constructed based on the sequence of the 2802/AL/2022 Italy isolate (Piedmont region) available on NCBI (ON108571.3) and was obtained from Macrogen Europe Milan Genome Center, Milan, Italy. Four different dilutions of the ASFV synthetic DNA were prepared, containing 6 \(\times {10}^{4}\), 6 \(\times {10}^{3}\), 6 \(\times {10}^{2}\), and 60 genome copies in a final volume of 10 µL of RNAase- and DNAase-free water and were designated as A–D, respectively. Each dilution was added to a separate water tank, generating samples subsequently labeled as WA1, WB2, WC3, and WD4, respectively, in correspondence to their sample type (W for water and S for soil), and synthetic DNA dilution concentration (A–D), and collection site in the LMRP (mudholes 1–). Soil samples were also labeled as SD1, SD2, SD3, and SD4 in the same manner mentioned before. The samples were kept at room temperature for 12 h before further processing.
Water samples were prepared filtering approximately 3 L of water using 0.1 µm Waterra filters (Argaly, Sainte-Helene Du Lac, France). Then, a minimum of 25 mL of AVL buffer was added to the filters’ capsule, which were closed and vortexed thoroughly. Waterra filters provide a long-surface (600 cm2) polyethersulfone membrane with a capacity of storing approximately 50 mL of liquid inside the filter capsule (Douchet et al. 2022). Therefore, a minimum of 25 mL buffer AVL was used to cover the entire membrane surface and ensure an adequate sample recovery. The liquid inside the filters was recovered and poured into a 50-mL falcon tube. RNA/DNAase-free water was added to all falcon tubes containing samples from filtered water or soil to reach a final volume of 50 mL.
eDNA extraction
The falcon tubes containing soil samples were vigorously shaken for 10 s, followed by a 10-s static phase to allow the sedimentation of heavy particles of soil. Subsequently, the supernatant was transferred into new 50 mL falcon tubes. All the tubes containing water or soil samples were centrifuged for 80 min at 4000 g and 18 °C. The supernatant was discarded, and 250 mg of sediment content was transferred into PowerBead Pro tubes. eDNA was purified using the DNeasy PowerSoil Pro Kit (Qiagen GmbH, Hilden, Germany). Negative controls were included in the extraction procedure.
qPCR assays for ASFV diagnosis and wild boar detection
The environmental diagnosis of ASFV from eDNA samples was performed using iTaq Universal SYBR Green Supermix (Bio-Rad Laboratories, CA, USA) and a primer set as described by Fernández-Pinero et al. 2013. For the environmental detection of wild boar DNA, TaqMan™ Universal PCR Master Mix (Life Technologies, CA, USA) and a primer set described by Williams et al. 2017 were used. All assays were performed in triplicate with a final volume of 25 µL, including 12.5 µL of TaqMan/iTaq SYBR Green master mix, 0.9 µM (2.25 µL) µM of each primer, 0.27 µM (0.675 µL) of TaqMan probe, and two µL of eDNA. The real-time thermocycling program involved a 10-min hold at 95 °C, followed by 45 cycles of 95 °C for 15 s and 60 °C for 1 min for both assays. A dissociation curve analysis was conducted at the end of the 45th cycle of the ASFV qPCR program. The absolute quantification method was used with six tenfold serial dilutions standard curves for both assays. The limit of quantification (LOQ) was 0.6 genome copies/µL for ASFV and 12 genome copies/µL for wild boar assays, corresponding to the cycle threshold at which the most diluted standard was quantified (\({R}^{2}=0.99\) for ASFV and \({R}^{2}=0.98\) for wild boar).
To determine a sample as positive, we established the minimum requirement of two replicates above the limit of quantification (LOQ) yielding positive results. In the case of ASFV analysis, we additionally considered the melting temperature analogy between the samples and standards.
