Abstract
Insecticide resistance in Aedes mosquitoes presents a major challenge to the control of arboviral diseases. However, resistance mechanisms for many of the insecticides remain unknown. A commonly used insecticide, deltamethrin, was used to select a resistance strain of the vector mosquito, Aedes albopictus, and we identified an F1534S substitution in the voltage-gated sodium channel (VGSC) gene product as the first event in generating resistance. Engineering an F1534S substitution using Cas9/gRNA technologies conferred deltamethrin resistance on a previously susceptible strain. Crosses that removed this mutation restored the susceptible phenotype. Predicted protein structural changes and differences in transcript accumulation levels were correlated with the resistance phenotype. Furthermore, F1534S mutations were detected in all resistant Ae. albopictus populations collected in the field. We conclude that the VGSC F1534S mutation is essential for resistance to deltamethrin in Ae. albopictus, and is a suitable molecular index for pyrethroid resistance detection and monitoring in this species.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Key message
-
Target-site resistance plays an initial and important role in the evolution of deltamethrin resistance in Aedes albopictus.
-
An F1534S substitution in the VGSC gene product is essential and sufficient in generating deltamethrin resistance in Aedes albopictus.
-
F1534S mutations were detected in the resistant Aedes albopictus populations collected in the field.
Introduction
Mosquito-borne diseases cause a significant burden to human health. The Asian tiger mosquito, Aedes albopictus, is the vector for several arboviruses, including dengue, Zika and chikungunya fever viruses (Paupy et al. 2009), and also is the most invasive mosquito species in the world (Bonizzoni et al. 2013). Within the past half-century, Ae. albopictus has expanded from its Southeast Asia origin to all continents except Antarctica (Chen et al. 2015), bringing with it a major threat to public health. Vector control, especially with synthetic insecticides, is the main strategy of mosquito-borne disease prevention and control (WHO 2011). However, wide and improper application of insecticides has led to ever-increasing insecticide resistance, especially against the most commonly used pyrethroids (Su et al. 2019), and this has compromised the effectiveness of many control programs (WHO 2012). Understanding the molecular mechanisms of resistance and resistance evolution under selection in Ae. albopictus is essential to the development of reliable resistance detection methods (Ishak et al. 2016) and resistance management (Shi et al. 2015).
Recognized mechanisms of pyrethroid insecticide resistance in mosquitoes include mutations in genes that affect the binding affinity of insecticides (target-site resistance) (Marcombe et al. 2019; Xu et al. 2016), such as knockdown resistance (kdr) caused by point mutations in the voltage-gated sodium channel (VGSC) genes, the targets of pyrethroids and DDT (Dong et al. 2014), and metabolic detoxification of insecticides prior to reaching their targets (metabolic resistance) (Liu 2015; Marcombe et al. 2019). In addition, mechanisms such as cuticle modification (Balabanidou et al. 2016, 2018), behavioral resistance (Chareonviriyaphap et al. 2013) and chemosensory protein protection (Ingham et al. 2020) have been proposed. The association of these mechanisms to insecticide resistance is often based on statistical relationships from field populations, which are prone to confounding factors such as the complex history of insecticide use, life history of the mosquito populations and fluctuating environmental conditions. Establishment of a causal relationship between specific mechanisms and insecticide resistance has been challenging due to the lack of tools for gene function confirmation and epistasis.
To date, most resistance detection methods are based on mutations in genes that cause target-site insensitivity (Saingamsook et al. 2017; Zhu et al. 2019). When insecticide target genes have multiple alternate alleles or are part of closely related gene families, the contribution to resistance of individual alleles or genes must be quantified. Du et al. (2013) expressed an Ae. aegypti sodium channel, AaNav1-1, in Xenopus oocytes and provided in vitro evidence for pyrethroid resistance conferred by mutations in target-site genes. For in vivo confirmation, recent studies using Cas9/guide RNA (gRNA) technologies have demonstrated that mutations in insecticide target sites can lead to increased resistance in Drosophila melanogaster (Douris et al. 2020; Samantsidis et al. 2020; Xue et al. 2021), Spodoptera exigua (Zuo et al. 2017) and Anopheles gambiae (Grigoraki et al. 2021). Resistance mechanisms in Aedes mosquito remain unclear due to the lack of individual studies, in vivo experiments and causal evidence.
We used deltamethrin, a common pyrethroid insecticide (WHO 2012; Godfray 2013), to screen a susceptible laboratory strain of Ae. albopictus to select for a resistant strain and explore the mechanism of deltamethrin resistance in this species.
Materials and methods
Mosquito strains and collection
This study used three laboratory Ae. albopictus strains (Supplementary Table 1): (1) a strain provided by the Shanghai Center for Disease Control and Prevention (Lab-S); (2) a strain provided by the Beijing Center for Disease Prevention and Control (Lab-BJ); and (3) a strain provided by Jiangsu Institute of Parasitic Diseases (Lab-WX). The Lab-S strain was used as a reference strain for the resistance bioassay, the initial strain for selection of the laboratory resistant colony and the construction of the genome-edited strain.
All mosquito strains were reared in the standard insectary conditions (26 ± 2 °C, 14 h light: 10 h dark, 70 ± 10% relative humidity) without insecticide exposure. Larvae were fed with turtle food (the main ingredient is yeast powder, Gold-inch, Huizhou, China). Adult mosquitoes were housed in 20 cm × 20 cm × 35 cm cages and fed with 10% glucose. Female adults were blood fed from defibrinated sheep blood (Solarbio Life Sciences, Beijing, China) using a Hemotek membrane feeding system (Discovery Workshops, Accrington, UK).
Aedes albopictus were collected in 2021 from 11 field sites in Guangdong Province, China, including three cities in Guangdong Province: Jiangmen (JM), Zhanjiang (ZJ) and Guangzhou (GZ), all in a subtropical area with a monsoon climate and where it is the predominant dengue vector (Supplementary Table 1). Field-collected larvae and pupae were reared to adults and preserved for subsequent experiments.
Bioassays
Larval bioassay
The larval resistance bioassays were conducted following WHO guidelines (WHO 2005). Industrial-grade deltamethrin (94.6%) was provided by the Chinese Centers for Disease Control and Prevention. Twenty-five 3rd–4th instar larvae were added to 99 mL of dechlorinated tap water and 1 mL of different concentrations of insecticide solution. Five to nine concentrations, providing a range of mortalities between 10 and 95%, were used to determine LC50 values, three replicates were performed per concentration. Control bioassays were conducted by adding 1 ml of acetone to 99 ml of distilled water. Larval mortality was recorded after 24 h exposure.
Adult bioassay
The adult resistance bioassays were performed using 0.05% deltamethrin insecticide-impregnated papers (Universiti Sains Malaysia, Penang, Malaysia) following the standard WHO tube test protocol (WHO 2016a, b). In each holding tube, 25 non-blood-fed female adults aged 3–5 days post-emergence were tested with five replicates of field mosquitoes and two replicates of controls. After 1 h of exposure, the mosquitoes were transferred to holding tubes and fed on a 10% sucrose solution. Mortality was recorded 24 h after exposure, and the Lab-S strain was used as a control.
Selection of deltamethrin resistance strain
The Lab-S strain was used for selection of a deltamethrin resistance strain. Briefly, selection was performed by exposing the 4th instar larvae for 24 h to a 50% lethal concentration (LC50) of deltamethrin as in previous studies (Deng et al. 2021; Shi et al. 2015; Thornton et al. 2020). The LC50 concentration was determined for each generation by the larval resistance bioassay described above. After 24 h, the surviving larvae were transferred to dechlorinated water for rearing to adults. During the selecting process, at least 1000 female adult mosquitoes were maintained at each generation to minimize the founder effect. Three to four days post-emergence, female adults of different generations were preserved for subsequent molecular genotyping. The selected resistant strain is named “Lab-R.”
