Abstract
Background
Mutation of the voltage-gated sodium channel (VGSC) gene, or knockdown resistance (kdr) gene, is an important resistance mechanism of the dengue vector Aedes aegypti mosquitoes against pyrethroids. In many countries in Asia, a valine to glycine substitution (V1016G) and a phenylalanine to cysteine substitution (F1534C) are common in Ae. aegypti populations. The G1016 and C1534 allele frequencies have been increasing in recent years, and hence there is a need to have a simple and inexpensive tool to monitor the alleles in large scale.
Methods
A multiplex PCR to detect V1016G and F1534C mutations has been developed in the current study. This study utilized primers from previous studies for detecting the mutation at position 1016 and newly designed primers to detect variants at position 1534. The PCR conditions were validated and compared with DNA sequencing using known kdr mutant laboratory strains and field collected mosquitoes. The efficacy of this method was also compared with allele-specific PCR (AS-PCR).
Results
The results of our multiplex PCR were in complete agreement with sequencing data and better than the AS-PCR. In addition, the efficiency of two non-toxic DNA staining dyes, Ultrapower™ and RedSafe™, were evaluated by comparing with ethidium bromide (EtBr) and the results were satisfactory.
Conclusions
Our multiplex PCR method is highly reliable and useful for implementing vector surveillance in locations where the two alleles co-occur.
Similar content being viewed by others
Background
Insecticide resistance of Aedes aegypti, the primary mosquito vector of dengue, chikungunya, Zika and yellow fever viruses, is known to be widely spread throughout the world. There are two major resistance mechanisms, i.e. metabolic enzyme-based resistance and target site insensitivity [1]. Metabolic enzyme-based resistance is principally associated with three major enzyme groups: cytochrome P450 monooxygenases (P450s), esterases and glutathione S-transferases. Target site insensitivity in mosquitoes and other insects is associated with single or multiple mutations of the voltage-gated sodium channel (VGSC) protein, commonly referred to as knockdown resistance (kdr). Knockdown resistance is the important mechanism for resistance to pyrethroids and DDT [2]. Several mutations in VGSC of Ae. aegypti have been reported, but only a few of them have been confirmed to be associated with pyrethroid resistance. With reference to the homologous house fly VGSC sequence, a valine to glycine substitution at position 1016 within domain II of the VGSC (V1016G) is associated with resistance to type I and II pyrethroids, such as permethrin and deltamethrin, respectively [3, 4]. The V1016G mutation has been found in many countries in Asia, i.e. Indonesia [3, 5, 6], Thailand [7], Vietnam [8], Taiwan [9], Bhutan [10], Myanmar [11], Singapore [12], Malaysia [13] and China [14]. Moreover, a valine to isoleucine substitution in domain II (V1016I), conferring pyrethroid resistance, occurs amongst Ae. aegypti populations in Latin America [15,16,17,18,19], and also in Vietnam [20].
A second mutation, involving a phenylalanine to cysteine substitution at position 1534 within domain III (F1534C), is associated with resistance to type I pyrethroids [21]. The F1534C mutation has been reported from many countries in Asia [4,5,6, 8, 11,12,13,14, 22, 23] and Latin America [15,16,17,18,19]. Recently, it has also been reported from Africa [24].
Continuous and heavy use of space sprays in mosquito control programs are considered to be the cause of a dramatic increase in resistance and kdr allele frequencies [5, 19, 25]. The occurrence of kdr mutations in wild populations is expected to reduce the efficacy of Ae. aegypti mosquito control programs using pyrethroid insecticides [26, 27]. Therefore, monitoring the frequency of kdr alleles in the Ae. aegypti mosquito populations is important in the surveillance system of vector control programs.
