Abstract
Disruption of naturally evolved spatial patterns of genetic variation and local adaptations is a growing concern in wildlife management and conservation. During the last decade, releases of native taxa with potentially non-native genotypes have received increased attention. This has mostly concerned conservation programs, but releases are also widely carried out to boost harvest opportunities. The mallard, Anas platyrhynchos, is one of few terrestrial migratory vertebrates subjected to large-scale releases for hunting purposes. It is the most numerous and widespread duck in the world, yet each year more than three million farmed mallard ducklings are released into the wild in the European Union alone to increase the harvestable population. This study aimed to determine the genetic effects of such large-scale releases of a native species, specifically if wild and released farmed mallards differ genetically among subpopulations in Europe, if there are signs of admixture between the two groups, if the genetic structure of the wild mallard population has changed since large-scale releases began in the 1970s, and if the current data matches global patterns across the Northern hemisphere. We used Bayesian clustering (Structure software) and Discriminant Analysis of Principal Components (DAPC) to analyze the genetic structure of historical and present-day wild (n = 171 and n = 209, respectively) as well as farmed (n = 211) mallards from six European countries as inferred by 360 single-nucleotide polymorphisms (SNPs). Both methods showed a clear genetic differentiation between wild and farmed mallards. Admixed individuals were found in the present-day wild population, implicating introgression of farmed genotypes into wild mallards despite low survival among released farmed mallards. Such cryptic introgression would alter the genetic composition of wild populations and may have unknown long-term consequences for conservation.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Translocation of wild and farmed individuals is an increasingly common procedure to support threatened populations (Champagnon et al. 2012a). Besides conservation objectives, restocking viable populations to sustain or increase harvest has long been practiced in forestry, fisheries, and game management (Laikre et al. 2006). The potential risks associated with restocking of native species have been overshadowed by problems of translocating non-native species, especially invasive ones (Champagnon et al. 2012a; Laikre et al. 2010; McGinnity et al. 2009). However, as pointed out by Champagnon et al. (2012a), “conspecifics can be aliens too,” which is to say that release of individuals of non-local provenance may lead to introgression that disrupts spatial genetic structure, alter local genetic diversity, and ultimately threaten local adaptations (Mank et al. 2004; McGinnity et al. 2003; Rhymer and Simberloff 1996; Sutherland et al. 2006). Hybridization between released farmed or escaped domestic individuals and their wild conspecifics is also a driver of biodiversity loss (Randi 2008; Sutherland et al. 2006), posing risks in both conservation and restocking practices.
The importance of relying on local provenance in restocking programs is widely recognized in management of anadromous fish such as salmonids (Ryman 1981). This is, however, not always the case in restocking programs involving other animals or plants (Laikre et al. 2010). Among non-migratory gallinaceous birds (e.g., ring-necked pheasant Phasianus colchicus, grey partridge Perdix perdix, and red-legged partridge Alectoris rufa), restocking for harvest purposes has been carried out for a long time (e.g., Hill and Robertson 1986), often using non-local stocks (Barbanera et al. 2010; Blanco-Aguiar et al. 2008).
The genetic aspects of restocking become crucial in migratory species, owing to possibly much wider spatial consequences. Among migratory vertebrates, common quail (Coturnix coturnix) has been extensively restocked for decades in Europe to increase harvestable populations. Despite this, it was not until recently that the genetic effects of the practice were studied (Amaral et al. 2007; Deregnaucourt et al. 2002; Puigcerver et al. 2014; Puigcerver et al. 2007; Sanchez-Donoso et al. 2012). The latter authors showed that released farmed common quails were mainly hybrids between common quail and the domesticated Japanese quail (C. japonica).
The mallard, Anas platyrhynchos, is one of the best model species to study genetic aspects of restocking a migratory vertebrate: it is the most widespread duck in the world, with an estimated global population of 19+ million individuals (Wetlands International 2015); it is also one of the most socio-economically important game birds globally, with an estimated annual harvest of about 4.5 million birds each in Europe and North America (Hirschfeld and Heyd 2005; Raftovich et al. 2015). Annual large-scale releases of farmed mallards are now a common practice in several European countries to increase the harvestable population (Guillemain et al. 2010). From a modest start in the 1970s, more than three million unfledged ducklings are currently released in the European Union for hunting purposes each year (Champagnon 2011), while the breeding population in the same area is 4.5 million pairs (Wetlands International 2015). In France alone, 1.4 million ducklings are released annually (Mondain-Monval and Girard 2000), to be compared to an estimated national winter population of 300,000 birds (Deceuninck et al. 2014). In Sweden, more than 250,000 ducklings are released annually (Söderquist 2015) compared to a breeding population of 200,000 pairs (Ottosson et al. 2012), and in the Czech Republic 200,000–300,000 released ducklings exceed the wild breeding population five- to tenfold (Hůda et al. 2001; Šťastný et al. 2009; Sýkora 2005). Farmed ducklings are released from May through July, and they are usually fed and protected from predators until the start of hunting season, which lasts from late August until the end of December or January. Although releases of this type have been carried out for about 40 years, the genetic consequences for the wild mallard population remain poorly investigated.
Released mallards have lower survival than wild-born conspecifics (Champagnon et al. 2012b; Söderquist et al. 2013), likely due to “burdens of captivity” such as a less developed digestive system (Champagnon et al. 2012b). Ducklings have an innate response to avoid predators (Dessborn et al. 2012), a behavior that is most crucial the first weeks after hatching. During this time, farmed ducklings to be released are still in captivity, protected from predators. This may result in a naïve behavior affecting their survival after release. Despite high mortality rates, tens of thousands of farmed mallards survive their first year, simply because such large numbers are released annually (Champagnon et al. 2016). These potentially mate with wild conspecifics in subsequent years, leading to introgression by non-local genotypes. Changes observed in wild mallard populations in recent decades, e.g., in bill morphology, likely result from genetic homogenization by farmed-released mallards (Champagnon et al. 2010; Söderquist et al. 2014). Still, such introgression can be termed “cryptic” due to the fact that individuals from the two groups cannot easily be distinguished (Saltonstall 2002), partly owing to strong counter-selection of obvious “unnatural” phenotypes by duck farmers before release.
The mallard ranges widely in the Northern Hemisphere and breeds in biomes from tundra to Mediterranean types (Scott and Rose 1996). Within the geographic range, natural spatial genetic structuring is nevertheless limited (Kraus et al. 2013; Kraus et al. 2011b; Kulikova et al. 2005). Due to founder effects, inbreeding, and genetic drift, a differentiation between wild and farmed mallards could be expected. However, studies thus far on genetic differentiation give ambiguous results; Baratti et al. (2009) did not find any differences using microsatellite markers, while Champagnon et al. (2013a), Čížková et al. (2012), and Baratti et al. (2014) detected genetic differences between regional wild and farmed populations in France, Czech Republic, and Italy, respectively. Thus, more studies of genetic structure across multiple flyways are needed to assess the degree of differentiation and potential genetic homogenization at a larger spatial scale, using a common and more powerful methodological framework.
