Abstract
The Department of Agricultural, Food, Environmental and Animal Sciences (DI4A) at the University of Udine, in collaboration with Friuli Venezia Giulia regional authorities, within northeastern Italy, set up a wildlife monitoring and surveillance regional network, named InfoFaunaFVG. Here we describe the development and application of this data repository system based on a novel progressive web application, and report the data gathered in the first two and a half years of its use. InfoFaunaFVG is made of a Web Database and an integrated WebGIS system. In particular, the following open source softwares are used: Apache HTTP Server, Oracle MySQL, Symfony, Apache Tomcat, GeoServer, OpenLayers. The web app can be accessed from any web browser or by installing the progressive web application in the desktop or mobile devices. In short, operating from November 2019, InfoFaunaFVG currently (April 2022) contains a total of 40,175 records, from 300 different users, from 16 institutions. Among all species recorded, mammals were 40% (16,018) of the total, whereas avian species represented 59% (23,741), and others (reptiles and amphibians) 1% (416), respectively. Two hundred twenty-six different species (175 avian and 51 mammals) were recorded. Details about causes of death and live animal rescue were reported. To date, InfoFaunaFVG has proven to be a successful wildlife data repository system providing high quality consistent, accurate and traceable data. These had a considerable impact on regional wildlife governance. In the authors’ knowledge, InfoFaunaFVG is the first example described in literature of such a progressive web application, coordinated on an institutional level, and not based on voluntary-citizen observations. InfoFaunaFVG has the potential to become the largest wildlife monitoring and surveillance data repository system on a national level.
Similar content being viewed by others
Explore related subjects
Find the latest articles, discoveries, and news in related topics.Avoid common mistakes on your manuscript.
Introduction
Importance of wildlife monitoring and surveillance activities is widely recognized in a global context, where the human–wildlife interactions have increased dramatically, carrying economic, health and environmental risks (OIE 2010).
The increased percentage of private or public metropolitan areas in developed countries has had a profound impact both on ecological landscape and balance between wildlife and humans’ activity (Mustățea and Patru-Stupariu 2021). Although the extention of urban areas into rural zones has been traditionally considered the main problem, in more recent years, the overabundant wildlife populations are also coming into contact with settled areas, through source–sink dynamics, especially towards human food sources (Messmer 2000; Konig et al. 2020). The growing interaction and human-wildlife closer contact can certainly generate conflicts; this leads to reciprocal negative outcomes, such as crops and residential or household damages, vehicle collisions, incidental dangerous human-wildlife encounters, introduction of diseases, social and economic costs associated with the elimination or restrictions placed on traditional wildlife management (Soulsbury and White 2015; Pooley 2021). Furthermore, public perception of human-wildlife interaction is becoming more and more complex (Frank et al., 2019). This is not solely based on anthropocentric views, but conveyed towards a more sustainable state of coexistence (Pooley 2021). On reflection, the strive towards a balanced coexistence has had an impact on wildlife governance, leading institutions to take into account investigations and monitoring of biodiversity, when making decisions about wildlife conservation. Governance models are now required to implement sustainable management plans, and wildlife policy instruments through the use of ecological knowledge, and organized comprehensive data acquisition (Corona et al. 2011; Waetje and Shilling 2017; Carter et al. 2020).
The use of information system databases has become an essential tool in a context of widespread use of portable devices, and online internet access. The importance of remote access resides in the possibility of real-time input, and retrieval of data without spatial and temporal restrictions (Lahoz-Monfort and Magrath 2021). Several information systems for wildlife monitoring through web and mobile applications have been reported and described in literature (Olson et al. 2014; Shilling et al. 2015; Waetje and Shilling 2017, Marvin et al. 2016; Duffy 2020, Gabriel and Ravindran 2021). Particular focus has been placed on systems to report wildlife near roadways and roadkills. Idaho Fish Wildlife Information System https://idfg.idaho.gov/species/roadkill), I-90 Wildlife watch (https://i90wildlifewatch.org/) in the Snoqualmie Pass region of Washington, the Belgian “Animals under wheels” (http://waarnemingen.be) and the “Taiwan Roadkill Observation Network” (https://roadkill.tw) are examples of large volume databases, which have been collecting observations from both institutional sources and on a voluntary basis. Spatial Monitoring and Reporting Tool (SMART) is a new technology that can be used to increase the temporal and spatial scale and dimensionality of ecological and conservation observations and research. This open source software has been developed through collaboration among conservation agencies, and organizations to improve site-based conservation area effectiveness. SMART allows Android and Windows Mobile–enabled smartphone, or tablet, uploading and managing data to be viewed by patrol teams (Gabriel et al. 2021).
Similar examples of information systems for wildlife monitoring on an Italian national level, reported by institutional observation centers, have been attempted, but with limited longevity.
Often, these databases originated spontaneously from social media groups to develop in more structured projects, gradually established through discussions and consensus among members.