Detection of kdr alleles in mosquitoes can be performed by several methods. Nucleotide sequencing is considered to be the most accurate method as a gold standard, but this method is expensive and not suitable for examining a large number of mosquitoes. A number of PCR-based techniques for detecting kdr alleles have been reported. An assay was optimized for use in a real time PCR machine, although the amplified products could also be detected via agarose gel electrophoresis [28]. An alternative technique using a heated oligonucleotide ligation assay (HOLA) was developed [7]. Although this assay does not involve the use of radioisotope or any specialized machine, it requires additional reagents that can contribute to increased costs. We recently developed allele-specific PCR-based assays (AS-PCR) to detect the F1534C [10] and V1016G mutations [4]. Although techniques are simpler and genotyping results can only be determined by gel electrophoresis, testing one sample requires two separate reactions, i.e. one for F1534C and the other V1016G. In addition, AS-PCR often shows a discrepancy with the DNA sequencing [10]. The purpose of this study was to develop a multiplex PCR to detect both F1534C and V1016G mutations in a single reaction. This technique can reduce the cost and time consumed in monitoring the mutant allele frequencies in many countries where both V1016G and F1534C mutations co-exist.
Methods
Mosquito samples
Five laboratory strains, two F1 hybrids, DNA samples and field-collected Ae. aegypti mosquitoes were used to develop, optimize and validate the multiplex PCR method. The three laboratory strains included a pyrethroid susceptible strain, PMD (homozygous wild type for both V1016 and F1534 alleles, V/V1016 + F/F1534), and two pyrethroid-resistant strains, PMD-R (homozygous wild type for the V1016 allele but homozygous mutant for the C1534 allele, V/V1016 + C/C1534) [21, 29], and UPK-R (homozygous mutant for G1016 allele but homozygous wild type for the F1534 allele, G/G1016 + F/F1534) [30]. The PMD and PMD-R strains originated from a rural area of Chiang Mai Province and have been maintained in our laboratory since 1997. The UPK-R strain was established from mosquitoes collected from Chiang Mai city and maintained since 2006. An F1 hybrid was derived from the cross between the PMD (male) and UPK-R (female) strains to produce heterozygous mutant for the G1016 allele but homozygous wild type for the F1534 allele, V/G1016 + F/F1534. Another F1 hybrid was derived from the cross between the PMD-R (male) and UPK-R (female) strains to produce heterozygous mutant for both G1016 and F1534 allele, V/G1016 + F/C1534.
The other two laboratory strains were Dagon Myothit North Yangon (YG) and Than Bya Zayet Monstate (MS) strains. These two strains originated from Myanmar, however, the kdr genotypes of each strain were not determined until the present study.
The DNA samples of Ae. aegypti collected from wild populations in Thailand, Myanmar, Cambodia, Bhutan and Pakistan were obtained from previous studies [4, 10].
Field-collected mosquito samples were obtained from larval surveys in various temples around Chiang Mai city, Thailand, as temples are numerous and readily accessible throughout Chiang Mai, and from households from several rural villages in Mae Taeng district. We also collected the immature stages from Myanmar (Yangon city) and Indonesia (Ternate Island and Soppeng Regency, South Sulawesi). The samples collected were reared to adulthood, identified morphologically and preserved in absolute ethanol until the multiplex PCR was performed.
Development of multiplex PCR method
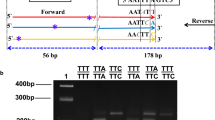
This assay was designed by combining two sets of primers. The first set was previously designed to genotype the V1016G mutation [28] and was used to develop an AS-PCR assay [4]. This set consists of three primers: a common forward primer (Gly1016f) and two specific reverse primers, Val1016r and Gly1016r, that differ in length and distinguish the V1016 and G1016 alleles, respectively (Fig. 1). In the case of a heterozygote, both products would be amplified.
Schematic of the AS-PCR assay for detection of the V1016G mutation as described in Stenhouse et al. [4]
The second set of primers was newly designed using the web-based Primer3Plus software [31]. The cDNA nucleotide sequences of VGSC domains IIIS4-IVS2 of the Ae. aegypti PMD strain (GenBank: EU259810.1) and PMD-R strain (GenBank: EU259811.1), which were submitted by Yanola et al. [21], were used as reference sequences. This set of primers were designed to genotype the F1534C mutation (Fig. 2) and consisted of four primers (tetra primer PCR assay). In this assay, the outermost primers (c1534-f and c1534-r) amplified a control band of 368 bp. Two internal allele-specific primers, Ae1534F-r and Ae1534C-f, were designed to work in conjunction with the external primers to give amplified products of either 232 bp for the F1534 allele or 180 bp for the C1534 allele, respectively. In the case of a heterozygote, all three products would be amplified. All primer sequences used in this study are shown in Table 1.