The aims of this study were to determine (a) if present-day wild (hereafter referred to as “wild”) and farmed mallards differ genetically among subpopulations in Europe, (b) if there are signs of previous or ongoing admixture between farmed and wild mallards, (c) if such admixture has led to a detectable, changed genetic structure in the wild population compared to the pre-release period, and (d) if the data collected in this study matches global patterns across the Northern hemisphere. To address these questions, we analyzed samples from six European countries using a set of 384 single-nucleotide polymorphism (SNPs) markers developed by Kraus et al. (2011a).
Material and methods
Biological sampling
A total of 656 samples were collected, of which 231 were from the wild (Sweden, 70; France, 50; the Czech Republic, 69; Norway, 21; Finland, 5; the Netherlands, 16), 212 from mallard farms (Sweden, 82; France, 50; the Czech Republic, 80), and 213 from museums (hereafter referred to as “historical,” i.e., samples collected 1831–1977, from before large-scale releases started in the study region; Sweden, 110; France, 27; the Czech Republic, 16; Norway, 15; Finland, 45. In historical samples, approximately 10 mm2 skin from the toe pad was cut with a scalpel. In (present-day) wild and farmed mallards, blood, muscle, egg membranes, or feathers (only in farmed mallards) were collected from individuals without any known kinship. Historical and present-day wild samples from Sweden, Finland, Norway, the Czech Republic, and the Netherlands all emanated from the breeding season (May to August) to minimize sampling of mixed provenances. Sampling adults, egg shells (only one per nest to avoid sampling relatives), or adult feathers from nests in the wild during breeding season excluded the risk of including released farmed mallards from the same year (as they are released as unfledged ducklings in summer) and thereby also minimized the overall risk of having farmed individuals in the wild sample. Historical and wild French samples were from the winter period, as they had been collected as part of an earlier study not limited to breeding birds. All samples from France as well as present-day samples from the Czech Republic had been used in precedent genetic studies. For further sample information and sampling protocols, see Table S1, Champagnon et al. (2013a) and Čížková et al. (2012).
DNA extraction and genotyping
DNA extraction of samples in this study was performed with the DNeasy Blood & Tissue kit (Qiagen, Hilden, Germany). To avoid contamination, extractions of historical samples were carried out in laboratories where no avian DNA had previously been processed. Samples with a wavelength absorption ratio of 260/230 nm below 1.7 on a NanoDrop 2000 (Thermo Fischer Scientific, Wilmington, USA) and with a low DNA concentration were cleaned with the DNA Clean & Concentrator-5 kit (Zymo Research, Irvine, USA) to obtain a final volume of 15 μl and a DNA concentration of at least 50 ng/μl.
A published set of 384 SNP markers was used for genotyping (Kraus et al. 2011a) using previously described protocols (Kraus et al. 2012). The SNP markers adhered to neutral expectations, e.g., Hardy Weinberg and linkage equilibria, and have previously proven useful for investigations of demography and genetic structure (Kraus et al. 2013) as well as for addressing evolutionary questions such as genetic introgression and speciation (Kraus et al. 2012).
As a genotyping platform, we used the Illumina GoldenGate Genotyping Assay on the Illumina BeadXpress, with accession numbers ss263068950–ss263069333 in dbSNP for the set of 384 SNPs. When possible, we put 5 μl of 75 ng/μl of DNA per genotyping reaction, and a minimum of 50 ng/μl. Samples at a lower concentration were checked for an excess of missing data and eventually excluded when required (see “Results” section for thresholds). Raw data were analyzed in GenomeStudio (Illumina Inc.). We also tested for a bias in homozygosity between historical museum samples versus contemporary ones that could arise through allelic dropout. No such bias was found, which is in concordance with previous applications of the methods in a study of museum samples of geese (Jonker et al. 2012). Genotype calls of all individuals are available at Dryad repository under https://doi.org/10.5061/dryad.74gd7 in Structure input file format.
Genetic diversity
The degree of observed and expected heterozygosity (H obs and H exp) along with values of allelic richness and tests for HWE at each locus, as well as inbreeding coefficients (as F IS values) were calculated in diveRsity (Keenan et al. 2013) v1.9.5 in R (R Development Core Team 2009), using function divBasic and 1000 bootstrap iterations. As no genetic population structure between countries was previously known (Kraus et al. 2013), nor found by us (see Figs. 1 and 4), all historical groups were combined into one. Moreover, due to their small number (n = 5), the Finnish wild sample was pooled with the Norwegian wild (Table 1). Both these groups represent biologically meaningful entities in the wild (see “Discussion” section).
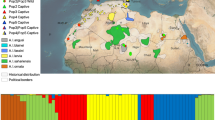
Structure assignments of 591 individual genotypes for K = 2 to K = 5. Each individual genotype is represented by one vertical bar. Black lines separate the 14 different groups of mallards included in the study. Dashed horizontal lines at assignment probability q = 0.2 and q = 0.8 show the thresholds for individuals considered as admixed. The most likely number of clusters is K = 2 (Fig. S2), where predominantly blue bars represent wild (historical and present-day) mallards and yellow bars farmed mallards. In addition, K = 3 show that Czech farmed mallards (red) are clustered separately from other farmed birds (yellow), and for K = 4, all farmed mallards (Swedish: green; French: yellow; Czech: red) are separate clusters. Throughout higher levels of K, the assignment of mallards from farms to the farm clusters remains stable, while the resolution of wild mallard clusters is obscured
Genetic population structure analysis
Using Structure v2.3.4 (Pritchard et al. 2000), the most likely number of clusters (K) was inferred with Evanno et al.’s (2005) method in Structure Harvester (Earl and vonHoldt 2012). All models of K = 1–10 were tested with 10 replicates each, using 2,000,000 steps in the Markov chain of which the first 400,000 were discarded as burn-in. Allele frequencies were assumed to be correlated and the admixture model set for genetic cluster assignment.
Clumpp v1.1.2 (Jakobsson and Rosenberg 2007) was used to achieve individual assignment convergence into one combined estimate of cluster ancestry for a given K. Depending on the quantity D (D < 10−13) described in the Clumpp manual, we selected the FullSearch algorithm for K = 1–2, Greedy for K = 3–5 (with 10,000 random input sequences for K = 3–4, and 5000 for K = 5), and LargeKGreedy for K = 6–10 (10,000 random input sequences for K = 6–7, and 5000 for K = 8–10). Distruct v1.1 (Rosenberg 2004) was used to graphically visualize the results.