DNA extraction and kdr mutation detection
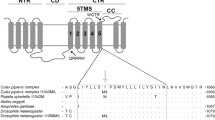
Genomic DNA was extracted from legs of individual female adults using the REDExtract-N-Amp™ Tissue PCR Kit (Sigma-Aldrich) following the manufacturer’s protocol. Portions of domains II (480 bp), III (346 bp) and IV (280 bp), covering 989, 1011, 1014, 1016, 1534 and 1763 sites of VGSC gene (codon nomenclature based on the Musca domestica VGSC gene, consistent a with previously established knockdown resistance (kdr) codon nomenclature method), were amplified by PCR, using published protocols and primers (Kasai et al. 2011). These sites were described in the literature as potentially associated with pyrethroid resistance in Aedes mosquitoes (Moyes et al. 2017). PCR products were purified with a MiniBEST DNA Fragment Purification Kit (Takara) and directly sequenced. Sequence analysis and alignments were performed with Chromas software and MEGA7 (version 7.1.0, http://www.megasoftware.net/).
CDC bottle bioassay and effect of synergists
The effect of cytochrome P450 monooxygenases (P450s), choline/carboxylesterases (CCEs) and the glutathione-S-transferase (GSTs) on the resistance of Lab-R strain to deltamethrin was tested using the synergistic agents piperonyl butoxide (PBO), S.S.S-tributlyphosphorotrithioate (DEF) and diethyl maleate (DM), respectively, according to the protocol described by CDC (Brogdon et al. 2011). Three- to five-day-old female Ae. albopictus without blood feeding were exposed to synergist-control bottles and synergist-exposure bottles for 1 h (25 mosquitoes per bottle with five replicates). After 1 h exposure, mosquitoes were transferred to holding cages for a 15-min recovery period. The CDC bottle bioassay was performed and mortality after 30 min (diagnostic time) was recorded.
Genomic engineering strategy
Design and preparation of sgRNA and ssODN
Guide Design Resources (http://crispor.tefor.net/) were used to design appropriate guide RNAs (sgRNA1 and sgRNA2) with PAM sequence close to the 1534 site and with few predicted genomic off-target sites (1 or 2 mismatches) that contain the T7 polymerase-binding site (Supplementary Table 2) and specifically target the sequences of VGSC locus 1534 (Fig. 1a). The template DNA for transcription in vitro of sgRNAs was prepared with PCR fusion of oligonucleotides by using Q5 HighFidelity DNAPolymerase (NEB Inc., MA, USA). The sgRNA templates were transcribed in vitro using the T7 RiboMAX Express Large Scale RNA Production System (Promega Corporation, Madison WI, USA) following the manufacturer's protocol. RNA transcripts were purified by phenol chloroform extraction and isopropanol precipitation, and then diluted to 1000 ng/µL in RNase-free water and stored at 80 °C. To introduce an F1534S mutation, a 121-bp single-stranded oligodeoxynucleotide (ssODN) (Supplementary Table 2) was designed to serve as a template for homology-directed repair (HDR) following the Cas9-induced double-strand breaks (DSBs) (Fig. 1b). The ssODN was synthesized by Thermo Fisher Scientific (Shanghai, China).
Cas9/gRNA-mediated F1534S mutation and resistance in relevant strains of Aedes albopictus. a Schematic structure of voltage-gated sodium channels (VGSC) in Ae. albopictus. b Design of a 121-bp ssODN as donor DNA for an F1534S substitution in VGSC. c Representative chromatograms of direct sequencing of the PCR products for genotyping 1534 site mutations. d Diagram detailing crossing schemes to obtain F1534S, S1534F, F′ and F1′ strains of Ae. albopictus. e Larval and adult bioassays of deltamethrin of the Lab-S, F1534S and S1534F strains, and F1 progeny from reciprocal crosses of Ae. albopictus. Error bars represent 95% CIs
Microinjection of embryos
Aedes albopictus embryo microinjection were performed using Eppendorf’s electronic microinjector FemtoJet® 4i (Eppendorf, Hamburg, Germany) with an ECLIPSE TS100 inverted microscope (Nikon, Tokyo, Japan) according to the previous protocol (Liu et al. 2019; 2020). The microcapillary needles (CAT# BF100-58-10, Sutter) were pulled with a Sutter P-97 Micropipette puller (Sutter, Novato, CA, USA) and were ground with a Narishige EG-400 Microgrinder (Setagaya-ku, Tokyo, Japan). Three to four days post-blood feeding, mated Ae. albopictus Lab-S female adults were allowed to lay eggs on a wet filter paper in a 300 mL plastic cup placed in the dark. After 20–30 min, the embryos were collected and were lined up on a filter paper immersed with microinjection buffer. The injection mixes contained 300 ng/µL NLS-Cas9-NLS Nuclease (Genscript, NJ, USA) with 100 ng/µL purified sgRNA and 100 ng/µL ssODN added to the injection buffer (Jasinskiene et al. 2007). The injection mixtures were incubated for 15 min at 37 °C to reconstitute active ribonucleoproteins (RNPs) and then were injected into embryos with suitable needles. All injected embryos were incubated for 3–5 days in insectary conditions for hatching, and the hatched larvae and emerged adults were designated G0.
Verification of Cas9/gRNA-induced mutations by high-resolution melting assay (HRMA)
The candidate sgRNAs (sgRNA1 and sgRNA2) were injected with Cas9 RNPs into Ae. albopictus embryos. Genomic DNA was extracted from Lab-S mosquitoes and injected embryos at 24 h post-injection using DNeasy Blood and Tissue Kits (Qiagen, Hilden, Germany). A high-resolution melting assay (HRMA) was conducted with an HRM Analysis Kit (EvaGreen, Tiangen, Beijing, China) on a LightCycler®96 System to identify successful substitution events (Supplementary Fig. 1b). Primers (HRM-F/R) were designed flanking the putative mutation site (Supplementary Fig. 1a and supplementary Table 2). PCR products were cloned into pMD18-T vector (Takara) and sequenced (Thermo Fisher Scientific, China). Alignment of amino acid sequences showed that the F1534S substitution occurred in the injected embryos, confirming that sgRNA1, sgRNA2 and ssODN could be used for subsequent embryonic injections (Supplementary Fig. 1c).
Genomic analysis of F1534S substitution mediated by Cas9/gRNA
Each G0 virgin female was mated with three wild-type (WT) males (Lab-S strain). In parallel, each G0 male was mated with three WT virgin females. Mated females were bloodfed twice, and subsequently, they laid eggs twice to ensure sufficient number of eggs produced. After laying eggs, genomic DNA samples from all G0 adults were extracted using DNeasy Blood and Tissue Kits (Qiagen, Hilden, Germany) for detection of F1534S mutations. To detect progeny genotypes, genomic DNA of mosquito legs was extracted using the Extract-N-Amp Tissue PCR Kit (Sigma-Aldrich) following the manufacturer’s protocol. The F1534S substitution in VGSC domains III of individuals were amplified using GoTaq® Green Master Mix (Promega) with PCR primers (1534-F/R) (Supplementary Table 2). The thermal conditions were 95 °C for 5 min, followed by 35 cycles of 95 °C for 30 s, 55 °C for 30 s and 72 °C for 30 s, with final extension step for 5 min at 72 °C. PCR products were directly sequenced with 1534-F primer by Thermo Fisher Scientific (Guangzhou, China). The sequences were analyzed using Chromas, and representative sequencing chromatograms are shown in Fig. 1c.