Since volunteers-based databases have become more common, the need for sophisticated tools and wider scope have become necessary (Bonney et al. 2009). Thus, not only roadkills-focused projects have developed through the years, but also more taxonomically focused databases such as eBird (Sullivan et al. 2014; ebird.org/home), and broader biodiversity repositories of data, like iNaturalist (Wittmann et al. 2019; inaturalist.org). iMammalia app represents a further example of this tendency (mammalnet.com): this system, has the goal to encourage recording of mammals, especially wild boars, in the natural environment by using smartphones (ENETWILD-consortium et al. 2022). Essential challenges of these systems are the lack of a commonly used set of rules for data collection, visualization and management, and most importantly the quality and accuracy of citizen-based observations (Waetjen and Shilling 2017). To overcome these challenges, most of these projects adopted a standard set of rules for reporting wildlife observations such as the Darwin Core standards and the use of open repositories like the Global Biodiversity Information Facility (GBIF), to share their data publicly.
Within the Italian context, national Italian legislation (Legislation n.157/1992) regulates the definition of wildlife, and delegates the responsability of its governance to regional and provincial authorities. The same legislation regulates selective culling for mammals and birds.
In 2017, the Autonomous Friuli Venezia Giulia (FVG) Region and the Department of Agricultural, Food and Animal Sciences of the University of Udine (UniUd-DI4A) signed a collaboration agreement (regional legislation 31/2017) to carry out scientific work regarding wildlife recovery and surveillance. The wildlife vertebrates included within this agreement were only debilitated and/or compromised or dead or selectively culled animals.
The scientific activity in the collaboration agreement included both support and consulting for the Regional Forestry Service and the Wildlife Rescue Centers (CRASs), and the design of a novel information-database system to organize data gathered during the course of the project. The DI4A developed a progressive web application (PWA), characterized by a WebDatabase and a WebGIS system, through the utilization of different open source software, and named it InfoFaunaFVG. The objectives of the present study are to describe the development and application of InfoFaunaFVG PWA, and to illustrate its recent use for wildlife surveillance on a regional level.
Material and methods
Development of the information system: InfoFaunaFVG
The information (IT) structure InfoFaunaFVG is essentially based on a server “Debian GNU/Linux” (https://www.debian.org/), a web server “Apache HTTP Server’’ (https://httpd.apache.org/), a relational database management system “Oracle MySQL’’ (https://www.mysql.com/), a PHP framework for web application “Symfony ™” (https://symfony.com/), a servlet container “Apache Tomcat” (https://tomcat.apache.org/), an open source server for sharing geospatial data “GeoServer” (https://geoserver.org/), an high-performance and feature-packed library for displaying interactive maps on the web “OpenLayers” (https://openlayers.org/) and finally an Open Source WebGIS framework called “MapStore” (https://mapstore.readthedocs.io/). A schematic representation of the Web Application Architecture is illustrated in Fig. 1.
“Oracle MySQL” is the chosen relational database management system (RDBMS). This is one of the most used and widespread Database Management Systems, thanks to its solidity, scalability and the potential data management from different web applications.
The PHP framework “Symfony” allows to develop web information systems based on a classic three levels MVC pattern: the model, the view and the controller. The MVC architecture separates the model and the view and is designed to optimize modularity, code reuse, scalability and to be easy to maintain. “Symfony” makes possible the integration of several bundles in order to extend its functions. As an example, FOSUserBundle is able to set up a very flexible and detailed system for client’s management.
Geolocalisation features were managed through the “Apache Tomcat” servlet container and its relative web server “GeoServer”. This server allows both direct connection to the database and to manage topographic data, particularly Web Feature Service—Transactional (WFS-T) and Web Map Service (WMS). These services have been integrated inside the web app through OpenLayers (a Geospatial JavaScript Library).
Mapstore has been used in order to seek, query and visualize spatial data as it offers all the features of a “Geo-portal” system, and it makes all data published through WFS and WMS available on the web within a flexible and personalized interface.
Furthermore, the georeferencing accuracy is recorded automatically by the GPS device or can be estimated by the user (under the field “coordinateUncertaintyInMetersProperty” — see Fig. 6, Appendix).
To improve user experience, especially when using the mobile devices, the web app has been designed to be a hybrid between a web and native mobile application; it is fully responsive, and the content visualization is optimized depending upon the type of users’ device. InfoFaunaFVG can be installed in any mobile device through PWA principles. A progressive web application (PWA) is a browser-based application that has become an alternative to a native mobile app, built using common web technologies including HTML, CSS, and JavaScript. While native apps are created to run on mobile devices, PWAs are designed to run inside a web browser. They are intended to work on any platform with a standard’s compliant browser, including desktop and mobile devices. Developers can simply publish the web application online, ensure that it meets baseline installation requirements, and users will be able to add the application to their home screen.
Traditional functions of management software were developed for this project: create, read, update, and delete (CRUD) operations, perform batch operations, create dynamic reports and filters and export data in Open Document format through dynamic reports.
Access to InfoFaunaFVG
The access to the web application is possible, upon authentication, through a web browser or by installing the PWA on the computer or mobile devices. In order to define detailed user profiles, and fulfill General Data Protection Regulation (GDPR), different levels of use are assigned to each user, depending on their operational level.