Discriminant Analysis of Principal Components (DAPC) (Jombart et al. 2010) from adegenet v1.4-1 (Jombart 2008) in R was used as an additional method to characterize the potential differences between historical, present-day wild, and farmed mallards. The function find.clusters determined the most likely number of genetic clusters in the data using all available principal components (PCs; Jombart et al. 2010). In order to calculate the posterior probability (q) of assignment of individuals to each of these clusters using DAPC, the optimal number of PCs was determined by the optim.a.score with 25 simulations. To avoid unstable assignments of individuals to clusters, only 120 PCs (sample size divided by three) were retained in a preliminary DAPC, yet using all the discriminant functions (Frosch et al. 2014; Jonker et al. 2013). The final DAPC analyses were carried out with the optimal number of PCs (1-24 depending on K).
Individuals with an assignment probability of q > 0.8 were defined as belonging either to the wild or farmed population (cf. Jonker et al. 2013), while those with q ≤ 0.8 were targeted as admixed.
In an attempt to evaluate the results in a wider geographical context, we combined the samples genotyped in the present study with the 709 samples from throughout the Northern Hemisphere and analyzed with the same SNP set by Kraus et al. (2013). A total of 32 SNPs were excluded to make the two datasets comparable (Table S2). This analysis was run in Structure with all samples pooled and the same settings as for the present analysis.
Results
Sampling and genotyping
By arbitrarily setting, a threshold of missing genotypes at 10%, a total of 24 SNP loci (out of 384) were excluded from further analysis. Next, individuals with more than 10% missing SNPs (65 samples; 10%) were also excluded. Of the remaining samples, 209 were present-day wild, 211 were farmed, and 171 were historical (final sample sizes for each country and additional information about samples, see Table 1 and S1). A total of 591 individual samples and 360 SNP loci were hence used in the subsequent population structure analyses (for excluded SNPs see Table S2).
Genetic diversity and inbreeding
Basic summary statistics including sample sizes for each SNP locus, allelic richness, heterozygosity, tests for HWE (considering a Bonferroni corrected p value of p < 0.004; i.e., 14 test populations), levels of missing data, and inbreeding coefficient (as F IS) can be found in the Electronic Supplementary file 1 (.xlsx file). Generally, loci were in HWE and showed no other conspicuous features confirming previous reports (cf. Kraus et al. 2011a; Kraus et al. 2012; Kraus et al. 2013). When summarized per group, observed and expected heterozygosity were similar among groups (Table 1). However, F IS values revealed significant deviations from zero in several cases (Fig. S1). The 95% confidence intervals (CIs) of F IS did not encompass the zero value (i.e., were significantly positive) in historical, Swedish wild, French wild, Czech wild, Swedish farmed, and Czech farmed individuals. F IS values significantly below zero were not found in any group (Fig. S1). However, Dutch wild individuals had a negative F IS point estimate, yet with the upper limit of the CI positive. The F IS of the combined Norwegian-Finnish wild and the French farmed groups were not different from zero (Fig. S1).
Genetic admixture analyses
Structure Harvester indicated that the most likely number of clusters was K = 2 for Structure (Fig. S2), that of DAPC being instead K = 3 (Fig. S3). Inferences of K = 2 to K = 4 were similar in both Structure and DAPC, with the difference that DAPC was more decisive and showed fewer admixed individuals than Structure (Figs. 1 and 2). For K = 2, one cluster consisted mainly of historical and wild individuals, and the other of farmed individuals. For K = 3, farmed individuals sampled in the Czech Republic formed a separate cluster and for K = 4 all three farmed groups formed separate clusters (N.B. in Structure most Swedish farmed individuals in K = 4 do not have a q > 0.8, as was the case for the same individuals in K = 2 and K = 3). Thus, K = 4 gives an informative picture of the potential situation when considering genetic results and sampling, but as the main aim of this study was to identify wild, farmed, and admixed individuals, the most probable number of clusters (K = 2) from Structure, was used in the subsequent analysis of admixed individuals. For higher K, the patterns were less clear and K = 6–10 were excluded from Figs. 1, 2, and 4. By arbitrarily defining an individual as belonging to a certain cluster when assignment probability (q) was above 0.8, 168 of 171 historical individuals clustered together for K = 2 in Structure. In wild birds, the proportion of admixed individuals was higher in present-day than in historical everywhere except for Norway (one admixed individual in the historical group, none in present-day wild) (Fig. 3). As no historical individuals could be an admixture between wild and farmed mallards, these three cases must be a result of interpretation bias or that they had genes from domesticated ducks in them. Note that Swedish, French, and Czech wild groups also included some individuals assigned to the farmed cluster other than the admixed. When considering the output from K = 4, it is evident that these latter individuals originated from farms in their respective country. The Swedish and French farmed groups consisted of farmed individuals from their respective country as well as individuals classified as admixed with wild, while the Czech farmed group consisted of farmed individuals only, except one assigned to the wild cluster (Fig. 3).
Discriminant Analysis of Principal Components (DAPC) assignment for K = 2 to K = 5 of 591 individual genotypes, each represented by a colored vertical bar. Black lines separate the 14 different groups of mallards included in the study. The most likely number of clusters is K = 3 (Fig. S3), where wild (historical and present-day) mallards form a blue cluster, farmed mallards from Sweden and France a yellow, and Czech farmed mallards a red cluster. At K = 4, Swedish farmed mallards (green) split from the farmed yellow cluster in K = 3 separating all farmed groups. As for the Structure results (Fig. 1), mallards from the wild cannot be assigned to separate clusters in a meaningful way for higher estimates of K
Proportion of historical, present-day wild, and farmed mallards from six countries as inferred from Structure K = 2 (historical/wild: blue; Czech farmed: red; French farmed: yellow; Swedish farmed: green), when applying an assignment probability limit at q > 0.8. Individuals with q ≤ 0.8 are considered admixed (orange). That farmed individuals in the wild originate from farms in their respective country becomes evident at K = 4 (Fig. 1). Sample sizes for each group are shown below each respective pie chart. N.B. low sample sizes for Finnish wild and Dutch wild may lead to non-representative results
Our analysis, adding previously published global mallard samples, indicated a continent-wide absence of geographical structure as a general pattern in this species (except for Greenland mallards, Fig. 4). An elevated rate of individuals targeted as farmed or admixed (less than 80% of the individuals assigned to the wild cluster) was observed also in samples from the Faroe Islands, Germany, Great Britain, Iran, and the Netherlands (Table 2). The above results were based on Structure, while DAPC yielded qualitatively the same results but underestimated admixture in the same way as explained above.
Structure assignment for K = 2 to K = 5 for the 591 mallards in this study combined with the 709 in Kraus et al. (2013), resulting in a total n = 1300. Each individual genotype is represented by a colored vertical bar. Black lines separate the 15 different groups of mallards. Dashed horizontal lines at assignment probability q = 0.2 and q = 0.8 show the threshold for individuals considered as admixed. For K = 3, the blue cluster comprises wild (historical and present-day) mallards, the yellow cluster the three farmed groups (Swedish, French, and Czech), and the red cluster wild mallards from Greenland. As for Figs. 1 and 2, in estimates of higher K individuals cannot be assigned in a meaningful way
Discussion
Our analyses of mallards from Europe show that farmed birds have a different genetic composition than both present-day and historical wild conspecifics. Moreover, historical specimens do not show any clear signs of genetic patterns. Both these results are consistent with studies on other game birds (e.g., Barbanera et al. 2015; Barbanera et al. 2010). Compared to earlier studies on mallards (Baratti et al. 2014; Champagnon et al. 2013ba; Čížková et al. 2012), the inclusion of samples from a wider geographical area enabled us to identify geographic regions where substantial genetic admixture of farmed genotypes with wild populations may have occurred. Accordingly, merging European with Asian and North American samples confirmed previous regional results from France (Champagnon et al. 2013ba), the Czech Republic (Čížková et al. 2012), and Italy (Baratti et al. 2014), i.e., occurrence of admixture of genetic material from farmed mallards in wild conspecifics.