After detection of the F1534S substitution, the G1 progeny of G0 adults with successful substitutions was hatched separately (G1 from the same G0/pool). The 4th instar larvae of each G1 pool were selected randomly and genomic DNA extracted to determine whether the G0 F1534S mutation was transmitted to the next generation. The eggs of positive G1 pools with the F1534S mutation were raised to adulthood, and each adult mosquito was genotyped using the genomic DNA from legs (Fig. 1c). The G1 adults heterozygous for F1534S insertion were crossed with each other to produce G2. Those F1534S homozygotes were selected from G2 through the genotyping method and mass-crossed to establish the F1534S strain, which is homozygous for the F1534S mutation (Fig. 1d).
Reverse mutation at 1534 site of VGSC gene in F1534S strain
We used a genetic backcrossing method (Brito et al. 2013; Smith et al. 2018, 2021; Zuo et al. 2020) to reverse the F1534S mutation in the F1534S strain (Fig. 1d). Virgin males from Lab-S were mass-crossed with virgin females from the F1534S strain; then, virgin male adults of F1 progeny were backcrossed with virgin female adults from the F1534S strain to produce BC1. Genomic DNA was extracted from legs of individual male adults of BC1 to detect heterozygous individuals. The heterozygous male adults from BC1 were backcrossed with virgin female adults from the F1534S strain to produce BC2. The heterozygous male adults from BC2 were backcrossed with virgin female adults from the F1534S strain to produce BC3. Heterozygous BC3male then were crossed with virgin heterozygous BC3 females to produce BC3F1. Finally, heterozygous BC3F1 males were crossed with heterozygous BC3F1 females to establish the S1534F strain with 1534F/F homozygous genotype.
Genetic analysis of deltamethrin resistance in the F1534S strain
To obtained F1 and F1′ strains, virgin female adults of the Lab-S strain were mass-crossed reciprocally with virgin male adults of F1534S strain (Fig. 1d). Toxicological responses of F1 offspring from the reciprocal crosses to deltamethrin insecticide were determined using the bioassay described above. The degree of dominance (D) was estimated using Stone’s formula (Stone 1968): D = (2X2−X1−X3)/(X1−X3), where X1, X2 and X3 are log LC50 values of the F1534S strain, F1 hybrids and Lab-S strain, respectively.
Quantitative real-time PCR detection of the mRNA accumulation levels of VGSC gene
The 3rd–4th instar larvae (8 larvae/pool) and non-blood-fed female adults aged 3–5 days (5 females/pool) raised under the same conditions (100 larvae of each strain raised in 300 ml water with the same amount of turtle food every day) were collected from each strain (Lab-S strain, Lab-R strain, F1534S strain, S1534F strain and F1′ strain) to represent the larval and adult stages. Total RNA of larvae and adults was extracted using TRIzol (Ambion, Life Technologies, Carlsbad, CA, USA). cDNA was prepared using GoScript Reverse Transcription System (Promega, Madison, WI, USA) and random primers from pooled RNA of mosquitoes, and three biological replicates were used. All quantitative real-time PCR (RT-qPCR) assays for the relative quantification of VGSC transcript levels were performed in a LightCycler 96 (Roche) with Hieff® qPCR SYBR Green Master Mix (Yeasen, China). The primers employed for RT-qPCR are listed in Table S1. The relative quantification of VGSC gene was normalized to the internal control rpS7 (ribosomal protein 7) gene and analyzed using the 2−ΔΔCT method (Livak et al. 2001).
Molecular modeling of wild type and mutated protein VGSC
Molecular modeling was conducted by using the SWISS-MODEL (Waterhouse et al. 2018) server homology modeling pipeline, which relied on ProMod3, a comparative modeling engine based on OpenStructure (Biasini et al. 2013). We built a homology model based on the 1201-1800 amino acid sequence of VGSC in Ae. albopictus, using hNav1.2 (PDB code 6J8E) with sequence identity of 59.6% as the template structure. The F1534S mutations were performed in MOE v2018.0101. The structures of above proteins (WT and F1534S) were optimized by MD simulation. Each protein structure was neutralized by adding sodium/chlorine counter ions and solvated in a cuboid box of TIP3P water molecules with solvent layers 10 Å between the box edges and solute surface.
All MD simulations were performed using AMBER16 (Case et al. 2005). The AMBER FF14SB force field was applied, and the SHAKE algorithm was used to restrict all covalent bonds involving hydrogen atoms with a time step of 2 fs. The particle mesh Ewald (PME) method was employed to treat long-range electrostatic interactions. For each solvated system, two steps of minimization were performed before the heating step. The first 4000 cycles of minimization were performed with all heavy atoms restrained with 50 kcal/(mol Å2), whereas solvent molecules and hydrogen atoms were free to move. Non-restrained minimization then was carried out involving 2000 cycles of steepest descent minimization and 2000 cycles of conjugated gradient minimization. Afterward, the system was first heated from 0 to 300 K in 50 ps using the Langevin dynamics at a constant volume and then equilibrated for 400 ps at a constant pressure of 1 atm. A weak constraint of 10 kcal/(mol·Å2) was used to restrain all the heavy atoms during the heating steps. Periodic boundary dynamics simulations were carried out for the system with an NPT (constant composition, pressure and temperature) ensemble at a constant pressure of 1 atm and 300 K in the production step. In production phase, 200 ns simulation was carried out.
MOE Dock was used for molecular docking of proteins with small molecules. The 2D structure of deltamethrin was downloaded from PubChem and converted to 3D in MOE through energy minimization. The binding site of VGSC was identified by the Site Finder module in MOE. The docking workflow followed the “induced fit” protocol. The best-ranked pose was selected as the final binding mode.
Statistical analysis
For larvae bioassays, the extent of resistance was measured by the resistance ratio (RR50), which is calculated as the ratio of LC50 for the Lab-R population at the specific test generation to the LC50 of the Lab-S strain. LC50 were estimated using the log-probit models. Larval resistance status was defined as susceptible if RR50 < 5, moderately resistant if 5 < RR50 < 10 and highly resistant if RR50 > 10 (WHO 2016b). For adult bioassays, resistant status definition follows the WHO classification criteria: resistant if mortality < 90%, probably resistant if mortality was between 90 and 98%, and susceptible if mortality > 98% (WHO 2016a, b). For CDC bottle bioassays, 98–100% mortality is classified as susceptible, 80–97% mortality as possible resistant, < 80% mortality as resistant (Brogdon et al. 2011). Linear and exponential regression analyses were used to determine correlations between deltamethrin susceptibility and generations under selection. The association between non-synonymous mutations and deltamethrin resistance was tested by Fisher’s exact test or the Chi-square test (when all n > 5), and the odds ratio (OR) was calculated for each mutation. A Chi-square test was used to examine differences in adult mortality rates between synergist-control groups and synergist-exposed groups. Significance tests for relative expression differences in gene expression data (ΔCT values) were compared using t-test. All statistical analyses were performed with SPSS 20.0 (IBM), and P < 0.05 was considered statistically significant.