The main users’ profiles and a description of their operational level are listed hierarchically here:
-
1.
Super administrator: able to visualize, enter and modify any type of data input (University of Udine and Friuli Venezia Giulia Regional authorities).
-
2.
Local Administrator: able to visualize, enter and modify any type of data input within the territory of competence (Inspectorates).
-
3.
Local user: able to visualize all data within the territory of competence, to enter and modify its own data input (National forestry services, Wildlife Rescue Centers (“Centri di recupero di Animali Selvatici’’- CRAS), Italian health authority and research organization for animal health and food safety (Istituto Zooprofilattico- IZS).
-
4.
Basic user: able to visualize, enter and modify only its own data input (third party contractors involved in animal rescue activity recognized by regional authorities).
InfoFaunaFVG has built in both a notification system, to send communications to every user at any level, and a log system, able to trace all the data input changes.
Workflow diagram [Entity Relationship Diagram (ERD)]
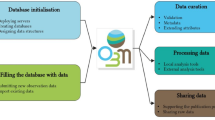
The classification of the information within the database is based on a relational model that organizes data through tables, rows and columns, as shown in Workflow diagram Fig. 2.
Entity-Relationship (ER) diagram: graphical representation illustrating the simplified database structure through entities (light blue), relationships (yellow) and attributes (light green). Roles and permissions for observers are defined by the site administrators and described in detail in the manuscript
To be able to use a common language for possible future data sharing, InfoFaunaFVG uses classes and terms defined by Darwin Core (Wieczorek et al. 2012). The Appendix describes all the occurrence datasets provided by InfoFaunaFVG.
The data input is entered into the system by using the “report section” (“scheda segnalazione”), which contains a detailed set of information including an unique identification code (occurrenceID/ recordNumber), type of individual record (dynamicProperties), date and time of the event (eventDate and eventTime), details of the user reporting the case (recordedBy), geolocalization (decimalLatitude, decimalLongitude, geodeticDatum), any video or photographic material, and documentation available through attachments (associatedMedia). The localization data can be recorded directly through mobile device GPS or by the WebGIS CRUD tools or specific geographic coordinates or addresses (Geocoding API) (see Appendix – Fig. 6 for more details).
At this stage of the project, basisOfRecords are all “human observations”. Further datasets are reported and described in details in the Appendix.
The “intervention request” (“Richiesta Intervento”) is the link between the “report section” and “intervention section”. Depending upon the type of intervention requested, the “intervention section” leads to the “rescue center section” (“scheda CRAS”), if live animals are rescued, otherwise the “necropsy section” can be accessed when protected dead animals are found.
Outcome of dead or selectively culled animals, not undergoing necropsy, can be entered directly in the “Result” section .
The “rescue center section” includes details about the characteristics of the center and hospitalization details (clinical data, veterinary assessments and treatment and follow up details). The “necropsy section” includes details about post-mortem examination, samples taken and their results.
Finally, the “result section” (“esito”) ends the process regardless of the outcome, and allows the user to understand the results of all the actions taken since the animal was found.
This type of workflow (illustrated in Fig. 2) can optimize traceability of input data from “report section” to “result section” in any direction. Furthermore, its solid structure and standard parameters through which data are inserted, allow the user to extrapolate information efficiently in a short period of time.
Appendix summarizes InfoFaunaFVG structure and scopes, and explains step by step the workflow by showing the user interfaces.
Results
In the present study, we analyzed all records from the InfoFaunaFVG database relative to wildlife observed on a regional level (Friuli Venezia Giulia), from November 2019 to April 2022. Main data are illustrated in Table n.1. A total of 40,175 records through the “report section” were entered as “human observation”. Among them, 32,176 are georeferenced data and a total 6014 were entered with attachments such as pictures and or videos. Georeferenced data and their specific distribution are illustrated in Fig. 3. The total record input inside InfoFaunaFVG was 4013 in 2019 (from November), 15,397 in 2020, 17,138 in 2021 and 3627 in 2022 (up to April), respectively. The users involved in the data input were 28 different national forestry centers (21,185 records), 6 different rescue centers (5681), 4 different third-party companies involved in wildlife rescue activity (9634 input data), Friuli Venezia Giulia (Region) 3555. IZS visualized 468 records from April 2021.
Distribution of georeferenced data extrapolated from InfoFaunaFVG within Friuli Venezia Giulia Region (red area in the center). Selectively culled animals (a, red dots), animals found dead (b, black dots) and rescued animals (c, green dots) are represented here. Roadkills and animals found dead in artificial water canals are reported within the bottom right picture (d, orange and blue dots respectively)
Among all species recorded, mammals were 40% (16,018), whereas avians represented 59% (23,741) and others (Reptiles and amphibians) were 1% (416).
The wildlife species reported to be selectively culled were 42% (16,732), 17% (7829) were found dead and 39% (15,614) were rescued alive.
The records of animals found dead were 1035 in 2019, 2252 in 2020, 3282 in 2021 and 1260 in 2022, respectively. Live animals rescued were 1412 in 2019, 5826 in 2020, 7158 in 2021 and 1218 in 2022, respectively. Animals selectively culled were 1576 in 2019, 7309 in 2020, 6690 in 2021 and 1157 in 2022.