Heterozygosity and inbreeding
Estimates of expected and observed heterozygosity did not reveal clear signs of a genetic diversity loss among wild or farmed mallards. Compared to other genetic markers, the use of SNPs to make such estimates might not always be reliable due to the low numbers of alleles considered. Often, the use of SNPs is also influenced by the respective allele frequencies and the prevalence of private or nearly private alleles. Indeed, most SNPs used in this study have rather intermediate allele frequencies (Kraus et al. 2011a), and comparisons with other marker sets are not straightforward. This could explain the inconsistent results regarding heterozygosity between the current study and those by Champagnon et al. (2013a) and Čížková et al. (2012), who found a greater difference between farmed and wild mallards based on microsatellite analyses. Even though it is commonly acknowledged that the intra-population genetic structure of migratory birds is largely protected by philopatric habits (Liu et al. 2012) or other migratory traditions (Jonker et al. 2013), there are also examples of species with no signs of genetic structure on the continental scale (Reudink et al. 2011). As a case in point, this is true for the mallard, which shows an almost total lack of global genetic structure (Delany and Scott 2006; Kraus et al. 2013). Panmixia among mallards makes them resilient to inbreeding, which indeed is confirmed by our combined Norwegian-Finnish wild group (indicated by F IS value close to zero). These are countries where no large-scale releases of farmed mallards have, to our knowledge, ever occurred. The low level of inbreeding at breeding facilities observed in France could be maintained by the continuous exchange of specimens between farms (Champagnon et al. 2013bb). In the Czech and Swedish farmed groups, this does not seem to be the case, as they both show significantly positive F IS.
The historical wild group showed a positive F IS, which is an indication of decreasing heterozygosity. This can be explained by global sampling of different subpopulations, i.e., a Wahlund effect (Wahlund 1928). The Wahlund effect can also explain the positive F IS observed in the Swedish, French, and Czech wild groups, since these wild caught mallards in fact may have consisted of both truly wild as well as released farmed mallards, and crosses between these groups.
Genetic structure
The coherence between Structure and DAPC outputs confirmed the robustness of our results, especially as the underlying methods rely on different models, with Structure requiring assumptions about population genetic parameters unlike DAPC. The latter tends to be overconfident and to assign more individuals to a given cluster. By retaining too many principal components in DAPC, there is a risk of over-fitting the discriminant functions and overestimating the posterior membership probability (q) which should be corrected for by using optim.a.score (Jonker et al. 2013). However, despite implementing this correction due to high values of q in this study, we believe that DAPC still overestimated q and thereby underestimated the genetic admixture in our sample (cf. Frosch et al. 2014). Structure provides more conservative estimates in terms of clear cluster assignment and is therefore less prone to underestimate the number of admixed individuals.
The output for K = 2 showed that farmed mallards were genetically different from their wild conspecifics (Fig. 1). Animals bred and raised in captivity are always at risk of being affected by inbreeding and genetic drift, resulting in a different genetic structure compared to wild populations (Price 1999). In addition, a change in genetic structure over time in farmed mallards is consistent with Lynch and O’Hely (2001), who argued that adaptation to captivity together with anthropogenic selection at breeding facilities will inevitably lead to a genetic alteration of the captive population. Such differences in genetics may, at least partly, explain discrepancies documented in captive ducks regarding morphology, e.g., brain volume (Guay and Iwaniuk 2008), digestive organs (Champagnon et al. 2012b; Moore and Battley 2006), feeding apparatus (Champagnon et al. 2010; Söderquist et al. 2014), sexual behavior (Desforges and Wood-Gush 1976), mate preferences (Cheng et al. 1978; Cheng et al. 1979), and habituation to humans (Desforges and Wood-Gush 1975).
When Structure and DAPC models for K = 3 and K = 4 were investigated, the farmed mallards appeared in three clusters, with the Czech being the first to emerge. The use of hybrids between wild and domesticated mallards in the founder population of the Czech farmed mallards, or stochastic processes (e.g., genetic drift), could explain this clear separation (Hůda et al. 2001). French and Swedish mallard farms are also believed to have used wild birds as founders (pers. comm., PS, MG; JC). Birds in the Swedish farmed group did not appear exclusively in one cluster for K = 4 and could have a common ancestry with individuals in the French farmed group. In fact, several Swedish duck breeders import eggs from abroad, mostly from Denmark (Swedish Board of Agriculture 2011), which in turn imports eggs from France. The reason for this practice is most likely just economical; however, it may also decrease risks of inbreeding and keep farmed populations in different countries genetically similar.
When looking at the proportion of individuals assigned to different clusters, mallards in the same groups as Swedish, French, and Czech farmed clusters were found within the respective wild groups (Fig. 3). This clearly indicates that farmed mallards live together with wild counterparts after release and perform very limited movements once in the wild (Champagnon 2011). The relatively large proportion of individuals assigned to the farmed cluster within the French wild group may be biased, since these samples were collected during winter and were at least partly from private estates where hunting and releases have occurred for many years (Table S1). Nevertheless, it is clear that the wild populations in the three countries where farmed mallards were sampled also comprise admixed individuals. We argue that these are the result of hybridization in the wild rather than being individuals already admixed at release (cf. Sanchez-Donoso et al. 2014). The main argument for this is that no admixed individuals were found in the Czech farmed population, although they could be found in the Czech wild group. Also, the relative proportion of admixed individuals is higher in Swedish wild than in Swedish farmed. In addition, although survival of released mallards is low, those that do survive can be assumed to be more likely to mate with wild mallards, as they outnumber farmed.
In Finland, no large-scale releases of mallards have ever occurred. However, farmed Swedish mallards surviving the hunting season may intermix, mate, and subsequently breed in Finland with wild mallards (Söderquist et al. 2013). Released mallards may also breed with wild ones in Sweden, and their admixed offspring may later end up in Finland. Such mechanisms could explain the relatively high proportion of admixed individuals found among wild mallards in Finland, but admittedly, the sample is too small to make reliable inferences. The opposite scenario was observed in the Norwegian wild group, whose samples were mainly collected in Tromsø, northern Norway, where mallards are mostly non-migratory (Nygård et al. 1988). All individuals in this group were assigned to the historical/wild cluster. It is likely that this remote population has little exchange with wild birds affected by releases from other parts of Europe.