For optimization, DNA samples extracted from the laboratory strains, PMD, PMD-R, UPK-R and (PMD-R × UPK-R) F1 hybrids were used. Our preliminary study revealed that the PCR conditions from previous reports [4, 28] were not suitable for our multiplex technique. Therefore, several PCR conditions were optimized for a total reaction volume of 10 μl: primer annealing temperature (50–65 °C), concentration of each primer (0.1–0.5 μM), Taq DNA polymerase concentration (0.05–0.5 unit), MgCl2 concentration (0.5–3.0 mM) and dNTP concentration (50–200 μM). Furthermore, the lowest amount of genomic DNA (DNA template) that still gave a clear result on agarose gel, i.e. the detection limit of this method, was also determined.
V1016G and F1534C genotyping by multiplex PCR
Genomic DNA from each alcohol-preserved mosquito was extracted using DNAzol® reagent (Invitrogen, Carlsbad, California, USA). After PCR reactions, the amplified products were analyzed on 3% agarose gel with a low molecular weight DNA ladder (New England Biolab, Ipswich, Massachusetts, USA) used to estimate the band size. The electrophoresis was run for 50 min at 100 V in TBE buffer. The gel was then submerged in 0.5 μg/ml ethidium bromide (EtBr) (Invitrogen) solution for 15 min, de-stained for 5 min in distilled water, and visualized in a UV transilluminator.
Since EtBr is known to be a strong mutagen and is treated as hazardous waste, as alternatives we tried using Ultrapower™ (BioTeke, Beijing, China) and RedSafe™ (iNtRON Biotechnology, Gyeonggi-do, Korea) dyes, which are advertised as non-toxic and have no hazard waste. For Ultrapower™ staining, the dye solution (10,000×) was diluted 100-fold in 6× loading dye (New England Biolab), then 1 μl of diluted dye was mixed with 5 μl of PCR product. 1 μl of diluted dye was also added to 5 μl of the DNA ladder before loading on the gel. For RedSafe™ staining, 5 μl of this dye (20,000×) was mixed in with 100 ml molten agarose gel prior to gel pouring. Visualization was done immediately after gel electrophoresis.
DNA sequencing
In order to validate the multiplex PCR method, the results of samples tested by this assay were compared by using DNA sequencing data obtained from previous studies [4, 10] as well as the present study. The IIS6 and IIIS6 regions of the VGSC gene, which encompass the V1016G and F1534C mutations, respectively, were amplified and purified. This method has been described previously [10]. For domain IIS6 amplification, each PCR was carried out in a 20 μl reaction volume, containing: 2 μl of DNA sample (50 ng), 0.4 units of Platinum Taq DNA polymerase (Invitrogen), 1.6 μl of 2.5 mM dNTPs mix (200 μM) (New England Biolabs), 0.6 μl of 50 mM MgCl2 (1.5 mM), 2 μl of 10× PCR buffer (1×) (Invitrogen), and 2 μl of 5 μM each of IIP-F (0.5 μM) and IIS6_R primers (0.5 μM) (Table 1), and made up to 20 μl with sterile water. The amplification consisted of 95 °C for a 2 min heat activation step, followed by 35 cycles of 95 °C for 30 s, 63 °C for 30 s and 72 °C for 30 s with a 2 min final extension step at 72 °C. Amplifying domain IIIS6 used the same conditions, but the primers were changed to Ge-IIIS6_F and IIIS6_R.
The amplified products were purified using Illustra™ ExoProStar™ 1-Step DNA purification reagent (GE Healthcare Life Sciences, Buckinghamshire, UK) and sent to Macrogen, Inc. (Seoul, Korea) for direct sequencing in both the forward and reverse directions. Sequence data were analyzed using Geneious software, version 5.3.6 (Biomatters Ltd., UK). Finally, for each mosquito sample, sequencing results were compared against the genotype previously obtained by the multiplex PCR method.
Allele-specific PCR (AS-PCR)
The efficacy of multiplex PCR was compared with the AS-PCR methods for detecting F1534C and V1016G mutations as described in previous studies [4, 10]. The tested materials included DNA samples from the previous studies and newly extracted field samples.