Figure 4 illustrates the details of records per month. Live wildlife animals were rescued mostly during April to August months with an evident peak in June (1230 in 2020 and 1639 in 2021), and lowest number in March 2020 (136) and February 171 (2021); the mean value of animals rescued per month was 485 in 2020 and 596 in 2021. The mean values of dead animals per month are 187 for 2020, and 273 for 2021.
The “rescue center” section records were 12,848 in total; from March 2022, 71 hospital sheets with details of therapies were registered.
Animals reintroduced in a natural environment were 61% of the total of records from rescue centers. Animals died at rescue centers in 31.5% of cases, whereas euthanasia was performed in 5% of cases. Animals still present in CRASs centers were 2.5% of the total.
From November 2020, a total of 104 necropsies were performed by veterinarians at the University of Udine in collaboration with IZS institute. The species subjected to post-mortem examination are reported in Table 1. Roadkills were the cause of death in about 92% of these cases, whereas poisoning was determined to be the most likely cause of death in about 5% of cases.
A detailed description of necroscopies and related post-mortem analysis is outside the scope of the present manuscript, and will be the subject of further studies.
The mammals belonged to 51 species. Coypu (Myocastor coypus) (4574, 27,8%), Roe deer (Capreolus capreolus) (3953, 24,6%), Wild boar (Sus scrofa) (1731, 10,7%), Red foxes (Vulpes vulpes) (1363, 8,4%) and hedgehog (Erinaceus europaeus and Erinaceus concolor) (1018, 6.3%) are the most frequent ones. Among the dead mammals recorded, Roe deer (Capreolus capreolus) (3953, 24.6%), Red foxes (Vulpes vulpes) (1361, 8.4%) and hedgehog (Erinaceus sp.) (1018, 6.3%) were the most common ones, whereas Coypu (4471, 81.5%), Wild boar (Sus scrofa) (945, 17.2%) and European hare (Lepus europaeus Pallas) (64, 1.1%) were the most commonly culled mammals.
Rescued live mammals were 3707 in total: 743 (20%) were in poor health general conditions, 515 (13.8%) were rescued for presumptive lack of parental care, 504 (13.5%) were involved in road traffic accidents, 295 (7.9%) were wounded, and trapped animals were 226 (6%).
Regarding avian species, 175 different species were recorded in total, being Feral pigeons (Columba livia domestica) (12,783, 53.8%), Yellow-legged gull (Larus michaellis) (1326, 10.5%), Common blackbird (Turdus merula) (1110, 8.8%), Common swift (Apus apus) (1084, 8.6%) and Hooded crow (Corvus cornix) (773 5.8%) the most common 5 ones (representing 87.5% of the total). Species more commonly found dead were Feral pigeons (Columba livia domestica) with 222 cases (28%), Yellow-legged gull (Larus michaellis) (82 cases, 10.8%), Common pheasant (Phasianus colchicus) (55 cases, 6.9%), Common buzzard (Buteo buteo) (34 cases, 4.2%) and Mallard (Anas platyrhynchos) (34 cases, 4.2%).
Feral pigeons (Columba livia domestica) (11,076, 98.4%), Cormorant (Phalacrocorax carbo) (76, 0.67%) and Hooded crow (Corvus cornix) (39, 0.34%) were the avian species had undergone most commonly to selective culling.
Reasons for rescuing live avian species were recorded in 12,658 cases. In these cases, where appropriate, more than one suspected reason could be recorded. Presumptive lack of parental care was entered in 1976 (15.6%) cases, poor health status in 1886 (14.8%) of cases, wounded or traumatized birds were 1649 (13%), suspected nest falling was recorded in 1599 (12.6%) cases, being unable to fly in 1276 (10%) cases and trapped in 697 (5.5%) of cases, respectively.
Apart from wildlife selective culling (11,249 avian and 5483 mammals), roadkills were the main cause of death both for mammals (4512) and avian species (226). Animals found dead for apparent unknown causes were 1094 in mammals, approximately 16.1% of the total of deaths and 225 in avian species (36.8% of the total deaths). Predation accounted for 207 cases in mammals and 51 in avian species. Mammals found dead in waterways and artificial water canals were 245. Railway accidents were the cause of 63 deaths in mammalian wildlife (0.9%) and only 1 case in avian species (0.1%).
Non-native species recorded were the Coypu with 4471 cases selectively culled, 86 found dead and 17 cases rescued alive, and the Eastern gray squirrel (Sciurus carolinensis) with 5 cases rescued alive and 1 case selectively culled. Euthanasia was performed in CRAS centers on all non-native species rescued.
Finally, 198 (0.49% of all records) taxonomical inaccuracies, which were both found and amended by the coordination center (UniUd- DI4A) at “rescue center section” and “necropsy section” levels. Among these, 186 (93.9%) cases regarded avian species and 12 (6.1%) were mammals.