By combining data presented here with global mallard data from Kraus et al. (2013), additional geographical regions in the expanded data set displayed admixed individuals (Fig. 4). At K = 2, historical and wild mallards were separated from the farmed clusters, and at K = 3, Greenland emerged as a separate cluster. The Greenland population has indeed been previously identified as genetically and geographically isolated (Kraus et al. 2013; Scott and Rose 1996). Signs of admixed farmed mallards were found among wild mallards from the Faroe Islands, Germany, Great Britain, Iran, and the Netherlands, but their genetic origin could not be ascertained, because there were no farmed mallards available for reference from these countries. Nevertheless, admixture in Germany and Great Britain could result from the annual releases of 100,000 and 500,000 farmed mallards in these two countries, respectively (Champagnon et al. 2013bb). In Iran, mallard hunting is considerable (Balmaki and Barati 2006), but we have no information about supplementary releases there or within the respective flyway that could explain why some individuals fell outside the wild cluster (Fig. 4 and Table 2).
Compared to Kraus et al. (2013), we found a lower proportion of admixed individuals in Norway, and a higher proportion of admixed individuals in France and Sweden. As far as Norway is concerned, the discrepancy between the two studies may be a sampling bias; birds were from the resident population in Tromsø in the present study and from near Stavanger (southern Norway) in Kraus et al. (2013). In the latter area, birds are more likely to breed with farmed-released mallards from Sweden and Great Britain. Likewise, the higher proportion of admixed individuals in France and Sweden can certainly be explained by sampling location, e.g., nature reserves versus hunting areas (Table S1 and Champagnon et al. 2013ba), and sampling season. Swedish mallards in Kraus et al. (2013) were sampled during autumn at a stopover site (when Finnish and Russian breeding mallards, potentially unaffected by releases, move through the area and are caught for ringing; Gunnarsson et al. 2012), while Swedish samples in this study comprised only mallards sampled during the breeding season. As the temporal patterns of sampling differ between this study and Kraus et al. (2013), comparisons between the two should be made with caution.
Conclusions and implications
Genetic differences between farmed and wild mallards, as well as signs of genetic admixture between these groups, suggest that released farmed mallards survive, intermix with wild conspecifics, breed, and thus may transfer genetic material to the wild population. However, introgression per se could not be proven by this study. Cryptic introgression of alleles from farmed mallards would alter the genetic composition of wild populations and may have unknown long-term consequences, also for populations geographically distant from release sites. The mallard is not a rare species, but modified genetic structure and loss of local adaptations are nevertheless of concern from a conservation perspective.
The rate of introgression due to genetic admixture is likely low due to the poor survival of released farmed mallards (Champagnon et al. 2012b) but, as suggested by this study, restocking practice involves so many birds that the “footprint” in the wild population may be detectable and potentially significant. With continued releases, the genetic load of farmed stocks among wild mallards will persist or grow, potentially leading to lower fitness in the latter due to introgression of genotypes that are inferior under natural selection regimes. If alleles are recessive, they can propagate in heterozygotes and thereby be less sensitive to genetic purging (Crnokrak and Barrett 2002).
Hybridization is common in birds in general (Grant and Grant 1992) and among ducks in particular (Kraus et al. 2012; Rhymer 2006; Tubaro and Lijtmaer 2002). Erosion of native genetic resources by hybridization with non-native ones is a conservation concern, as it may eventually lead to extinction of rare taxa (Muñoz-Fuentes et al. 2007; Rhymer and Simberloff 1996). Likewise, released mallards may threaten native populations of American black duck Anas rubripes in North America (Mank et al. 2004) and Pacific black duck A. superciliosa in New Zealand and Australia (Taysom et al. 2014).
Genetic monitoring and ringing can serve to evaluate the status of both wild and farmed populations and would also meet the need for better knowledge about numbers, origin, and future fate of farmed individuals after release (e.g., Champagnon et al. 2016). We encourage further genetic studies on released mallards with a more targeted molecular marker set (e.g., SNP sets specifically developed to determine hybrid class assignment: Nussberger et al. 2013), or full genomic re-sequencing (Kraus and Wink 2015), to better evaluate the rate of hybridization and introgression.
References
Amaral AJ, Silva AB, Grosso AR, Chikhi L, Bastos-Silveira C, Dias D (2007) Detection of hybridization and species identification in domesticated and wild quails using genetic markers. Folia Zool 56:285–300
Balmaki B, Barati A (2006) Harvesting status of migratory waterfowl in northern Iran: a case study from Gilan Province. In: Boere GC, Galbraith CA, Stroud DA (eds) Waterbirds around the world. The Stationary Office, Edingburgh, pp 868–869
Baratti M, Baccetti N, Cordaro M, Mori A, Dessì-Fulgheri F (2014) Investigating the puzzling genetic structure of mallard populations (Anas platyrhynchos L.) in Italy. Eur J Wildl Res 61(1):1–9. https://doi.org/10.1007/s10344-014-0876-2
Baratti M, Cordaro M, Dessi-Fulgheri F, Vannini M, Fratini S (2009) Molecular and ecological characterization of urban populations of the mallard (Anas platyrhynchos L.) in Italy. Italian J Zool 76(3):330–339. https://doi.org/10.1080/11250000802566624
Barbanera F, Forcina G, Cappello A, Guerrini M, Grouw H, Aebischer NJ (2015) Introductions over introductions: the genomic adulteration of an early genetically valuable alien species in the United Kingdom. Biol Invasions 17(1):409–422. https://doi.org/10.1007/s10530-014-0739-5
Barbanera F, Pergams ORW, Guerrini M, Forcina G, Panayides P, Dini F (2010) Genetic consequences of intensive management in game birds. Biol Conserv 143(5):1259–1268. https://doi.org/10.1016/j.biocon.2010.02.035