Results
Development of multiplex PCR method
Tests for the optimization of PCR conditions resulted in the following multiplex PCR protocol. Each PCR reaction was performed in a 10 μl volume containing: 1 μl of DNA sample (25 ng), 0.4 units of Platinum Taq DNA polymerase (Invitrogen), 0.8 μl of 2.5 mM dNTPs mix (200 μM) (New England Biolabs), 0.3 μl of 50 mM MgCl2 (1.5 mM), 1 μl of 10× PCR buffer (1×) (Invitrogen), and primer concentrations: Gly1016f (0.5 μM), Val1016r (0.25 μM), Gly1016r (0.5 μM), c1534-f (0.25 μM), c1534-r (0.25 μM), Ae1534F-r (0.1 μM) and Ae1534C-f (0.5 μM), and made up to 10 μl with sterile water. The amplification consisted of 95 °C for a 2 min heat activation step, followed by 35 cycles of 95 °C for 30 s, 55 °C for 30 s and 72 °C for 30 s with a 2 min final extension step at 72 °C.
For these two polymorphic sites, there are nine possible genotypes, all of which were present in the laboratory and field-collected strains (Table 2, Fig. 3). The detection limit of this method was also evaluated by testing with a set of DNA dilutions of each genotype. The double heterozygous patterned sample (V/G1016 + F/C1534) needed the highest amount of DNA template (2 ng) to get a reliable result. Thus, the detection limit of this method was 2 ng of genomic DNA.
Gel electrophoresis results. a Stained with ethidium bromide. b Stained with Ultrapower™ dye. c Stained with RedSafe™ dye. All panels represent each of the nine possible genotypes. Lane L: contains low molecular weight DNA ladder. Lanes 1–9: contain PCR products by using a single mosquito DNA sample as template. Lane 1: VV/FF; Lane 2: VV/FC; Lane 3: VV/CC; Lane 4: VG/FF; Lane 5: VG/FC; Lane 6: VG/CC; Lane 7: GG/FF; Lane 8: GG/FC; Lane 9: GG/CC; Lane 10: negative control in which distilled water was used as the template in the PCR reaction
To compare the efficiency of three DNA staining dyes on this multiplex PCR, the PCR products from the same set of DNA templates were stained by EtBr (Fig. 3a), Ultrapower™ (Fig. 3b) and RedSafe™ (Fig. 3c), and the gels were visualized (with the best adjustment) under UV light. The EtBr and RedSafe™ staining methods gave clear and accurate results and the PCR product band sizes were correct, when determined by DNA Ladder. The RedSafe™ staining dye had a more faded result than EtBr; however, it had fewer non-specific bands. Ultrapower™ staining gave the brightest result, but bands were oversized when determined by DNA ladder. The C1534 (180 bp), F1534 (232 bp) and common 1534 (368 bp) bands were estimated as 200, 250 and 400 bp, respectively. The results were still interpretable by using these oversized bands, instead of original sizes.
Comparison of DNA sequencing with the multiplex PCR method
A total of 151 samples from laboratory and field materials covering the nine genotypes were tested; all were successfully amplified by the multiplex PCR and sequenced. The sequences of all samples were in agreement with the multiplex PCR (Table 2). Thus, this multiplex PCR method has 100% specificity. However, there were limited numbers in a few genotypes (VV/FC, VG/CC and GG/CC) because they were rare in populations. The sequences of representative genotypes have been deposited in GenBank with accession numbers as follows: MF794972–3 (VV/FF), MF794974–5 (VV/FC), MF794976–7 (VV/CC), MF794978–9 (VG/FF), MF794980–1 (VG/FC), MF794982–3 (VG/CC), MF794984–5 (GG/FF), MF794986–7 (GG/FC) and MF794988–9 (GG/CC).
Comparison of AS-PCR with the multiplex PCR method
A total of 169 field samples tested for F1534, C1534, V1016 and G1016 alleles by the AS-PCR were in agreement with the multiplex PCR, except one sample from Mae Sariang district which showed homozygous C/C1534 by AS-PCR, but heterozygous F/C1534 by the multiplex PCR (Table 3); the sequence of this sample (accession number MF794990) agrees with the multiplex PCR.