Results
An F1534S substitution in VGSC in Ae. albopictus is linked to deltamethrin resistance
Selection of a laboratory deltamethrin-resistant Ae. albopictus strain
To establish a stable resistant strain de novo, we used the most commonly used pyrethroid, deltamethrin, to select Ae. albopictus larvae for increased resistance in a laboratory colony (Lab-S). According to the WHO classification criteria, Ae. albopictus resistance status was defined as resistant if the larval resistance ratio RR50 > 5 and adult mortality < 90%. The resistance level of larvae increased slowly in the first 12 generations following exposure to the insecticide (Fig. 2a and supplementary Table 3). The resistance ratio RR50 was 5.5 at the 12th generation, indicating moderate resistance (Fig. 2a and supplementary Table 3). Subsequently, resistance increased rapidly, and the population became highly resistant by the 18th generation, with an RR50 value of 11.75 and 83.0% adult mortality (Fig. 2a, b and supplementary Table 3). Resistance increased exponentially in larvae during the selection process (Fig. 2a), resulting in a gradual increase of resistance in adults (Fig. 2b). These data support the conclusion that an Ae. albopictus laboratory deltamethrin-resistant strain (Lab-R) could be derived successfully from the laboratory susceptible strain (Lab-S).
Dynamics of deltamethrin resistance and an F1534S mutation in Aedes albopictus. a Scatter plot and curve fitting of larval LC50 in different generations. Error bars represent 95% CIs. b Scatter plot and curve fitting of adult mortality in different generations. Error bars represent 95% CIs. c Genotype proportions of homozygous wild-type (SS) and knockdown resistance (kdr) heterozygous (RS) and homozygous (RR) mutations in different generations. d Metabolic resistance in different generations. Synergistic agent piperonyl butoxide (PBO), S.S.S-tributlyphosphorotrithioate (DEF) and diethyl maleate (DM) are respective inhibitors of cytochrome P450 monooxygenases (P450s), choline/carboxylesterases (CCEs) and the glutathione-S-transferase (GSTs). Vertical bars represent the standard error. *P < 0.05; **P < 0.01; ***P < 0.005
Target-site resistance and metabolic resistance in different selected generations
As the primary target of deltamethrin (Dong et al. 2014), mutations in VGSC gene were associated with the knockdown resistance (kdr) phenotype assessed throughout the whole screening process. The knockdown resistance phenotype is characterized by the ability of a mosquito to move after exposure to an insecticide. Heterozygous 1534F/S (TTC/TCC) mutations in domain III of the VGSC gene were detected in the 6th generation (RR50 = 2.50), and homozygous 1534S/S (TCC/TCC) mutants were detected in the 12th generation (RR50 = 5.50) (Fig. 2c and supplementary Table 4). The 1532I/T (ATC/ACC) and 1532T/T (ACC/ACC) mutations were detected in the 24th generation (Fig. 2c and supplementary Table 4). No non-synonymous mutations were found in domains II and IV of the Lab-R strain, including any at previously reported mutation sites (Moyes et al. 2017) (S989, I1011, L1014, V1016 and D1763). F1534S and I1532T mutations were associated significantly with the resistance to deltamethrin, with an odds ratios > 1 (Supplementary Table 4). The frequencies of F1534S mutation in the 1st–24th generations was correlated significantly with the LC50 of Lab-R strain (r = 0.975, P = 005, Spearman correlation analysis), indicating that the deltamethrin resistance level of the population was correlated with the increase in frequency of F1534S. For metabolic resistance, cytochrome P450 monooxygenase (P450s) activity was detected in the 24th generation as the first metabolic enzyme to reduce the deltamethrin resistance of adults, followed by choline/carboxylesterases (CCEs) and glutathione-S-transferases (GSTs) (Fig. 2d). These data indicate that target-site resistance played an earlier role than metabolic resistance in the phenotype and that the VGSC F1534S mutation is the first recorded event in the development of deltamethrin resistance in Ae. albopictus.
The F1534S mutation is essential and sufficient for the generation of deltamethrin resistance in Ae. albopictus
Cas9/gRNA-mediated F1534S substitution in VGSC of Ae. Albopictus
We hypothesized based on the previous results that Ae. albopictus deltamethrin resistance is initiated by the F1534S mutation in VGSC gene (Fig. 1a). In order to induce an HDR-mediated F1534S allele (Fig. 1b), a mixture of Cas9 protein, ssODN template and the specific sgRNAs were co-injected into 2651 fresh Ae. albopictus eggs. A total of 170 larvae hatched and 137 of these developed into adults in a generation designated G0 (Supplementary Table 5). Samples of genomic DNA of all G0 adults were extracted for gene amplification (PCR) detection after they were mated and females laid eggs. Sequencing of amplicons of G0 individuals revealed that 13 of them had successful insertions of the HDR-mediated F1534S mutation (9.5% efficiency). As detailed in the schematic diagram in Fig. 1d, we successfully established the F1534S strain with a homozygous mutation of F1534S, and sequencing chromatograms for genotyping wild type, heterozygotes and homozygotes at nucleotide 1534 are shown in Fig. 1c. Following backcrossing of the F1534S strain with the Lab-S strain, a “restored” susceptible strain, S1534F, was recovered (Fig. 1d).
The impact of F1534S on the susceptibility of Ae. albopictus to deltamethrin
The genome-edited F1534S strain showed high resistance to deltamethrin with a larval RR50 of 17.0 and 71.93% adult mortality (Fig. 1e and supplementary Table 6). A larval RR50 of 1.5 and 100.0% adult mortality in S1534F showed that lack of an F1534S mutation restores the susceptibility of Ae. albopictus to deltamethrin (Fig. 1e and supplementary Table 6). Heterozygosity of the mutation at the 1534 site in the F1 and F1′ offspring also can result in high deltamethrin resistance (Fig. 1e and supplementary Table 6). The selected resistance to deltamethrin in the F1534S strain is autosomal, and there was no evidence of a sex bias in the phenotype. The degrees of dominance (D) of the F1 and F1′ offspring were 0.625 and 0.875, respectively (Supplementary Table 6), indicating that deltamethrin resistance imparted by the F1534S mutation is incompletely dominant. These results show that the F1534S mutation can confer high levels of resistance to deltamethrin within a controlled genetic background of Ae. albopictus.
Predicted protein structural changes and differences in transcript abundance of VGSC are correlated with resistance
Molecular modeling and molecular docking of deltamethrin to VGSC proteins
The structure of the VGSC_WT and VGSC_F1534S was simulated to determine the effect of F1534S mutation on the VGSC protein. The modeling structure of VGSC_WT (codons 1201-1800) with multiple α-helices and β-sheets is depicted in Supplementary Fig. 2a, and a Ramachandran plot for VGSC_WT showed that 99% of the residues are in allowed regions (Supplementary Fig. 2b). Molecular dynamic (MD) simulations were performed to search for the stable structure of mutated VGSC_F1534S. For VGSC_WT and VGSC_F1534S, the root means square deviation (RMSD) from each MD simulations tended to converge after 50 ns and reached equilibrium after 200 ns MD simulation (Fig. 3b). The final stable structure of VGSC_WT and VGSC_F1534S at 200 ns simulation time from MD trajectory showed that the overall conformations of VGSC_F1534S (RMSD = 5.383 Å) was similar with VGSC_WT (Fig. 3a). The docking score of final stable structure of two proteins with deltamethrin showed that VGSC_WT was more sensitive to deltamethrin than VGSC_F1534S (Fig. 3c). Deltamethrin interacted with Arg1405 and Lys1341 via hydrogen bonding and formed H−π stacking with the sidechain of Gln1402 (Supplementary Fig. 2c, e), which mainly contribute to the binding energy between deltamethrin with the VGSC_WT protein. When binding to the VGSC_F1534S, only a hydrogen bond was formed between deltamethrin and Arg1405 (Supplementary Fig. 2d, f). These interactions of deltamethrin with VGSC proteins are consistent with the docking scores. The results showed that the F1534S mutation did not significantly change the structure of VGSC protein, but could induce deltamethrin resistance by reducing the binding affinity of deltamethrin to the protein.