Blanco-Aguiar J, González-Jara P, Ferrero M, Sánchez-Barbudo I, Virgós E, Villafuerte R, Dávila J (2008) Assessment of game restocking contributions to anthropogenic hybridization: the case of the Iberian red-legged partridge. Anim Conserv 11(6):535–545. https://doi.org/10.1111/j.1469-1795.2008.00212.x
Champagnon J (2011) Conséquences des introductions d'individus dans les populations exploitées: l'example du canard colvert Anas platyrhynchos. PhD Thesis, University Montpellier II, France
Champagnon J, Crochet PA, Kreisinger J, Čížková D, Gauthier-Clerc M, Massez G, Söderquist P, Albrecht T, Guillemain M (2013a) Assessing the genetic impact of massive restocking on wild mallard. Anim Conserv 16(3):295–305. https://doi.org/10.1111/j.1469-1795.2012.00600.x
Champagnon J, Elmberg J, Guillemain M, Gauthier-Clerc M, Lebreton J-D (2012a) Conspecifics can be aliens too: a review of effects of restocking practices in vertebrates. J Nat Conserv 20(4):231–241. https://doi.org/10.1016/j.jnc.2012.02.002
Champagnon J, Gauthier-Clerc M, Lebreton J-D, Mouronval J-B, Guillemain M (2013b) Les canards colverts lâchés pour la chasse interagissent-ils avec les populations sauvages? (In french) Faune Sauvage 298:4-9
Champagnon J, Guillemain M, Elmberg J, Folkesson K, Gauthier-Clerc M (2010) Changes in Mallard Anas platyrhynchos bill morphology after 30 years of supplemental stocking. Bird Study 57:1–8
Champagnon J, Guillemain M, Elmberg J, Massez G, Cavallo F, Gauthier-Clerc M (2012b) Low survival after release into the wild: assessing the burden of captivity on Mallard physiology and behaviour. Eur J Wildl Res 58(1):255–267. https://doi.org/10.1007/s10344-011-0573-3
Champagnon J, Legagneux P, Souchay G, Inchausti P, Bretagnolle V, Bourguemestre F, van Ingen L, Guillemain M (2016) Robust estimation of survival and contribution of captive-bred Mallards Anas platyrhynchos to a wild population in a large-scale release programme. Ibis 158(2):343–352. https://doi.org/10.1111/ibi.12341
Cheng KM, Shoffner RN, Phillips RE, Lee FB (1978) Mate preference in wild and domesticated (game-farm) mallards (Anas platyrhynchos): I. Initial preference. Animal Behaviour 26. Part 4:996–1003
Cheng KM, Shoffner RN, Phillips RE, Lee FB (1979) Mate preference in wild and domesticated (game-farm) mallards: II. Pairing success. Animal Behaviour 27. Part 2:417–425
Čížková D, Javůrková V, Champagnon J, Kreisinger J (2012) Duck’s not dead: does restocking with captive bred individuals affect the genetic integrity of wild mallard (Anas platyrhynchos) population? Biol Conserv 152:231–240. https://doi.org/10.1016/j.biocon.2012.04.008
Crnokrak P, Barrett SCH (2002) Perspective: purging the genetic load: a review of the experimental evidence. Evol Int J Org Evol 56(12):2347–2358. https://doi.org/10.1111/j.0014-3820.2002.tb00160.x
Deceuninck B, Quaintenne G, Ward A, Dronneau C, Mahéo R (2014) Synthèse des dénombrements d'anatidés et de foulques hivernant en France à la mi-Janvier 2013. Ligue pour la Protection des Oiseaux, Wetlands International, Ministère de l'Ecologie, du Développement durable, des Transports et du Logement, Rochefort, France (in French, summary in English)
Delany S, Scott DA (2006) Waterbird population estimates, 4th edn. Wageningen, Wetlands International
Deregnaucourt S, Guyomarc'h JC, Aebischer NJ (2002) Hybridization between European Quail Coturnix coturnix and Japanese Quail Coturnix japonica Ardea 90:15–21
Desforges MF, Wood-Gush DGM (1975) A behavioural comparison of domestic and mallard ducks. Habituation and flight reactions. Animal Behaviour 23, Part 3:692–697. https://doi.org/10.1016/0003-3472(75)90145-1
Desforges MF, Wood-Gush DGM (1976) Behavioural comparison of aylesbury and mallard ducks: sexual behaviour. Anim Behav 24(2):391–397. https://doi.org/10.1016/S0003-3472(76)80049-8
Dessborn L, Elmberg J, Englund G, Arzél C (2012) Innate responses of mallard ducklings towards aerial, aquatic and terrestrial predators. Behaviour 149(13-14):1299–1317. https://doi.org/10.1163/1568539x-00003014
Earl DA, vonHoldt BM (2012) STRUCTURE HARVESTER: a website and program for visualizing STRUCTURE output and implementing the Evanno method. Conservation Genet Resour 4(2):359–361. https://doi.org/10.1007/s12686-011-9548-7
Evanno G, Regnaut S, Goudet J (2005) Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol Ecol 14(8):2611–2620. https://doi.org/10.1111/j.1365-294X.2005.02553.x
Frosch C, Kraus RHS, Angst C, Allgöwer R, Michaux J, Teubner J, Nowak C (2014) The genetic legacy of multiple beaver reintroductions in Central Europe. PLoS One 9(5):e97619. https://doi.org/10.1371/journal.pone.0097619
Grant PR, Grant BR (1992) Hybridization of bird species. Science 256(5054):193–197. https://doi.org/10.1126/science.256.5054.193
Guay P-J, Iwaniuk AN (2008) Captive breeding reduces brain volume in waterfowl (Anseriformes). Condor 110(2):276–284. https://doi.org/10.1525/cond.2008.8424
Guillemain M, Elmberg J, Gauthier-Clerc M, Massez G, Hearn R, Champagnon J, Simon G (2010) Wintering French mallard and teal are heavier and in better body condition than 30 years ago: effects of a changing environment? Ambio 39(2):170–180. https://doi.org/10.1007/s13280-010-0020-9
Gunnarsson G, Latorre-Margalef N, Hobson KA, van Wilgenburg SL, Elmberg J, Olsen B, Fouchier RAM, Waldenström J (2012) Disease dynamics and bird migration—linking mallards Anas platyrhynchos and subtype diversity of the influenza A virus in time and space. PLoS One 7(4):e35679. https://doi.org/10.1371/journal.pone.0035679
Hill DA, Robertson P (1986) Hand reared pheasants: how do they compare with wild birds? Game Conser Ann Rep 17:76–84
Hirschfeld A, Heyd A (2005) Mortality of migratory birds caused by hunting in Europe: bag statistics and proposals for the conservation of birds and animal welfare Berichte zum Vogelschutz 42:47–74
Hůda J, Hanzal V, Kunitzká E, Plaňanská J Chov kachny divoké v honitbách Rybářství Třeboň. In: Conference Pernatá zvěř 2001. In: Sborník referátů z celostátní konference s mezinárodní účastí. Konopiště u Benešova 7.-8. září 2001: pp. 141–142. Česká lesnická společnost, Praha (abstract book, in Czech). 2001
Jakobsson M, Rosenberg NA (2007) CLUMPP: a cluster matching and permutation program for dealing with label switching and multimodality in analysis of population structure. Bioinformatics 23(14):1801–1806. https://doi.org/10.1093/bioinformatics/btm233