Discussion
We have successfully developed the multiplex PCR method to detect both V1016G and F1534C kdr mutations in Ae. aegypti in a single reaction. The results of samples tested by our multiplex PCR method and DNA sequencing were in complete agreement to detect all nine possible kdr genotypic patterns derived from V1016G and F1534C mutations: VV/FF, VV/FC, VV/CC, VG/FF, VG/FC, VG/CC, GG/FF, GG/FC and GG/CC. According to previous studies, only three patterns, VV/CC, VG/FC and GG/FF, were found in Chiang Mai city [27] and several provinces throughout Thailand [4], which is in agreement with the current study. While VV/FC can be found in other provinces i.e. Mae Hong Son, Song Khla and Ubon Ratchathani, the wild type (VV/FF) and double homozygous mutant (GG/CC) were rarely detected, and only found in Myanmar. For VG/CC and GG/FC genotypes, these patterns have never been reported in Thailand, but have been found in Myanmar and elsewhere [5, 11]. Hence, we found these genotypic patterns in YG, MS and field-collected Yangon strains from Myanmar. The samples from Cambodia and Pakistan had only C1534 mutant allele (with VV/CC pattern), while Bhutan had only G1016 mutant allele (with GG/FF pattern).
In a previous study, we developed an AS-PCR assay to detect the F1534C mutation, but there were slight discrepancies between the AS-PCR results and those from sequencing [10]. A similar situation was found in the current study when we performed the AS-PCR. However, our multiplex PCR method showed no discrepancy with the sequence data; hence, this method is as good as DNA sequencing for both V1016G and F1534C mutations. Our multiplex PCR method is simple, has a lower cost and needs less special equipment compared to other molecular techniques, e.g. DNA sequencing, Taqman assay and Heated Oligonucleotide Ligation assay.
In this study, we used two alternative safe nucleic acid stains to compare with the traditional EtBr stain when determining the multiplex PCR results. EtBr has high sensitivity for DNA staining, provides accurate band sizes, and is cheaper than the safe dyes used. However, due to its carcinogenic property, it is preferable to use alternative safe stains, such as Ultrapower™ and RedSafe™, which are non-toxic, non-mutagenic, non-carcinogenic, and leave no hazardous waste. However, both have some disadvantages. Although the prestain safe dye Ultrapower™ staining had higher sensitivity than EtBr and RedSafe™, it gave oversized bands when determined by the DNA ladder. This may be because the dye bound to the double strand DNA PCR product before gel electrophoresis and thus reduced product mobility. This problem has been investigated with other prestain safe dyes, SYBR® Gold and SYBR® Green I [32]. Nonetheless, for this multiplex PCR method, the results were still interpretable by using these oversized bands, instead of true sizes. For RedSafe™ staining, which was mixed into the gel, the band sizes were accurate and had less non-specific bands than EtBr, but this had the lowest sensitivity. Following the manufacturer protocols, 1 μl RedSafe™ can stain 10 samples, while 1 μl Ultrapower™ can stain 100 samples. Because the price per volume is similar, Ultrapower™ is more cost effective by 10-fold. We therefore prefer to use Ultrapower™, particularly when testing a large number of samples.
Further development of multiplex PCR to include the serine to proline mutation (S989P) in domain II of VGSC is challenging, since P989 allele has a synergistic effect with G and C alleles in reducing the sensitivity of VGSC [30, 33]. However, adding an additional mutation would increase the number of possible genotypes and exponentially increase the complexity of banding patterns, potentially making the method unreliable due to difficulty in interpretation of the banding patterns. At present, detection of S989P may be less important in some countries where this mutation is known to co-occur or be highly associated with V1016G, such as Thailand, Singapore, Myanmar and China [4, 11, 12, 14]. However, in countries where P989 allele has not been detected or has a low frequency [5, 6, 9, 10, 13], monitoring this mutation in wild populations by an AS-PCR [14] may be a necessary component of the surveillance system.
Conclusions
The multiplex PCR developed has high specificity and sensitivity to detect V1016G and F1534C kdr mutations in Ae. aegypti that allows all possible genotypes to be identified in a single step. This method was proved to be highly reliable and will be useful for monitoring mutant allele and genotype frequencies in wild populations throughout Thailand, and many other disease endemic countries in Asia, where these two alleles are prevalent.