Comparisons of VGSC product structural changes and mRNA accumulation levels caused by an F1534S mutation. a Final stable structure at 200 ns simulation time of VGSC_WT superposed over VGSC_F1534S. VGSC_WT was colored in green and VGSC_F1534S in purple. The residue Phe1534 and mutated Ser1534 are shown in stick view with correspondent colored. b System flexibility analysis of VGSC_WT and VGSC_F1534S. The root means square deviation (RMSD) of the backbone of VGSC_WT is less than 8.0 angstrom and that of VGSC_F1534S is less than 11.0 angstrom, and the system achieves equilibrium within the simulation time. c Docking scores of deltamethrin molecules with VGSC_WT and VGSC_F1534S proteins. The more negative the docking score, the better the binding of ligand with protein. d Relative mRNA accumulation levels of the VGSC gene at larval and adult stages in Lab-S, F1534S, F1’and S1534F strains. e Relative mRNA accumulation level of VGSC genes in four strains at larval and adult stages. The fold change (FC) value of VGSC genes at larval stage in d was set to 1, and the FC value of VGSC genes in the Lab-S strain of e was set to 1. The results were normalized to Ae. albopictus Rps7 (AalRpS7). The results are represented as the mean ± standard error. ns: not significant. *P < 0.05, **P < 0.01, ***P < 0.005
Reduced accumulation of VGSC transcripts in mutant strains
VGSC gene expression profiles as indexed by mRNA accumulation levels in Lab-S, F1534S, F1′, S1534F and Lab-R strains at larval and adult stages were detected by RT-qPCR. The accumulation levels of VGSC gene mRNA in adult females of all strains were significantly higher than that in 3rd–4th instar larvae (Fig. 3d and supplementary Fig. 3a). Compared to the Lab-S strain, VGSC transcript levels in the F1534S strain, F1′ strain and Lab-R strain were lower in both larval and adult stages, while the abundance in S1534F strain showed no difference (Fig. 3e and supplementary Fig. 3b). These results support the interpretation that the VGSC gene is gradually up-regulated with the development of mosquitoes (Dong et al. 2014) and plays a key role in the resistant phenotype (based on the assumption that the mRNAs all have the same stability profile). The gene-edited strain provides the first evidence that the F1534S resistance phenotype is correlated with a low level of VGSC mRNA accumulation.
F1534S mutations are detected in deltamethrin-resistant Ae. albopictus populations collected in the field
A multiplex PCR for F1534S mutation detection in Ae. albopictus
An allele-specific PCR-based assays (AS-PCR) was designed to genotype the F1534S mutation in Ae. albopictus (Fig. 4). All primer sequences used in this AS-PCR are shown in Supplementary Table 2. Sequenced, genotyped samples for the 1534 alleles in the Lab-R strain were used to validate the multiplex PCR method. A total of nine samples covering three genotypes (three individuals each of FF wild-type homozygotes, FS heterozygotes and SS mutant homozygotes) were tested, and a total of three predicted amplicons for each genotype are shown in Supplementary Fig. 4. The largest amplicon is 589 base-pairs in length (bp) and represents the positive control. The moieties at 240 and 589 bp verify the wild-type FF, while those at 390 and 589 bp indicate homozygous mutant SS, and the presence of all three amplicons represent heterozygous FS. Surveillance of Ae. albopictus field populations for F1534S-mediated deltamethrin resistance in genomic DNA samples of multiple larvae or adult mosquitoes can be done quickly using this multiplex PCR.
Schematic diagram showing primer locations and amplicon product sizes of the AS-PCR assay for F1534S mutation in Aedes albopictus. A pair of common primers (Aal1534F and Aal1534R) were designed to amplify a control amplicon of 589 bp to verify the availability of the template. Two specific primers (AalF1534R and AalS1534F) were designed to amplify two amplicons of different lengths to distinguish the F1534 (240 bp) and S1534 alleles (390 bp). Genomic DNA for mosquito samples were extracted from legs of individual adults using the REDExtract-N-AmpTM Tissue PCR Kit (Sigma-Aldrich) or from the larval or adult tissue using DNeasy Blood and Tissue Kits (Qiagen, Hilden, Germany). The multiplex PCR conditions were optimized for a total reaction volume of 25 μL:12.5 μL of DreamTaq Green PCR Master Mix (Thermo Fisher), 1.0 μL (10 μM) Aal1534F and Aal1534R primers, 0.5 μL (10 μM) AalF1534R and AalS1534F primers, 2 μL of DNA template, and made up to 25 μL with ddH2O. The amplification consisted of 95 °C for 3 min pre-denaturation step, followed by 35 cycles of 95 °C for 30 s, 58 °C for 30 s and 72 °C for 30 s, and a final extension step at 72 °C for 5 min. The amplified PCR products were stored at 4 °C. After PCRs, the amplified products were analyzed on 1% agarose gel with a low molecular weight DNA ladder to estimate the band size and F1534S genotyping
Detection of deltamethrin resistance and F1534S mutations in laboratory and field populations
We tested multiple populations of Ae. albopictus (Supplementary Table 1) to explore the relationship between deltamethrin resistance and the 1534 site mutation. The results of larval and adult bioassays of laboratory strains (Lab-S, Lab-BJ and Lab-WX) showed that both of developmental stages were susceptible to deltamethrin (Fig. 5b, c) and the genotyping of the 1534 VGSC gene site were wild type (TTC) (Fig. 5a and Supplementary Table 7). With the exception of the susceptible JMHS strain (RR50 = 3.0), all other field populations were highly resistant to deltamethrin (RR50 > 10). No mutations of the 1534 site were detected in the field-derived susceptible population, JMHS, while the F1534S mutation (TCC) and other types of mutations were detected in all of the resistant populations (Fig. 5 and Supplementary Table 7). These data verify that there is no mutation at 1534 site in the susceptible population, while in contrast, F1534S mutations were correlated with populations that exhibited the deltamethrin resistance.
Deltamethrin resistance and kdr mutations at 1534 site in laboratory- and field-derived populations of Aedes albopictus. a Frequency of kdr alleles in different Ae. albopictus populations. "Other" amino acid substitutions include 1534L and 1534C. b Adult mortality of different populations. c Larval LC50 of different populations. Error bars represent 95% CIs
Discussion
We show here that deltamethrin resistance in Ae. albopictus progresses from a slow to rapid phase during selection, and this is consistent with previous studies of field-collected populations of Culex pipiens pallens (Shi et al. 2015) and A. aegypti (Kumar et al. 2002). We also discovered that F1534S substitution in the voltage-gated sodium channel, appearing early during the exposure to deltamethrin, is essential to deltamethrin resistance in Ae. albopictus. These findings support the conclusion that target-site resistance plays an initial and important role in the evolution of deltamethrin resistance in Ae. albopictus. It is worth mentioning that the classical kdr mutation (L1014F) is one of the most widely distributed resistance alleles in pyrethroid-resistant Anopheles mosquitoes and is used as a marker of pyrethroid resistance in malaria vectors (Koukpo et al. 2019; Zhang et al. 2021; Cruz et al. 2021). Different from the mutation L1014F in Anopheles mosquitoes, the kdr mutation at locus 1534 has been widely reported in pyrethroid-resistant Ae. albopictus in the field, especially the high frequency of F1534S mutation (Chen et al. 2021; Wei et al. 2021; Wu et al. 2021; Abernathy et al. 2022; Li et al. 2021; Zheng et al. 2022). These field studies support the conclusion that F1534S plays an important role in pyrethroid resistance of Ae. albopictus.
kdr mutations with adaptive potential come mainly from de novo mutations or from standing variation (Hawkins et al. 2018). In this study, we were able to reveal in Ae. albopictus the regularity and mechanism of pyrethroid resistance using a strain selected for resistance from a susceptible laboratory strain rather than field populations whose source of resistance alleles could not be determined (Melo-Santos et al. 2010; Shi et al. 2015; Thornton et al. 2020; Yang et al. 2021). The de novo mutation F1534S we found in the laboratory screening process of Ae. albopictus also was widely reported in the wild populations. This repeatability may be due to the selection pressure of deltamethrin not just on a single molecular target (VGSC) and even specific insecticide-binding residues (two pyrethroid receptor sites) (Du et al. 2013; Hawkins et al. 2018). However, it should be emphasized that more samples and studies are needed to verify the universality of the results due to the homogeneous genetic background of single replicate populations and differences in the selection process used here and what may result from the actual use of insecticides in the field.