Jombart T (2008) adegenet: a R package for the multivariate analysis of genetic markers. Bioinformatics 24(11):1403–1405. https://doi.org/10.1093/bioinformatics/btn129
Jombart T, Devillard S, Balloux F (2010) Discriminant analysis of principal components: a new method for the analysis of genetically structured populations. BMC Genet 11(1):94. https://doi.org/10.1186/1471-2156-11-94
Jonker RM, Kraus RHS, Zhang Q, van Hooft P, Larsson K, van der Jeugd HP, Kurvers RHJM, van Wieren SE, Loonen MJJE, Crooijmans RPMA, Ydenberg RC, Groenen MAM, Prins HHT (2013) Genetic consequences of breaking migratory traditions in barnacle geese Branta leucopsis. Mol Ecol 22(23):5835–5847. https://doi.org/10.1111/mec.12548
Jonker RM, Zhang Q, van Hooft P, Loonen MJJE, van der Jeugd HP, Crooijmans RPMA, Groenen MAM, Prins HHT, Kraus RHS (2012) The development of a genome wide SNP set for the Barnacle goose Branta leucopsis. PLoS One 7(7):e38412. https://doi.org/10.1371/journal.pone.0038412
Keenan K, McGinnity P, Cross TF, Crozier WW, Prodöhl PA (2013) diveRsity: an R package for the estimation and exploration of population genetics parameters and their associated errors. Methods Ecol Evol 4(8):782–788. https://doi.org/10.1111/2041-210X.12067
Kraus RH et al (2011a) Genome wide SNP discovery, analysis and evaluation in mallard (Anas platyrhynchos). BMC Genomics 12(1):150. https://doi.org/10.1186/1471-2164-12-150
Kraus RH et al (2012) Widespread horizontal genomic exchange does not erode species barriers among sympatric ducks. BMC Evol Biol 12(1):45. https://doi.org/10.1186/1471-2148-12-45
Kraus RH, van Hooft P, Megens HJ, Tsvey A, Fokin SY, Ydenberg RC, Prins HH (2013) Global lack of flyway structure in a cosmopolitan bird revealed by a genome wide survey of single nucleotide polymorphisms. Mol Ecol 22(1):41–55. https://doi.org/10.1111/mec.12098
Kraus RH, Zeddeman A, van Hooft P, Sartakov D, Soloviev SA, Ydenberg RC, Prins HH (2011b) Evolution and connectivity in the world-wide migration system of the mallard: inferences from mitochondrial DNA. BMC Genet 12(1):99. https://doi.org/10.1186/1471-2156-12-99
Kraus RS, Wink M (2015) Avian genomics: fledging into the wild. J Ornithol 156(4):1–15. https://doi.org/10.1007/s10336-015-1253-y
Kulikova IV, Drovetski SV, Gibson DD, Harrigan RJ, Rohwer S, Sorenson MD, Winker K, Zhuravlev YN, McCracken KG (2005) Phylogeography of the mallard (Anas playrhynchos): hybridization, dispersal, and lineage sorting contribute to complex geographic structure. Auk 122(3):949–965. https://doi.org/10.1642/0004-8038(2005)122[0949:POTMAP]2.0.CO;2
Laikre L, Palmé A, Josefsson M, Utter F, Ryman N (2006) Release of alien populations in Sweden. Ambio 35(5):255–261. https://doi.org/10.1579/05-A-060R.1
Laikre L, Schwartz MK, Waples RS, Ryman N, Group GW (2010) Compromising genetic diversity in the wild: unmonitored large-scale release of plants and animals. Trends Ecol Evol 25(9):520–529. https://doi.org/10.1016/j.tree.2010.06.013
Liu Y, Keller I, Heckel G (2012) Breeding site fidelity and winter admixture in a long-distance migrant, the tufted duck (Aythya fuligula). Heredity 109(2):108–116. https://doi.org/10.1038/hdy.2012.19
Lynch M, O’Hely M (2001) Captive breeding and the genetic fitness of natural populations. Conserv Genet 2(4):363–378. https://doi.org/10.1023/A:1012550620717
Mank JE, Carlson JE, Brittingham MC (2004) A century of hybridization: decreasing genetic distance between American black ducks and mallards. Conserv Genet 5(3):395–403. https://doi.org/10.1023/B:COGE.0000031139.55389.b1
McGinnity P, Jennings E, deEyto E, Allott N, Samuelsson P, Rogan G, Whelan K, Cross T (2009) Impact of naturally spawning captive-bred Atlantic salmon on wild populations: depressed recruitment and increased risk of climate-mediated extinction. Proc R Soc B Biol Sci 276(1673):3601–3610. https://doi.org/10.1098/rspb.2009.0799
McGinnity P, Prodohl P, Ferguson A, Hynes R, Maoileidigh N, Baker N, Cotter D, O'Hea B, Cooke D, Rogan G, Taggart J, Cross T (2003) Fitness reduction and potential extinction of wild populations of Atlantic salmon, Salmo salar, as a result of interactions with escaped farm salmon. Proc R Soc Lond Ser B Biol Sci 270(1532):2443–2450. https://doi.org/10.1098/rspb.2003.2520
Mondain-Monval J-Y, Girard O (2000) Le canard colvert, la sarcelle d’hiver et autres canards de surface Faune Sauvage 251:124-139
Moore SJ, Battley PF (2006) Differences in the digestive organ morphology of captive and wild Brown Teal Anas chlorotis and implications for releases. Bird Conserv Int 16(03):253. https://doi.org/10.1017/s0959270906000396
Muñoz-Fuentes V, Vilà C, Green AJ, Negro JJ, Sorenson MD (2007) Hybridization between white-headed ducks and introduced ruddy ducks in Spain. Mol Ecol 16(3):629–638. https://doi.org/10.1111/j.1365-294X.2006.03170.x
Nussberger B, Greminger MP, Grossen C, Keller LF, Wandeler P (2013) Development of SNP markers identifying European wildcats, domestic cats, and their admixed progeny. Mol Ecol Resour 13(3):447–460. https://doi.org/10.1111/1755-0998.12075
Nygård T, Larsen BH, Follestad A, Strann K-B (1988) Numbers and distribution of wintering waterfowl in Norway
Ottosson U et al (2012) Fåglarna i Sverige - antal och förekomst. Sveriges Ornitologiska Förening, Halmstad
Price EO (1999) Behavioral development in animals undergoing domestication. Appl Anim Behav Sci 65(3):245–271. https://doi.org/10.1016/S0168-1591(99)00087-8
Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics 155(2):945–959
Puigcerver M, Sanchez-Donoso I, Vila C, Sarda-Palomera F, Garcia-Galea E, Rodriguez-Teijeiro JD (2014) Decreased fitness of restocked hybrid quails prevents fast admixture with wild European quails. Biol Conserv 171:74–81. https://doi.org/10.1016/j.biocon.2014.01.010
Puigcerver M, Vinyoles D, Rodriguez-Teijeiro JD (2007) Does restocking with Japanese quail or hybrids affect native populations of common quail Coturnix coturnix? Biol Conserv 136(4):628–635. https://doi.org/10.1016/j.biocon.2007.01.007
R Development Core Team (2009) R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna
Raftovich RV, Chandler SC, Wilkins KA (2015) Migratory bird hunting activity and harvest during the 2013-14 and 2014-15 hunting seasons. U.S. Fish and Wildlife Service, 68pp, Laurel, Maryland, USA
Randi E (2008) Detecting hybridization between wild species and their domesticated relatives. Mol Ecol 17(1):285–293. https://doi.org/10.1111/j.1365-294X.2007.03417.x