Abbreviations
- AS-PCR:
-
Allele-specific PCR
- C1534:
-
Cysteine allele at position 1534
- DDT:
-
Dichlorodiphenyltrichloroethane
- EtBr:
-
Ethidium bromide
- F1:
-
First generation of progeny
- F1534:
-
Phenylalanine allele at position 1534
- F1534C:
-
Phenylalanine to cysteine substitution at position 1534
- G1016:
-
Glycine allele at position 1016
- GG/CC:
-
G/G1016 + C/C1534
- GG/FC:
-
G/G1016 + F/C1534
- GG/FF:
-
G/G1016 + F/F1534
- HOLA:
-
Heated oligonucleotide ligation assay
- IIIS4:
-
Domain III segment 4 of VGSC gene
- IIIS6:
-
Domain III segment 6 of VGSC gene
- IIS6:
-
Domain II segment 6 of VGSC gene
- IVS2:
-
Domain IV segment 2 of VGSC gene
- kdr :
-
Knockdown resistance
- MS:
-
Than Bya Zayet Monstate strain
- P450s:
-
Cytochrome P450 monooxygenases
- S989P:
-
Serine to proline substitution at position 989
- TBE:
-
Tris-Borate-EDTA buffer
- V1016:
-
Valine allele at position 1016
- V1016G:
-
Valine to glycine substitution at position 1016
- V1016I:
-
Valine to isoleucine substitution at position 1016
- VG/CC:
-
V/G1016 + C/C1534
- VG/FC:
-
V/G1016 + F/C1534
- VG/FF:
-
V/G1016 + F/F1534
- VGSC:
-
Voltage-gated sodium channel
- VV/CC:
-
V/V1016 + C/C1534
- VV/FC:
-
V/V1016 + F/C1534
- VV/FF:
-
V/V1016 + F/F1534
- YG:
-
Dagon Myothit North Yangon strain
References
Hemingway J, Ranson H. Insecticide resistance in insect vectors of human disease. Annu Rev Entomol. 2000;45:371–91.
Hemingway J, Hawkes NJ, McCarroll L, Ranson H. The molecular basis of insecticide resistance in mosquitoes. Insect Biochem Mol Biol. 2004;34:653–65.
Brengues C, Hawkes NJ, Chandre F, McCarroll L, Duchon S, Guillet P, et al. Pyrethroid and DDT cross-resistance in Aedes aegypti is correlated with novel mutations in the voltage-gated sodium channel gene. Med Vet Entomol. 2003;17:87–94.
Stenhouse SA, Plernsub S, Yanola J, Lumjuan N, Dantrakool A, Choochote W, et al. Detection of the V1016G mutation in the voltage-gated sodium channel gene of Aedes aegypti (Diptera: Culicidae) by allele-specific PCR assay, and its distribution and effect on deltamethrin resistance in Thailand. Parasit Vectors. 2013;6:253.
Sayono S, Hidayati AP, Fahri S, Sumanto D, Dharmana E, Hadisaputro S, et al. Distribution of voltage-gated sodium channel (Nav) alleles among the Aedes aegypti populations in central Java Province and its association with resistance to pyrethroid insecticides. PLoS One. 2016;11:e0150577.
Wuliandari J, Lee S, White V, Tantowijoyo W, Hoffmann A, Endersby-Harshman N. Association between three mutations, F1565C, V1023G and S996P, in the voltage-sensitive sodium channel gene and knockdown resistance in Aedes aegypti from Yogyakarta, Indonesia. Insects. 2015;6:658–85.
Rajatileka S, Black WC IV, Saavedra-Rodriguez K, Trongtokit Y, Apiwathnasorn C, McCall PJ, et al. Development and application of a simple colorimetric assay reveals widespread distribution of sodium channel mutations in Thai populations of Aedes aegypti. Acta Trop. 2008;108:54–7.
Kawada H, Higa Y, Komagata O, Kasai S, Tomita T, Thi Yen N, et al. Widespread distribution of a newly found point mutation in voltage-gated sodium channel in pyrethroid-resistant Aedes aegypti populations in Vietnam. PLoS Negl Trop Dis. 2009;3:e527.