Discussion
The development and application of a PWA for wildlife management and surveillance on a regional level named InfoFaunaFVG is described here. From the results of the present study, it seems evident that, to date (April 2022), this project has proven to be successful gathering about 40 k records; the database has been regularly used by approximately 300 different users, including veterinarians, forestry guards, veterinary technicians and specialized personnel in animal rescue belonging to 16 different institutions. The users were all specifically trained to use the PWA proficiently, and they were supported by a central coordination/patrol team at the University of Udine. In contrast with volunteer/citizen-based projects, InfoFaunaFVG was designed to be used solely on an institutional level. To the authors’ knowledge, it is the first example of a European standardized institutional wildlife data repository system reported in literature. At the present stage of the project, only data from debilitated and/or compromised or dead or selectively culled animals were included.
The idea of developing a PWA was born out of the need of using web-based functions and, at the same time, ensuring the most intuitive and smooth use in every device. From a technical point of view, InfoFaunaFVG has proven to be compatible with all device systems where it was installed and used, even though iOS does not support native push notifications and background sync for offline use. Most common requests for technical support were regarding accuracy of georeferenced data, and about adapting system functionalities to the need of single institutions, such as printing of personalized reports and labels.
The common challenges that wildlife data repository systems face regard data accuracy, consistency and traceability (Olson et al. 2014; Shilling et al. 2015; Ratnieks et al. 2016; Waetje and Shilling 2017; Duffy 2020).
Only in limited studies, volunteer-based projects have shown to produce comparable quality data to institutional researchers (Ryder et al. 2010; Kremen et al. 2011; Ratnieks et al. 2016, Waetje and Shilling 2017). When compared with other volunteer-based systems, where the accuracy of data are evaluated by the same users or in a subset of photographs (Waetje and Shilling 2017), InfoFaunaFVG seems superior in terms of observations accuracy, especially from a taxonomical point of view. Since November 2019, 226 wildlife animals have been reported at a species level with an accuracy of 99.51%. Most taxonomical mistakes involved the recognition of avian species, which seem to present the most variability (175 versus 51 mammals species) and challenges (especially passerines that made 67% of total inaccuracies). Among mammals, Chiroptera represented the most common source of taxonomical inaccuracies (12 out of 19). We believe that InfoFaunaFVG has overcome the problem of data accuracy by its intrinsic section-based structure, and the fact that users were all trained wildlife professionals. Specific training was aimed to make all users proficient with the PWA use and its terminology within Darwin core standards: this has been certainly labor intensive and time consuming, considering the large number of professionals involved. In the future, the use of remote training materials, such as video and pre-recorded remote presentations, would be helpful, especially when several training sessions are needed and new functions are added to the PWA. The section-based structure of InfoFaunaFVG made it possible to trace, control and confirm records at different levels. As an example, once report and intervention sections are entered, these must flow through a rescue center section, necropsy section or directly to the result section, where the details of each animal are verified and confirmed (Fig. 2).
Compared to recent database systems, where a maximum of 13% of observations contained photographs (Waetje and Shilling 2017), here about 15% of data were combined with pictures and videos. The amount of supplementary material increased considerably from November 2019 to 2022 thanks to training and increased users' confidence with InfoFaunaFVG’s functions. However, this will certainly need improvement in the future.
Localization accuracy was ensured by the georeferencing system, which located 80% of records through geographic coordinates. InfoFaunaFVG used a combination of GPS-enabled smartphones and confirmation using online maps as recently suggested in literature (Waetje and Shilling 2017); unfortunately, even though GPS satellites broadcast their signals in space with a certain accuracy, the quality of the georeferentiation depends on additional factors, including atmospheric conditions, satellite geometry, receiver design features/quality and signal blockage. In the remaining 20% of data, the municipal area was identified.
Data consistency and traceability were possible through regular data quality checks performed by the centralized coordination/patrol team at the University of Udine. Consistency is ensured using parameterized data through drop-down menus and the use of Darwin Core terms and classes; this can help to avoid redundancy of observations and inappropriate terminology. Traceability appears to be particularly important in cases of wildlife affected by dangerous diseases with serious zoonotic or epidemic potential (OIE 2010). Considering that wildlife monitoring across broad taxonomies is seldom carried out at the European and US scale (Waetje and Shilling 2017), InfoFaunaFVG can be a powerful tool to prevent and detect emerging disease in their early stages and survey and monitor existing ones. This can promote timely decisions and responses from governance structures. In our case, traceability is ensured, both for live rescued and dead animals, through all phases of intervention. This is true from rescuing to rehabilitation and reintroduction to natural environment, and for necropsy and related diagnostic analysis. When using InfoFaunaFVG, traceability is always possible anytime as every section is linked one to another so can be viewed retrospectively.
Another quality of InfoFaunaFVG is to enable institutions to estimate costs related to wildlife management. The majority of the studies in literature calculate losses from carnivore predation on livestock or damage to crops, while a few consider dispersal of diseases and the cost of traffic collisions (Gren et al. 2018). The database described here has the potential to bring all these factors together, and produce realistic and complex estimations of the total costs related to human-wildlife coexistence. Eventually, monthly data of animal deaths and rescues can help regional institutions to predict and focus their efforts in particular times of the year (for example June months in FVG region for rescuing alive animals).