Reudink MW, Kyle CJ, Nocera JJ, Oomen RA, GREEN M, Somers CM (2011) Panmixia on a continental scale in a widely distributed colonial waterbird. Biol J Linn Soc 102(3):583–592. https://doi.org/10.1111/j.1095-8312.2010.01608.x
Rhymer JM (2006) Extinction by hybridization and introgression in anatine ducks. Acta Zool Sin 52:583–585
Rhymer JM, Simberloff D (1996) Extinction by hybridization and introgression. Annu Rev Ecol Syst 27(1):83–109. https://doi.org/10.1146/annurev.ecolsys.27.1.83
Rosenberg NA (2004) DISTRUCT: a program for the graphical display of population structure. Mol Ecol Notes 4(1):137–138. https://doi.org/10.1046/j.1471-8286.2003.00566.x
Ryman N (1981) Conservation of genetic resources: experiemces from the brown trout (Salmo trutta) Ecological. Bulletin 34:61–74
Saltonstall K (2002) Cryptic invasion by a non-native genotype of the common reed, Phragmites australis, into North America. Proc Natl Acad Sci 99(4):2445–2449. https://doi.org/10.1073/pnas.032477999
Sanchez-Donoso I, Huisman J, Echegaray J, Puigcerver M, Rodríguez-Teijeiro JD, Hailer F, Vilà C (2014) Detecting slow introgression of invasive alleles in an extensively restocked game bird. Front Ecol Evol 2:15
Sanchez-Donoso I, Vila C, Puigcerver M, Butkauskas D, Caballero de la Calle JR (2012) Are farm-reared quails for game restocking really common quails (Coturnix coturnix)?: a genetic approach. PLoS One 7(6):e39031. https://doi.org/10.1371/journal.pone.0039031
Scott DA, Rose P (1996) Atlas of anatidae populations in Africa and western Eurasia. Wetland International Publication No.41, Wageningen, The Netherlands
Šťastný K, Bejček V, Hudec K (2009) Atlas hnízdního rozšíření ptáků v České republice: 2001–2003. Aventinum, Prague, Czech Republic
Sutherland WJ et al (2006) The identification of 100 ecological questions of high policy relevance in the UK. J Appl Ecol 43(4):617–627. https://doi.org/10.1111/j.1365-2664.2006.01188.x
Swedish Board of Agriculture (2011) Imported eggs, E-mail conversation from Swedish Board of Agriculture to Söderquist, P. edn.,
Sýkora I (2005) Bažant a kachna – Umělý odchov a stavy v přírodě Myslivost 7:34
Söderquist P (2015) Large-scale releases of native species: the mallard as a predictive model system. Swedish University of Agricultural Sciences
Söderquist P, Gunnarsson G, Elmberg J (2013) Longevity and migration distance differ between wild and hand-reared mallards Anas platyrhynchos in Northern Europe. Eur J Wildl Res 59(2):159–166. https://doi.org/10.1007/s10344-012-0660-0
Söderquist P, Norrström J, Elmberg J, Guillemain M, Gunnarsson G (2014) Wild mallards have more “goose-like” bills than their ancestors: a case of anthropogenic influence? PLoS One 9(12):e115143. https://doi.org/10.1371/journal.pone.0115143
Taysom A, Johnson J, Guay PJ (2014) Establishing a genetic system to distinguish between domestic Mallards, Pacific Black Ducks and their hybrids. Conservation Genet Resour 6(1):197–199. https://doi.org/10.1007/s12686-013-0054-y
Tubaro PL, Lijtmaer DA (2002) Hybridization patterns and the evolution of reproductive isolation in ducks. Biol J Linn Soc 77(2):193–200. https://doi.org/10.1046/j.1095-8312.2002.00096.x
Wahlund S (1928) Zusammensetzung von populationen und korrelationserscheinungen vom standpunkt der vererbungslehre aus betrachtet. Hereditas 11(1):65–106. https://doi.org/10.1111/j.1601-5223.1928.tb02483.x
Wetlands International (2015) Waterbird population estimates. wpewetlandsorg Accessed 26 August 2015 2015
Acknowledgements
We are grateful to The Swedish Museum of Natural History in Stockholm, The Museum of Evolution in Uppsala, The Natural History Museum in Gothenburg, The Bird Museum in Jönköping, The Museum of Zoology in Lund, Tromsø University Museum, The Natural History Museum in Helsinki, Kuopio Natural History Museum, The natural history museums in Paris, Toulon, Avignon, Aix en Provence, Nîmes, Marseille, Istres, Gap, Digne, Saint Gilles, Montpellier, Salon de Provence, Port St-Louis du Rhône, Nantes, La Rochelle, to the Department of Zoology of the National Museum in Prague, The Moravian Museum in Brno, The Čáslav museum, The Regional Beskyd Museum in Frýdek-Místek, The Museum of Ostrava, The Regional Museum in Teplice, The Regional Museum in Olomouc, The Ornithological Station of the Comenius Museum in Přerov, The Museum in Ústí nad Labem, The Museum of South-East Moravia in Zlín, The East Bohemian Museum in Pardubice, The Museum of the Labe Region in Poděbrady and their respective curators, as well as to owners and curators of private collections: Tour du Valat and Restaurant “le Flamant rose” for access to their collections. We are also grateful to hunters in Dalarna, Norrbotten, and Västerbotten, The Swedish Association for Hunting and Wildlife Management, Swedish Gamekeepers Association, Agusa hunting estate, game estate managers in France, wardens in Marais du Vigueirat, F. Cavallo, J.-B. Mouronval, M. Vittecoq, U. Haapaniemi, A. Glavhammar, R. Dahlström, V.-M. Väänänen, J. van Dijk, M. Wille, J. Chapman, D. Bengtsson, and to breeding facilities in Sweden and France for the help with collecting samples. Veronika Javůková Dagmar Čížková and Tomáš Albrecht participated in collecting and DNA extractions of Czech samples. The Czech part of the project was supported by VAV grant no. SP2D 3-60-08. Work by Robert Kraus was financially supported by the KNJV (Royal Netherlands Hunters Association). A special thanks to P. Sernert and Osby Naturbruksgymnasium, Å Gunnarsson, and R. Hessel for the support and help with the field work. We also thank C. Halldén, A. Lidén, H. Königsson, M. Groenen, and the Animal Breeding and Genomics laboratory, Wageningen University, and especially B. Dibbits for the technical assistance. The Structure calculations were performed on the CSC HPC cluster FUCHS of J. W. Goethe University of Frankfurt, Germany. The project was funded by grant V-205-09 from the Swedish Environmental Protection Agency.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Söderquist, P., Elmberg, J., Gunnarsson, G. et al. Admixture between released and wild game birds: a changing genetic landscape in European mallards (Anas platyrhynchos). Eur J Wildl Res 63, 98 (2017). https://doi.org/10.1007/s10344-017-1156-8
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s10344-017-1156-8