Results and discussion
Following the above mentioned criteria, all synthetic DNA spiked water and soil samples tested positive for ASFV (Fig. 1). However, the difference in results from the water tank samples is not proportional to the initial spiked synthetic DNA inputs. This inconsistency might be attributed to the degradation of synthetic DNA in the muddy water between the time of synthetic DNA addition and the filtration process (12 h). On the other hand, soil samples exhibited more consistent results, possibly due to the synthetic DNA being added directly to the soil buffer AVL mixture. Extracted soil samples demonstrated higher eDNA concentrations; however, a lower wild boar eDNA copy number was detected compared to the water samples. This latter could suggest that filtration of water could better capture eDNA rather than soil samples that are representative of a small spot on soil surface. Interestingly, all water and soil samples resulted positive for the presence of wild boar eDNA except for soil sample SD4 which failed to meet LOQ in two out of three replicates (Fig. 1). It is worth to mention that based on camera trap photos, the presence of at least one wild boar was confirmed on the day before water/soil collection in all mudhole sites, except for mudhole 1 which last recorded the presence of one wild boar 5 days before sampling. This did not influence eDNA detection of wild boar confirming the good persistence of eDNA in the environment (Williams et al. 2018).
Considering our strict inclusion criteria, results from this study serve as a proof of concept for the effective application of eDNA techniques for the rapid and safe detection of AFSV when adapted for sampling from turbid water or soil and combined with viral DNA/RNA lysis and preservation buffer (such as buffer AVL). In this study, due to experimental procedures, water samples were transferred to laboratory, while, in a real-world sampling setting, water samples would be filtered and both sample type would be treated with AVL buffer in situ. To our knowledge, it is the first effort to co-detect ASFV and wild boar’s DNA directly from soil or water samples. The absence of domestic pigs in the study area is known and confirmed; however, further research is necessary to develop molecular tests capable of discerning wild boar’s DNA from domestic pigs. Additionally, following our workflow, eDNA extraction was performed using a common extraction kit and some shared sample preparation steps for two initial different sample matrices (water and soil). This has further accelerated laboratory procedure in a cost-effective manner.
The aim of the present work was to present technical protocols and measures to exploit eDNA detection from highly turbid water and soil samples for environmental surveillance of ASFV. A full-range field application of the presented methods is still required to fully evaluate its applicability both as early-detection method in disease-free areas and as a tool to monitor both the extent of the outbreak and the progression of the infection in ASF affected areas. Nevertheless, our findings represent a foundation for further exploration of eDNA application in wildlife health management. In particular, ASF control poses severe challenges related to wild boar management and to rapid and effective pathogen detection. The collection of accurate and comparable data is a prerequisite for informed disease management decisions. eDNA has the potential for becoming an effective diagnostic tool in wildlife disease management (Bass et al. 2023).
Availability of data and materials
The datasets analyzed during the current study are available to readers and could be accessed as supplementary information (Supplementary material B).
References
Alfano N, Dayaram A, Axtner J, Tsangaras K, Kampmann M-L, Mohamed A, Wong ST, Gilbert MTP, Wilting A, Greenwood AD (2021) Non-invasive surveys of mammalian viruses using environmental DNA. Methods Ecol Evol 12:1941–1952. https://doi.org/10.1111/2041-210X.13661