Chang C, Shen WK, Wang TT, Lin YH, Hsu EL, Dai SM. A novel amino acid substitution in a voltage-gated sodium channel is associated with knockdown resistance to permethrin in Aedes aegypti. Insect Biochem Mol Biol. 2009;39:272–8.
Yanola J, Somboon P, Walton C, Nachaiwieng W, Somwang P, Prapanthadara LA. High-throughput assays for detection of the F1534C mutation in the voltage-gated sodium channel gene in permethrin-resistant Aedes aegypti and the distribution of this mutation throughout Thailand. Trop Med Int Health. 2011;16:501–9.
Kawada H, Oo SZ, Thaung S, Kawashima E, Maung YN, Thu HM, et al. Co-occurrence of point mutations in the voltage-gated sodium channel of pyrethroid-resistant Aedes aegypti populations in Myanmar. PLoS Negl Trop Dis. 2014;8:e3032.
Kasai S, Komagata O, Itokawa K, Shono T, Ng LC, Kobayashi M, et al. Mechanisms of pyrethroid resistance in the dengue mosquito vector, Aedes aegypti: target site insensitivity, penetration, and metabolism. PLoS Negl Trop Dis. 2014;8:e2948.
Ishak IH, Jaal Z, Ranson H, Wondji CS. Contrasting patterns of insecticide resistance and knockdown resistance (kdr) in the dengue vectors Aedes aegypti and Aedes albopictus from Malaysia. Parasit Vectors. 2015;8:181.
Li CX, Kaufman PE, Xue RD, Zhao MH, Wang G, Yan T, et al. Relationship between insecticide resistance and kdr mutations in the dengue vector Aedes aegypti in Southern China. Parasit Vectors. 2015;8:325.
Alvarez LC, Ponce G, Oviedo M, Lopez B, Flores AE. Susceptibility status of Aedes aegypti (L.) (Diptera: Culicidae) to temephos in Venezuela. Pest Manag Sci. 2014;70:1262–6.
Deming R, Manrique-Saide P, Medina Barreiro A, Cardena EU, Che-Mendoza A, Jones B, et al. Spatial variation of insecticide resistance in the dengue vector Aedes aegypti presents unique vector control challenges. Parasit Vectors. 2016;9:67.
Harris AF, Rajatileka S, Ranson H. Pyrethroid resistance in Aedes aegypti from Grand Cayman. Am J Trop Med Hyg. 2010;83:277–84.
Linss JG, Brito LP, Garcia GA, Araki AS, Bruno RV, Lima JB, et al. Distribution and dissemination of the Val1016Ile and Phe1534Cys k dr mutations in Aedes aegypti Brazilian natural populations. Parasit Vectors. 2014;7:25.
Vera-Maloof FZ, Saavedra-Rodriguez K, Elizondo-Quiroga AE, Lozano-Fuentes S, Black WC IV. Coevolution of the Ile1,016 and Cys1,534 Mutations in the voltage gated sodium channel gene of Aedes aegypti in Mexico. PLoS Negl Trop Dis. 2015;9:e0004263.
Bingham G, Strode C, Tran L, Khoa PT, Jamet HP. Can piperonyl butoxide enhance the efficacy of pyrethroids against pyrethroid-resistant Aedes aegypti? Trop Med Int Health. 2011;16:492–500.
Yanola J, Somboon P, Walton C, Nachaiwieng W, Prapanthadara L. A novel F1552/C1552 point mutation in the Aedes aegypti voltage-gated sodium channel gene associated with permethrin resistance. Pestic Biochem Phys. 2010;96:127–31.
Kushwah RB, Dykes CL, Kapoor N, Adak T, Singh OP. Pyrethroid-resistance and presence of two knockdown resistance (kdr) mutations, F1534C and a novel mutation T1520I, in Indian Aedes aegypti. PLoS Negl Trop Dis. 2015;9:e3332.
Muthusamy R, Shivakumar MS. Involvement of metabolic resistance and F1534C kdr mutation in the pyrethroid resistance mechanisms of Aedes aegypti in India. Acta Trop. 2015;148:137–41.