Records from dead animals highlight the importance of roadkills as a main cause of death in wildlife species. This is a common finding and has been extensively studied and reported in literature (Olson et al. 2014; Shilling et al. 2015; Waetje and Shilling 2017). On the other hand, animals found dead in artificial water canals are rarely, but increasingly reported (Peris and Morales 2004). The data reported here may lead to further specific studies, and will help in developing mitigation strategies both for wildlife-vehicle collision hotspots and for impact of artificial hydraulic infrastructure on terrestrial mammals. Furthermore, 92% of necropsies on protected species (recognized by Italian legislation 157/1992) were also compatible with roadkills.
The essential role of rescue centers is evident from the results presented in this study. Records from CRAS were 31.2% of the total. The rescue center sections were entered for avian species in 80% of admissions and for mammals in 18% of cases. These data seem similar to what is observed in literature (Kirkwood 2003). It is evident here that CRASs play key roles in animal welfare, supporting law enforcement, raising awareness, and nature conservation, and biodiversity. Thanks to the support of InfoFaunaFVG, individual animals’ history can be followed from rescue to possible reintroduction to the natural habitat (in 61% of the cases). Hospital records were 71, but this section was implemented only from March 2022, thus has the potential to collect a much larger number of data in the next future. The authors did not describe details of these data here, as it is outside the scope of the present manuscript, and will be the subject of future studies. The additional sections regarding hospitalisation, clinical data and treatment will allow veterinarians to improve the quality of the triage, and have a comprehensive knowledge about each wildlife patient, knowing its history from rescue activity (Mullineaux 2014).
Furthermore, InfoFaunaFVG made it possible to recognize non-native species (for example Coypu and Eastern gray squirrel), which can be extremely important to understand regional biodiversity and potential emerging risks for native species survival.
In conclusion, during the first 2 years and a half of its use, InfoFaunaFVG allowed the achievement of several objectives including the recording of consistent, accurate and traceable data regarding wildlife, the optimization of regional investment, and resources and improvement of regional wildlife knowledge. In this regard, the data can be used to define trends and species distributions, non-native species invasions, novel animal behavioral patterns and the recognition of ‘animal sentinels’, which can be used for monitoring of environmental contaminants and diseases (Schwartz et al. 2020). Here we describe data, which can be used for longterm studies, and can have an impact on specific regional or even national projects, such as improvement of transportation systems to reduce impacts of wildlife and culling regional programmes, to optimize public resources. Given the results of the first 2 years of its use, we conclude that InfoFaunaFVG has the potential to become the largest data repository for wildlife monitoring and surveillance in Italy. The application of such integrated information systems on a larger scale at a national level is auspicable to best coordinate and design sustainable management plans and policy instruments to improve human-wildlife coexistence.
Data availability
The datasets generated during the current study are available from the corresponding author upon reasonable request.
Change history
26 June 2023
A Correction to this paper has been published: https://doi.org/10.1007/s10344-023-01703-0
References
Bonney R, Cooper CB, Dickinson J, Kelling S, Phillips T, Rosenberg KV et al (2009) Citizen science: a developing tool for expanding science knowledge and scientific literacy. Bioscience 59:977–984. https://doi.org/10.1525/bio.2009.59.11.9
Carter N, Williamson MA, Gilbert S, Lischka SA, Prugh LR, Lawler JJ, Metcalf LA, Jacob AL, Beltrán BJ, Castro AJ et al (2020) Integrated spatial analysis for human-wildlife coexistence in the American West. Environ Res Lett 15:021001
Corona P, Chirici G, McRoberts RE, Winter S, Barbarti A (2011) Contribution of large-scale forest inventories to biodiversity assessment and monitoring. For Ecol Manage 262:2061–2069. https://doi.org/10.1016/j.foreco.2011.08.044
Duffy MM (2020) Wildlife rehabilitation datasets as an underutilized resource to understand avian threats, mortality, and mitigation opportunities. Electronic Theses and Dissertations 3301. https://digitalcommons.library.umaine.edu/etd/3301
ENETWILD-consortium, Blanco-Aguiar, J. A., Smith, G. C., & Vicente, J. (2022) Update on the development of an app (adaptation of iMammalia) for early reporting of wild boar carcasses by a warning system. EFSA Supporting Publications 19(2):7192E