Bass D, Christison KW, Stentiford GD, Cook LSJ, Hartikainen H (2023) Environmental DNA/RNA for pathogen and parasite detection, surveillance, and ecology. Trends Parasitol 39:285–304. https://doi.org/10.1016/j.pt.2022.12.010
Blow JA, Mores CN, Dyer J, Dohm DJ (2008) Viral nucleic acid stabilization by RNA extraction reagent. J Virol Methods 150:41–44. https://doi.org/10.1016/j.jviromet.2008.02.003
Cukor J, Linda R, Václavek P, Šatrán P, Mahlerová K, Vacek Z, Kunca T, Havránek F (2020) Wild boar deathbed choice in relation to ASF: are there any differences between positive and negative carcasses? Prev Vet Med 177:104943. https://doi.org/10.1016/j.prevetmed.2020.104943
Dixon LK, Stahl K, Jori F, Vial L, Pfeiffer DU (2020) African swine fever epidemiology and control. Annu Rev Anim Biosci 8:221–246. https://doi.org/10.1146/annurev-animal-021419-083741
Douchet P, Boissier J, Mulero S, Ferté H, Doberva M, Allienne JF, Toulza E, Bethune K, Rey O (2022) Make visible the invisible: optimized development of an environmental DNA metabarcoding tool for the characterization of trematode parasitic communities. Environ DNA. https://doi.org/10.1002/edn3.273
EFSA (European Food Safety Authority), Baños JV, Boklund A, Gogin A, Gortázar C, Guberti V, Helyes G, Kantere M, Korytarova D, Linden A, Masiulis M, Miteva A, Neghirla I, Oļševskis E, Ostojic S, Petr S, Staubach C, Thulke H-H, Viltrop A, Wozniakowski G, Broglia A, Abrahantes Cortiñas J, Dhollander S, Mur L, Papanikolaou A, Van der Stede Y, Zancanaro G, Ståhl K (2022) Epidemiological analyses of African swine fever in the European Union. EFSA J 20:e07290. https://doi.org/10.2903/j.efsa.2022.7290
EFSA (European Food Safety Authority), Ståhl K, Boklund A, Podgórski T, Vergne T, Abrahantes JC, Papanikolaou A, Zancanaro G, Mur L (2023) Epidemiological analysis of African swine fever in the European Union during 2022. EFSA J 21:e08016. https://doi.org/10.2903/j.efsa.2023.8016
ENETWILD-consortium, Alves PC, Gavier-Widen D, Ferroglio E, Queirós J, Rafael M, Santos N, Silva T, Gonçalves C, Vada R, Zanet S, Smith G, Gethöffer F, Keuling O, Staubach C, Sauter-Louis C, Blanco J, Podgorski T, Larska M, Richomme C, Knauf S, Rijks JM, Pasetto C, Benatti F, Poncina M, Gómez A, Dups-Bergmann J, Neimanis A, Vicente J (2022) Literature review on the main existing structures and systematic/academic initiatives for surveillance in the EU for zoonoses in the environment and the methods for surveillance of pathogens in the environment. EFSA Support Publ 19:7792E. https://doi.org/10.2903/sp.efsa.2022.EN-7792
ENETWILD-consortium, Guerrasio T, Pelayo Acevedo P, Apollonio M, Arnon A, Barroqueiro C, Belova O, Berdión O, Blanco-Aguiar JA, Bijl H, Bleier N, Bučko J, Elena Bužan E, Carniato D, Carro F, Casaer J, Carvalho J, Csányi S, Lucía del Rio L, Aliaga HDV, Ertürk A, Escribano F, Duniš L, Fernández-Lopez J, Ferroglio E, Fonseca C, Gačić D, Gavashelishvili A, Giannakopoulos A, Gómez-Molina A, Gómez-Peris C, Gruychev G, Gutiérrez I, Veith Häberlein V, Hasan SM, Hillström L, Hoxha B, Iranzo M, Mihael Janječić M, Jansen P, Illanas S, Kashyap B, Keuling O, Laguna E, Lefranc H, Licoppe A, Liefting Y, Martínez-Carrasco C, Mrđenović D, Nezaj M, Xosé Pardavila X, Palencia P, Pereira G, Pereira P, Pinto N, Plhal R, Plis K, Podgórski T, Pokorny B, Preite L, Radonjic M, Marcus Rowcliffe M, Ruiz-Rodríguez C, Santos J, Rodríguez O, Scandura M, Sebastián M, Sereno J, Šestovic B, Shyti I, Somoza E, Soriguer R, de la Torre JS, Soyumert A, Šprem N, Stoyanov S, Smith GC, Sulce M, Torres RT, Trajçe A, Urbaitis G, Urbani N, Uguzashvili T, Vada R, Zanet S, Vicente J (2023) Wild ungulate density data generated by camera trapping in 37 European areas: first output of the European Observatory of Wildlife (EOW). EFSA Support Publ 20:7892E. https://doi.org/10.2903/sp.efsa.2023.EN-7892
Fernández-Pinero J, Gallardo C, Elizalde M, Robles A, Gómez C, Bishop R, Heath L, Couacy-Hymann E, Fasina FO, Pelayo V, Soler A, Arias M (2013) Molecular diagnosis of African swine fever by a new real-time PCR using universal probe library: UPL PCR for African swine fever virus. Transbound Emerg Dis 60:48–58. https://doi.org/10.1111/j.1865-1682.2012.01317.x