Mutations in the VGSC gene in mosquitoes have proved by in vitro expression systems (Du et al. 2013; Yan et al. 2020) to be related to sensitivity to insecticides. However, some mutations, such as F1534S, were not able to confer resistance to Type II pyrethroids based on the electrophysiological data of an Ae. aegypti sodium channel AaNav1-1 (Yan et al. 2020), which was inconsistent with field research observations (Xu et al. 2016; Gao et al. 2018) because of the limitations of experiments in vitro and the complex systemic response in vivo. With Cas9/gRNA and backcrossing techniques, we introduced and recovered an F1534S mutation in the Ae. albopictus VGSC gene providing the first in vivo functional validation that an F1534S substitution is essential and sufficient for the resistance to deltamethrin, and demonstrating individually the causal relationship between a kdr phenotypes and resistance generation. Based on the resistance phenotype of the F1 and F1′ strains, we found that mosquitoes heterozygous for F1534S also exhibit a high level of resistance to deltamethrin, which differs from the recessive genetic trait of some target-site mutations in previous reports showing no or low level of resistance, such as V1016I and F1534C in mosquitoes (Saavedra-Rodriguez et al. 2007; Thornton et al. 2020), L1014S/F in Phlebotomus argentipes (Gomes et al. 2017), L1014F in house flies (Huang et al. 2004), and G4946E and I4743M in S. exigua (Zuo et al. 2017, 2020).
VGSC is the direct target of pyrethroid insecticides and mutations in it have been confirmed to reduce pyrethroid binding to sodium channels (Dong et al. 2014), leading to the resistance phenotype, which was also confirmed in our study by computer modeling. Yan et al. (2020) used computational modeling to explain the effects of kdr mutations on the interactions of pyrethroids with the wild-type sodium channel, but the mutation simulation of sodium channels was not carried out. Unlike the Nav1.4-based homology model of the AaNav1-1 channel built by Yan et al. (2020), the Nav1.2-based homology model built in this study was only based on the 1201-1800 amino acid sequence of VGSC in Ae. albopictus, and the mutant model was simulated to partly explain the change of interaction between deltamethrin and sodium channel after mutation. Interestingly, we found that the reduced accumulation of VGSC transcripts in the F1534S mutant strain and the normal levels in the S1534F restored strain were correlated with the status of deltamethrin resistance. This supports the hypothesis that pyrethroid resistance conferred by the mutation was associated not only with the decreased binding affinity, but also with the decrease of VGSC transcripts, which could reduce pyrethroid–target binding opportunities in the resistant phenotype (Kubik et al. 2021).
Based on previous studies (Chen et al. 2021; Wei et al. 2021; Wu et al. 2021; Abernathy et al. 2022; Li et al. 2021; Zheng et al. 2022) and the results of this study, we selected the F1534S mutation in VGSC as a molecular marker for screening deltamethrin resistance in Ae. albopictus and developed an AP-PCR method to detect the mutation in mosquitoes. The detections in Ae. albopictus samples derived either from laboratory or field supported the hypothesis that there is F1534S mutation in deltamethrin-resistant populations, and no F1534S mutations in the susceptible populations. These findings indicate that the detection of F1534S mutations in the VGSC gene can basically determine the generation of deltamethrin resistance in Ae. albopictus and could be used as a reference to develop a monitoring and warning system for Aedes mosquitoes to pyrethroid resistance.
Conclusions
With the serious insecticide resistance problems, studying the mechanisms of the generation and development of insecticide resistance is crucial for vector control strategies. Here, we found that F1534S mutation in VGSC gene, evolving early during the exposure to deltamethrin, is essential to deltamethrin resistance in Ae. albopictus. Using CRISPR/Cas9 techniques, we functionally characterize the F1534S substitution, showing that it confers high levels of deltamethrin resistance by altering the structure and expression levels of VGSC. Based on laboratory validation and field population detection, a diagnostic gene amplification tool was developed for surveillance of wild and laboratory populations. These latest findings reveal cause-and-effect mechanisms of pyrethroid resistance in mosquitoes, establishing a link between mechanistic studies and vector control strategies.
Author contributions
XGC and YJG conceived and designed the study. YJG, JNZ, YJZ, JLD and XHS performed the field collection and resistance bioassays. YJG and JLD conducted laboratory analyses. JXT, GDZ, XJZ and JBG processed the data. YJG wrote the manuscript with assistance from GYY, AAJ and XGC. All authors reviewed and approved the final draft of the paper.