Frank B, Glikman JA, Marchini S (2019) Human–wildlife interactions: turning conflict into coexistence. Cambridge University Press, Cambridge, United Kingdom
Gabriel MJ, Ravindran KM (2021) Spatial Monitoring and Reporting Tool (SMART) for innovative forest governance: Insights from Asia-Pacific countries. Asia-Pacific Forest Sector Outlook: Innovative forestry for a sustainable future. Youth contributions from Asia and the Pacific 49
Gren IM, Häggmark-Svensson T, Elofsson K et al (2018) Economics of wildlife management—an overview. Eur J Wildl Res 64:22. https://doi.org/10.1007/s10344-018-1180-3
Kirkwood JK (2003) Introduction: wildlife casualties and the veterinary surgeon. In: Mullineaux E, Best D, Cooper JE (eds) BSAVA Manual of Wildlife Casualties. BSAVA Publications, Gloucester, MA, USA, pp 1–5
König HJ, Kiffner C, Kramer-Schadt S, Fürst C, Keuling O, Ford AT (2020) Human-wildlife coexistence in a changing world. Conserv Biol 34:786–794
Kremen C, Ullmann KS, Thorp RW (2011) Evaluating the quality of citizen-scientist data on pollinator communities. Conserv Biol 25:607–617. https://doi.org/10.1111/j.1523-1739.2011.01657.x
Lahoz-Monfort JJ, Magrath M (2021) A comprehensive overview of technologies for species and habitat monitoring and conservation. Bioscience 71(10):1038–1062. https://doi.org/10.1093/biosci/biab073
Marvin DC, Koh LP, Lynam AJ, Wich S, Davies AB, Krishnamurthy R, Stokes E, Starkey R, Asner GP (2016) Integrating technologies for scalable ecology and conservation. Glob Ecol Conserv 7:262–275
Messmer T (2000) The emergence of human-wildlife conflict management: turning challenges into opportunities. Int Biodeterior Biodegradation 45(3–4):97–102. https://doi.org/10.1016/S0964-8305(00)00045-7
Mullineaux E (2014) Veterinary treatment and rehabilitation of indigenous wildlife. J Small Anim Pract 55(6):293–300. https://doi.org/10.1111/jsap.12213. Epub 2014 Apr 12 PMID: 24725160
Mustățea M, Patru-Stupariu I (2021) Using landscape change analysis and stakeholder perspective to identify driving forces of human–wildlife interactions. Land 10:146. https://doi.org/10.3390/land10020146
OIE (2010) Training manual on wildlife diseases and surveillance: Workshop for OIE National Focal Points for Wildlife. World Organisation for Animal Health, Paris, France. https://www.oie.int/app/uploads/2021/03/a-training-manual-wildlife.pdf
Olson DD, Bissonette JA, Cramer PC, Green AD, Davis ST et al (2014) Monitoring wildlife-vehicle collisions in the information age: how smartphones can improve data collection. PLoS ONE 9(6):e98613. https://doi.org/10.1371/journal.pone.0098613
Peris S, Morales J (2004) Use of passages across a canal by wild mammals and related mortality. Eur J Wildl Res 50(2):67–72. https://doi.org/10.1007/s10344-004-0045-0
Pooley S (2021) Coexistence for Whom? Front Conserv Sci 2:726991. https://doi.org/10.3389/fcosc.2021.726991
Ratnieks FLW, Schrell F, Rebecca C, Sheppard EB, Bristow OE, Garbuzov M (2016) Data reliability in citizen science: learning curve and the effects of training method, volunteer background and experience on identification accuracy of insects visiting ivy flowers. Methods Ecol Evol 7(122):6–1235. https://doi.org/10.1111/2041-210X.12581
Ryder TB, Reitsma R, Evans B, Marra PP (2010) Quantifying avian nest survival along an urbanization gradient using citizen- and scientist-generated data. Ecol Appl 20:419–426. https://doi.org/10.1890/09-0040.1
Schwartz ALW, Shilling FM, Perkins SE (2020) The value of monitoring wildlife roadkill. Eur J Wildl Res 66:18. https://doi.org/10.1007/s10344-019-1357-4
Shilling FM, Perkins S, Collinson W (2015) Wildlife/roadkill observation and reporting systems. In Handbook of Road Ecology, eds R. van der Ree, D. J. Smith, and C. Grilo (Oxford: John Wiley & Sons) 552
Soulsbury CD, White PCL (2015) Human-wildlife interactions in urban areas: a review of conflicts, benefits and opportunities. Wildl Res 42:541–553
Sullivan BL, Aycrigg JL, Barry JH, Bonney RE, Bruns N, Cooper CB, Kelling S (2014) The eBird enterprise: an integrated approach to development and application of citizen science. Biol Cons 169:31–40
Waetje DP, Shilling FM (2017) Large extent volunteer roadkill and wildlife observation systems as sources of reliable data. Front Ecol Evol 5:89. https://doi.org/10.3389/fevo.2017.00089
Wieczorek J, Bloom D, Guralnick R, Blum S, Döring M et al (2012) Darwin Core: an evolving community-developed biodiversity data standard. PLoS ONE 7(1):e29715. https://doi.org/10.1371/journal.pone.0029715
Wittmann J, Girman D, Crocker D (2019) Using iNaturalist in a coverboard protocol to measure data quality: suggestions for project design. Citizen Science: Theory and Practice 4(1)
Acknowledgements
A special acknowledgement goes to the "Direzione centrale risorse agroalimentari, forestali e ittiche" of the Autonomous Region of Friuli Venezia Giulia for their invaluable and continuous financial support.
Funding
Open access funding provided by Università degli Studi di Udine within the CRUI-CARE Agreement.