Iacolina L, Pernith ML, Bellini S, Chenais E, Jori F, Montoya M, Ståhl K, Gavier-Widén D (Eds.) (2021) Understanding and combatting African swine fever. Wageningen Academic Publishers. https://doi.org/10.3920/978-90-8686-910-7
Jo Y-S, Gortázar C (2021) African swine fever in wild boar: assessing interventions in South Korea. Transbound Emerg Dis 68:2878–2889. https://doi.org/10.1111/tbed.14106
Kosowska A, Barasona JA, Barroso-Arévalo S, Rivera B, Domínguez L, Sánchez-Vizcaíno JM (2021) A new method for sampling African swine fever virus genome and its inactivation in environmental samples. Sci Rep 11:21560. https://doi.org/10.1038/s41598-021-00552-8
Leempoel K, Hebert T, Hadly EA (2020) A comparison of eDNA to camera trapping for assessment of terrestrial mammal diversity. Proc Royal Soc B Biol Sci 287:20192353. https://doi.org/10.1098/rspb.2019.2353
Lim J-S, Andraud M, Kim E, Vergne T (2023) Three years of African swine fever in South Korea (2019–2021): a scoping review of epidemiological understanding. Transbound Emerg Dis 2023:e4686980. https://doi.org/10.1155/2023/4686980
Linden A, Licoppe A, Volpe R, Paternostre J, Lesenfants C, Cassart D, Garigliany M, Tignon M, van den Berg T, Desmecht D, Cay AB (2019) Summer 2018: African swine fever virus hits north-western Europe. Transbound Emerg Dis 66:54–55. https://doi.org/10.1111/tbed.13047
McCleary S, McCarthy RR, Strong R, Edwards J, Crooke H (2021) Inactivation of African swine fever virus by reagents commonly used in containment laboratories. J Virol Methods 295:114203. https://doi.org/10.1016/j.jviromet.2021.114203
Morelle K, Bubnicki J, Churski M, Gryz J, Podgórski T, Kuijper DPJ (2020) Disease-induced mortality outweighs hunting in causing wild boar population crash after African swine fever outbreak. Front Vet Sci 7:378. https://doi.org/10.3389/fvets.2020.00378
Sánchez-Cordón PJ, Montoya M, Reis AL, Dixon LK (2018) African swine fever: a re-emerging viral disease threatening the global pig industry. Vet J 233:41–48. https://doi.org/10.1016/j.tvjl.2017.12.025
Sauter-Louis C, Conraths FJ, Probst C, Blohm U, Schulz K, Sehl J, Fischer M, Forth JH, Zani L, Depner K, Mettenleiter TC, Beer M, Blome S (2021) African swine fever in wild boar in Europe—a review. Viruses 13:1717. https://doi.org/10.3390/v13091717
Thomsen PF, Willerslev E (2015) Environmental DNA – an emerging tool in conservation for monitoring past and present biodiversity. Biological Conservation, Special Issue: Environmental DNA: a Powerful New Tool for Biological Conservation 183:4–18. https://doi.org/10.1016/j.biocon.2014.11.019
Williams KE, Huyvaert KP, Piaggio AJ (2017) Clearing muddied waters: capture of environmental DNA from turbid waters. PLoS One 12:e0179282. https://doi.org/10.1371/journal.pone.0179282
Williams KE, Huyvaert KP, Vercauteren KC, Davis AJ, Piaggio AJ (2018) Detection and persistence of environmental DNA from an invasive, terrestrial mammal. Ecol Evol 8:688–695. https://doi.org/10.1002/ece3.3698
Funding
This work has been part of Enetwild activities. Enetwild is a consortium funded by EFSA through the OC/EFSA/BIOHAW/2022/01 call.
Author information
Authors and Affiliations
Contributions
A.R.V., S.Z., P.B.S., and E.F. contributed to conceptualization and methodology. A.R.V., S.Z., P.B.S., F.O., R.V., F.B., and P.B.M. contributed to investigation and analysis. A.R.V. contributed to writing the manuscript’s original draft. S.Z. and E.F. contributed to the supervision, review, and editing of the manuscript. All authors reviewed the manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
No approval of research ethics committees was required to accomplish the goals of this study.
Competing interests
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Varzandi, A.R., Zanet, S., Seano, P.B. et al. Detection of African swine fever virus and wild boar eDNA in soil and turbid water samples: towards environmental surveillance. Eur J Wildl Res 70, 4 (2024). https://doi.org/10.1007/s10344-023-01758-z
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s10344-023-01758-z