References
Abernathy HA, Hollingsworth BD, Giandomenico DA et al (2022) Prevalence of knock-down resistance F1534S mutations in Aedes albopictus (Skuse) (Diptera: Culicidae) in North Carolina. J Med Entomol 59:1363–1367. https://doi.org/10.1093/jme/tjac054
Balabanidou V, Kampouraki A, MacLean M et al (2016) Cytochrome P450 associated with insecticide resistance catalyzes cuticular hydrocarbon production in Anopheles gambiae. Proc Natl Acad Sci USA 113:9268–9273. https://doi.org/10.1073/pnas.1608295113
Balabanidou V, Grigoraki L, Vontas J (2018) Insect cuticle: a critical determinant of insecticide resistance. Curr Opin Insect Sci 27:68–74. https://doi.org/10.1016/j.cois.2018.03.001
Biasini M, Schmidt T, Bienert S et al (2013) OpenStructure: an integrated software framework for computational structural biology. Acta Crystallogr D Biol Crystallogr 69:701–709. https://doi.org/10.1107/S0907444913007051
Bonizzoni M, Gasperi G, Chen X, James AA (2013) The invasive mosquito species Aedes albopictus: current knowledge and future perspectives. Trends Parasitol 29:460–468. https://doi.org/10.1016/j.pt.2013.07.003
Brito LP, Linss JG, Lima-Camara TN et al (2013) Assessing the effects of Aedes aegypti kdr mutations on pyrethroid resistance and its fitness cost. PLoS ONE 8:e60878. https://doi.org/10.1371/journal.pone.0060878
Brogdon W, Chan T (2011). Guideline for evaluating insecticide resistance in vectors using the CDC bottle bioassay. Insect Control/methods. Available from: https://stacks.cdc.gov/view/cdc/21777
Case DA, Cheatham TE 3rd, Darden T et al (2005) The Amber biomolecular simulation programs. J Comput Chem 26:1668–1688. https://doi.org/10.1002/jcc.20290
Chareonviriyaphap T, Bangs MJ, Suwonkerd W et al (2013) Review of insecticide resistance and behavioral avoidance of vectors of human diseases in Thailand. Parasit Vectors 6:280. https://doi.org/10.1186/1756-3305-6-280
Chen XG, Jiang X, Gu J et al (2015) Genome sequence of the Asian Tiger mosquito, Aedes albopictus, reveals insights into its biology, genetics, and evolution. Proc Natl Acad Sci U S A 112:E5907-5915. https://doi.org/10.1073/pnas.1516410112
Chen H, Zhou Q, Dong H et al (2021) The pattern of kdr mutations correlated with the temperature in field populations of Aedes albopictus in China. Parasit Vectors 14:406. https://doi.org/10.1186/s13071-021-04906-z
Cruz DLD, Paiva MHS, Guedes DRD et al (2021) First report of the L1014F kdr mutation in wild populations of Anopheles arabiensis in Cabo Verde. West Africa Parasit Vectors 14:582. https://doi.org/10.1186/s13071-021-05088-4
Deng J, Guo Y, Su X et al (2021) Impact of deltamethrin-resistance in Aedes albopictus on its fitness cost and vector competence. PLoS Negl Trop Dis 15:e0009391. https://doi.org/10.1371/journal.pntd.0009391
Dong K, Du Y, Rinkevich F et al (2014) Molecular biology of insect sodium channels and pyrethroid resistance. Insect Biochem Mol Biol 50:1–17. https://doi.org/10.1016/j.ibmb.2014.03.012
Douris V, Denecke S, Van Leeuwen T (2020) Using CRISPR/Cas9 genome modification to understand the genetic basis of insecticide resistance: Drosophila and beyond. Pestic Biochem Physiol 167:104595. https://doi.org/10.1016/j.pestbp.2020.104595
Du Y, Nomura Y, Satar G et al (2013) Molecular evidence for dual pyrethroid-receptor sites on a mosquito sodium channel. Proc Natl Acad Sci U S A 110:11785–11790. https://doi.org/10.1073/pnas.1305118110
Gao JP, Chen HM, Shi H et al (2018) Correlation between adult pyrethroid resistance and knockdown resistance (kdr) mutations in Aedes albopictus (Diptera: Culicidae) field populations in China. Infect Dis Poverty 7:86. https://doi.org/10.1186/s40249-018-0471-y
Godfray HC (2013) Mosquito ecology and control of malaria. J Anim Ecol 82:15–25. https://doi.org/10.1111/1365-2656.12003
Gomes B, Purkait B, Deb RM et al (2017) Knockdown resistance mutations predict DDT resistance and pyrethroid tolerance in the visceral leishmaniasis vector Phlebotomus argentipes. PLoS Negl Trop Dis 11:e0005504. https://doi.org/10.1371/journal.pntd.0005504
Grigoraki L, Cowlishaw R, Nolan T et al (2021) CRISPR/Cas9 modified An. gambiae carrying kdr mutation L1014F functionally validate its contribution in insecticide resistance and combined effect with metabolic enzymes. PLoS Genet 17:e1009556. https://doi.org/10.1371/journal.pgen.1009556
Hawkins NJ, Bass C, Dixon A et al (2018) The evolutionary origins of pesticide resistance. Biol Rev Camb Philos Soc 94:135–155. https://doi.org/10.1111/brv.12440
Huang J, Kristensen M, Qiao CL, Jespersen JB (2004) Frequency of kdr gene in house fly field populations: correlation of pyrethroid resistance and kdr frequency. J Econ Entomol 97:1036–1041. https://doi.org/10.1093/jee/97.3.1036
Ingham VA, Anthousi A, Douris V (2020) A sensory appendage protein protects malaria vectors from pyrethroids. Nature 577:376–380. https://doi.org/10.1038/s41586-019-1864-1
Ishak IH, Riveron JM, Ibrahim SS et al (2016) The cytochrome P450 gene CYP6P12 confers pyrethroid resistance in kdr-free Malaysian populations of the dengue vector Aedes albopictus. Sci Rep 6:24707. https://doi.org/10.1038/srep24707
Jasinskiene N, Juhn J, James AA (2007) Microinjection of Aedes aegypti embryos to obtain transgenic mosquitoes. J vis Exp. https://doi.org/10.1016/j.ibmb.2019.103311
Kasai S, Ng LC, Lam-Phua SG et al (2011) First detection of a putative knockdown resistance gene in major mosquito vector, Aedes albopictus. Jpn J Infect Dis 64:217–221. https://doi.org/10.7883/yoken.64.217
Koukpo CZ, Fassinou AJYH, Ossè RA et al (2019) The current distribution and characterization of the L1014F resistance allele of the kdr gene in three malaria vectors (Anopheles gambiae, Anopheles coluzzii, Anopheles arabiensis) in Benin (West Africa). Malar J 18:175. https://doi.org/10.1186/s12936-019-2808-9
Kubik TD, Snell TK, Saavedra-Rodriguez K et al (2021) Aedes aegypti miRNA-33 modulates permethrin induced toxicity by regulating VGSC transcripts. Sci Rep 11:7301. https://doi.org/10.1038/s41598-021-86665-6
Kumar S, Thomas A, Sahgal A et al (2002) Effect of the synergist, piperonyl butoxide, on the development of deltamethrin resistance in yellow fever mosquito, Aedes aegypti L. (Diptera: Culicidae). Arch Insect Biochem Physiol 50:1–8. https://doi.org/10.1002/arch.10021
Li Y, Zhou G, Zhong D et al (2021) Widespread multiple insecticide resistance in the major dengue vector Aedes albopictus in Hainan province, China. Pest Manag Sci 77:1945–1953. https://doi.org/10.1002/ps.6222
Liu N (2015) Insecticide resistance in mosquitoes: impact, mechanisms, and research directions. Annu Rev Entomol 60:537–559. https://doi.org/10.1146/annurev-ento-010814-020828
Liu T, Yang WQ, Xie YG et al (2019) Construction of an efficient genomic editing system with CRISPR/Cas9 in the vector mosquito Aedes albopictus. Insect Sci 26:1045–1054. https://doi.org/10.1111/1744-7917.12645
Liu P, Jin B, Li X et al (2020) Nix is a male-determining factor in the Asian tiger mosquito Aedes albopictus. Insect Biochem Mol Biol 118:103311. https://doi.org/10.1016/j.ibmb.2019.103311
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using realtime quantitative PCR and the 2−ΔΔCT method. Methods 25:402–408. https://doi.org/10.1006/meth.2001.1262
Marcombe S, Fustec B, Cattel J et al (2019) Distribution of insecticide resistance and mechanisms involved in the arbovirus vector Aedes aegypti in Laos and implication for vector control. PLoS Negl Trop Dis 13:e0007852. https://doi.org/10.1371/journal.pntd.0007852