Author information
Authors and Affiliations
Contributions
PT: Conceptualization, Writing—original draft, Methodology, Writing—review & editing and web application design. SP: Conceptualization, Methodology, Manuscript revision, Supervision. MO: Conceptualization, Methodology, Writing—original draft, Writing—review & editing. EP: Conceptualization, Supervision. AC: Supervision. MG: Supervision.
Corresponding author
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Keypoints
• InfoFaunaFVG is a novel progressive web application and data repository system for wildlife monitoring and surveillance on a institutional level in Friuli Venezia Giulia (Italy).
• During the first two years of its use, the system gathered 40175 records from 300 different users, from 16 institutions, becoming the largest italian wildlife repository system reported in literature.
• InfoFaunaFVG has the potential to have a considerable impact on regional wildlife governance, providing high quality consistent, accurate and traceable data on wildlife species.
The original version of this article was revised: This article was originally published with a still a needed corrections in the body text.
Appendix. Synopsis of InfoFaunaFVG
Appendix. Synopsis of InfoFaunaFVG
InfoFaunaFVG and its scopes
-
What is InfoFaunaFVG?
It is a data repository system based on a novel PWA developed at the University of Udine (Department DI4A) in collaboration with regional and local authorities appointed to monitor, rescue, and to provide veterinary care to debilitated and/or compromised or dead or selectively culled wildlife species. InfoFaunaFVG adheres to Darwin Core set of terms and classes to standardize the data recorded in the system.
The following Darwin Core occurrence data are included: occurrenceID/ recordNumber, dynamicProperties, basisOfRecord, scientificName, eventDate, eventTime, fieldNotes, decimalLatitude, decimalLongitude, geodeticDatum, organism Quantity, organismQuantityType, Sex, Age, lifeStage, coordinateUncertaintyInMeters, individualCount, eventTime, and associatedMedia, recordedBy.
-
What is the goal of InfoFaunaFVG?
The scopes of this database are:
-
To facilitate and give a scientific support to wildlife monitoring and surveillance in Friuli Venezia Giulia, by providing high quality consistent, accurate and traceable data.
-
To guide evidence-based decisions with the aim of reducing wildlife health risk and improve wildlife governance.
-
To better understand the influence of human activities on changes in distribution patterns and the presence of introduced or invasive species.
-
To monitor transmissible diseases from wild animals to livestock and humans.
-
How InfoFaunaFVG works
InfoFaunaFVG is used only at institutional level and data are not shared publicly at this stage. Therefore, only local wildlife institutions and centres recognized and licensed by regional authorities can use InfoFaunaFVG. These insitutions are Inspectorates, National forestry services, Wildlife Rescue Centers (“Centri di recupero di Animali Selvatici’’- CRAS), Italian health authority and research organizations for animal health and food safety (Istituto Zooprofilattico—IZS). Third party contractors involved in animal rescue activity and recognized by regional authorities can use the system as well. InfoFaunaFVG has been structured hierarchically: each user is assigned a level of use, based on their operational needs and role.
At this stage of the project, all occurrence events are based on “human observations’’. In the event of a wildlife animal’s observation, users can open a “report section’’. Figure 5 shows dataset fields for reporting the occurrence event. From this section, multimedia data, such as video or pics, can be added to the report.
After the report section is completed, the users can georeference the occurrence event (Fig. 6).
Essentially, the user can: use the GPS, geocode, and/or use the WebGIS CRUD tool to point the location of the observation on the map.
At this point, the user can proceed with further sections of InfoFaunaFVG, depending if the wildlife animal is dead or alive (debilitated or compromised). If alive, the wildlife can be transferred to a rescue centre (the users will then open the “rescue section’’ – Fig. 7). Outcome of dead or selectively culled animals, not undergoing necropsy, can be entered directly in the result section (Fig. 9).
For dead protected wildlife (listed in the Italian regulation 157/1992), the user can access to “necropsy section’’ (Fig. 8) if the dead animal is going through post-mortem examination.
Regardless the outcome, the user will finalize the record by filling in the result section (Fig. 9).
How InfoFaunaFVG data recorded are stored and shared
At this stage of the project the data are, available on demand through https://web.infofaunafvg.com/richiesta_accesso_dati.pdf.
UniUd-DI4A intends to request endorsement from the Global Biodiversity Information Facility (GBIF) community to make this data widely available. Regional authorities evaluated positively the intention to request endorsement from Global Biodiversity Information Facility (GBIF) community to make this data widely available.
The authors are pleased to communicate that InfoFauna FVG was officially endorsed by GBIF and at the time of publication of this manuscript 22272 were already shared within the platform.
GBIF (https://www.gbif.org) is an international network and a research foundation funded by governments around the world and aimed at providing anyone, anywhere open access to data about all types of life on the Earth.
We strongly believe this could be advantageous so researchers can work together on related tasks with a common goal on a global level.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Tomè, P., Pesaro, S., Orioles, M. et al. InfoFaunaFVG: a novel progressive web application for wildlife surveillance. Eur J Wildl Res 69, 38 (2023). https://doi.org/10.1007/s10344-023-01664-4
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s10344-023-01664-4