Kawada H, Higa Y, Futami K, Muranami Y, Kawashima E, Osei JH, et al. Discovery of point mutations in the voltage-gated sodium channel from African Aedes aegypti populations: Potential phylogenetic reasons for gene introgression. PLoS Negl Trop Dis. 2016;10:e0004780.
Garcia GP, Flores AE, Fernandez-Salas I, Saavedra-Rodriguez K, Reyes-Solis G, Lozano-Fuentes S, et al. Recent rapid rise of a permethrin knock down resistance allele in Aedes aegypti in Mexico. PLoS Negl Trop Dis. 2009;3:e531.
Marcombe S, Carron A, Darriet F, Etienne M, Agnew P, Tolosa M, et al. Reduced efficacy of pyrethroid space sprays for dengue control in an area of Martinique with pyrethroid resistance. Am J Trop Med Hyg. 2009;80:745–51.
Plernsub S, Saingamsook J, Yanola J, Lumjuan N, Tippawangkosol P, Walton C, et al. Temporal frequency of knockdown resistance mutations, F1534C and V1016G, in Aedes aegypti in Chiang Mai city, Thailand and the impact of the mutations on the efficiency of thermal fogging spray with pyrethroids. Acta Trop. 2016;162:125–32.
Saavedra-Rodriguez K, Urdaneta-Marquez L, Rajatileka S, Moulton M, Flores AE, Fernandez-Salas I, et al. A mutation in the voltage-gated sodium channel gene associated with pyrethroid resistance in Latin American Aedes aegypti. Insect Mol Biol. 2007;16:785–98.
Somboon P, Prapanthadara LA, Suwonkerd W. Insecticide susceptibility tests of Anopheles minimus s.l., Aedes aegypti, Aedes albopictus, and Culex quinquefasciatus in northern Thailand. Southeast Asian J Trop Med Public Health. 2003;34:87–93.
Plernsub S, Saingamsook J, Yanola J, Lumjuan N, Tippawangkosol P, Sukontason K, et al. Additive effect of knockdown resistance mutations, S989P, V1016G and F1534C, in a heterozygous genotype conferring pyrethroid resistance in Aedes aegypti in Thailand. Parasit Vectors. 2016;9:417.
Untergasser A, Cutcutache I, Koressaar T, Ye J, Faircloth BC, Remm M, et al. Primer3 - new capabilities and interfaces. Nucleic Acids Res. 2012;40:e115.
Huang Q, Fu WL. Comparative analysis of the DNA staining efficiencies of different fluorescent dyes in preparative agarose gel electrophoresis. Clin Chem Lab Med. 2005;43:841–2.
Hirata K, Komagata O, Itokawa K, Yamamoto A, Tomita T, Kasai S. A single crossing-over event in voltage-sensitive Na+ channel genes may cause critical failure of dengue mosquito control by insecticides. PLoS Negl Trop Dis. 2014;8:e3085.
Acknowledgements
We thank Yan Naung Maung Maung and Rusdiyah Sudirman Made Ali for providing mosquitoes from Myanmar and Indonesia, respectively.
Funding
This work was funded by the Thailand Research Fund through the Royal Golden Jubilee PhD Program to PS and JS (PHD/0073/2556), and the Korean International Cooperation for Infectious Diseases (KOICID). This study was partially supported by the Diamond Research Grant from the Faculty of Medicine, Chiang Mai University to PS. The Research Administration Office of Chiang Mai University provided the budget to our Center of Excellence in Insect Vector Studies.
Availability of data and materials
All data analyzed during this study are included in this published article. The sequences are submitted in the GenBank database under the accession numbers MF794972–MF794990.
Author information
Authors and Affiliations
Contributions
PS conceived the study. AS, JY, NL and CW supervised the study. JS designed the multiplex PCR method and performed lab experiments. JY gave advice about primer design and provided some mosquito samples. AS and NL provided and gave suggestions on using safe nucleic acid stains. JS and AS collected mosquito samples. JS, JY and NL analyzed the data and interpreted the results. JS, CW and PS drafted the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Saingamsook, J., Saeung, A., Yanola, J. et al. A multiplex PCR for detection of knockdown resistance mutations, V1016G and F1534C, in pyrethroid-resistant Aedes aegypti . Parasites Vectors 10, 465 (2017). https://doi.org/10.1186/s13071-017-2416-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13071-017-2416-x