Melo-Santos MA, Varjal-Melo JJ, Araujo AP et al (2010) Resistance to the organophosphate temephos: mechanisms, evolution and reversion in an Aedes aegypti laboratory strain from Brazil. Acta Trop 113:180–189. https://doi.org/10.1016/j.actatropica.2009.10.015
Moyes CL, Vontas J, Martins AJ et al (2017) Contemporary status of insecticide resistance in the major Aedes vectors of arboviruses infecting humans. PLoS Negl Trop Dis 11:e0005625. https://doi.org/10.1371/journal.pntd.0005625
Paupy C, Delatte H, Bagny L et al (2009) Aedes albopictus, an arbovirus vector: from the darkness to the light. Microbes Infect 11:1177–1185. https://doi.org/10.1016/j.micinf.2009.05.005
Saavedra-Rodriguez K, Urdaneta-Marquez L, Rajatileka S et al (2007) A mutation in the voltage-gated sodium channel gene associated with pyrethroid resistance in Latin American Aedes aegypti. Insect Mol Biol 16:785–798. https://doi.org/10.1111/j.1365-2583.2007.00774.x
Saingamsook J, Saeung A, Yanola J et al (2017) A multiplex PCR for detection of knockdown resistance mutations, V1016G and F1534C, in pyrethroid-resistant Aedes aegypti. Parasit Vectors 10:465. https://doi.org/10.1186/s13071-017-2416-x
Samantsidis GR, Panteleri R, Denecke S et al (2020) “What I cannot create, I do not understand”: functionally validated synergism of metabolic and target site insecticide resistance. Proc Biol Sci 287:20200838. https://doi.org/10.1098/rspb.2020.0838
Shi L, Hu H, Ma K et al (2015) Development of resistance to pyrethroid in Culex pipiens pallens population under different insecticide selection pressures. PLoS Negl Trop Dis 9:e0003928. https://doi.org/10.1371/journal.pntd.0003928
Smith LB, Kasai S, Scott JG (2018) Voltage-sensitive sodium channel mutations S989P + V1016G in Aedes aegypti confer variable resistance to pyrethroids, DDT and oxadiazines. Pest Manag Sci 74:737–745. https://doi.org/10.1002/ps.4771
Smith LB, Silva JJ, Chen C et al (2021) Fitness costs of individual and combined pyrethroid resistance mechanisms, kdr and CYP-mediated detoxification, in Aedes aegypti. PLoS Negl Trop Dis 15:e0009271. https://doi.org/10.1371/journal.pntd.0009271
Stone BF (1968) A formula for determining degree of dominance in cases of monofactorial inheritance of resistance to chemicals. Bull World Health Organ 38:325–326 https://www.ncbi.nlm.nih.gov/pmc/articles/PMC2554319/pdf/bullwho00235-0164.pdf
Su X, Guo Y, Deng J et al (2019) Fast emerging insecticide resistance in Aedes albopictus in Guangzhou, China: alarm to the dengue epidemic. PLoS Negl Trop Dis 13:e0007665. https://doi.org/10.1371/journal.pntd.0007665
Thornton J, Gomes B, Ayres C et al (2020) Insecticide resistance selection and reversal in two strains of Aedes aegypti. Wellcome Open Research. https://doi.org/10.12688/wellcomeopenres.15974.1
Waterhouse A, Bertoni M, Bienert S et al (2018) SWISS-MODEL: homology modelling of protein structures and complexes. Nucleic Acids Res 46:W296–W303. https://doi.org/10.1093/nar/gky427
Wei Y, Zheng X, He S et al (2021) Insecticide susceptibility status and knockdown resistance (kdr) mutation in Aedes albopictus in China. Parasit Vectors 14:609. https://doi.org/10.1186/s13071-021-05095-5
WHO (2005) Guidelines for laboratory and field testing of mosquito larvicides. World Health Organization. https://apps.who.int/iris/handle/10665/69101.
WHO (2011) Global insecticide use for vector-borne disease control: a 10-year assessment (2000–2009). World Health Organization. Available from: https://apps.who.int/iris/handle/10665/44670.
WHO (2012) Global strategy for dengue prevention and control 2012–2020. World Health Organization. https://apps.who.int/iris/handle/10665/75303
WHO (2016a) Test procedures for insecticide resistance monitoring in malaria vector mosquitoes (2nd edn) World Health Organization. https://apps.who.int/iris/handle/10665/250677.
WHO (2016b) Monitoring and managing insecticide resistance in Aedes mosquito populations: interim guidance for entomologists. World Health Organization. https://apps.who.int/iris/handle/10665/204588.
Wu Y, Liu Q, Qi Y et al (2021) Knockdown resistance (kdr) mutations I1532T and F1534S were identified in Aedes albopictus field populations in Zhejiang province. Central China Front Cell Infect Microbiol 11:702081. https://doi.org/10.3389/fcimb.2021.702081
Xu J, Bonizzoni M, Zhong D et al (2016) Multi-country survey revealed prevalent and novel F1534S mutation in voltage-gated sodium channel (VGSC) gene in Aedes albopictus. PLoS Negl Trop Dis 10:e0004696. https://doi.org/10.1371/journal.pntd.0004696
Xue W, Mermans C, Papapostolou KM (2021) Untangling a Gordian knot: the role of a GluCl3 I321T mutation in abamectin resistance in Tetranychus urticae. Pest Manag Sci 77:1581–1593. https://doi.org/10.1002/ps.6215
Yan R, Zhou Q, Xu Z et al (2020) Three sodium channel mutations from Aedes albopictus confer resistance to type I, but not type II pyrethroids. Insect Biochem Mol Biol 123:103411. https://doi.org/10.1016/j.ibmb.2020.103411
Yang X, Zhou Y, Sun Y et al (2021) Multiple insecticide resistance and associated mechanisms to volatile pyrethroid in an Aedes albopictus population collected in southern China. Pestic Biochem Physiol 174:104823. https://doi.org/10.1016/j.pestbp.2021
Zhang H, Li M, Tan R et al (2021) Presence of L1014F knockdown-resistance mutation in Anopheles gambiae s.s. from São Tomé and Príncipe. Front Cell Infect Microbiol 11:633905. https://doi.org/10.3389/fcimb.2021.633905
Zheng X, Zheng Z, Wu S et al (2022) Spatial heterogeneity of knockdown resistance mutations in the dengue vector Aedes albopictus in Guangzhou. China Parasit Vectors 15:156. https://doi.org/10.1186/s13071-022-05241-7
Zhu CY, Zhao CC, Wang YG et al (2019) Establishment of an innovative and sustainable PCR technique for 1534 locus mutation of the knockdown resistance (kdr) gene in the dengue vector Aedes albopictus. Parasit Vectors 12:603. https://doi.org/10.1186/s13071-019-3829-5
Zuo Y, Wang H, Xu Y et al (2017) CRISPR/Cas9 mediated G4946E substitution in the ryanodine receptor of Spodoptera exigua confers high levels of resistance to diamide insecticides. Insect Biochem Mol Biol 89:79–85. https://doi.org/10.1016/j.ibmb.2017.09.005
Zuo YY, Ma HH, Lu WJ et al (2020) Identification of the ryanodine receptor mutation I4743M and its contribution to diamide insecticide resistance in Spodoptera exigua (Lepidoptera: Noctuidae). Insect Sci 27:791–800. https://doi.org/10.1111/1744-7917.12695
Acknowledgements
We thank Dr. Hongxia Liu from Shanghai Center for Disease Control and Prevention for providing the susceptible strain of Aedes albopictus for our study and Dr. Zhihong Liu from Sun Yat-sen University for providing the aforementioned modeling software and Wecomput Technology for providing computation consulting.
Funding
This work was supported by grants from the National Natural Science Foundation of China (81829004, 31830087), the National Key Research and Development Program of China (2020YFC1200100) and the National Institutes of Health, USA (AI136850), to XGC. AAJ is a Donald Bren Professor at the University of California, Irvine.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors have not disclosed any competing interests.
Additional information
Communicated by Ruth Muller and Antonio Biondi.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Guo, Y., Zhou, J., Zhao, Y. et al. CRISPR/Cas9-mediated F1534S substitution in the voltage-gated sodium channel reveals its necessity and sufficiency for deltamethrin resistance in Aedes albopictus. J Pest Sci 96, 1173–1186 (2023). https://doi.org/10.1007/s10340-022-01557-6
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10340-022-01557-6