Abstract
Metabolites provide a rich source of information regarding individual physiological states influenced by differences in diet, disease, health, and lifestyle. Effective sample preparation is a crucial step for the development of a sensitive, effective, and robust analytical method. In the present study, the metabolite extraction and identification protocol was optimized to achieve a reliable sample preparation technique for human plasma metabolites using gas chromatography–mass spectrometry (GC–MS). We compared the response of different sample preparation techniques with several extraction solvents [chloroform (CHCl3), methanol (MeOH), acetonitrile (ACN), MeOH and ACN (50:50), MeOH/MeOH/water (MMW)] coupled with N, O-Bis (trimethylsilyl) trifluoroacetamide with 1% trimethylchlorosilane (BSTFA + 1% TMCS), and N-Methyl-N-(trimethylsilyl) trifluoroacetamide with 1% trimethylchlorosilane (MSTFA + 1% TMCS) derivatizing reagents. The suitability of the extraction solvent and derivatizing reagent was assessed for human plasma metabolite identification. We conclude that extraction of metabolites using methanol/methanol/water and 50 min silylation at 70 °C with N-methyl-N-(trimethylsilyl) trifluoroacetamide with 1% trimethylchlorosilane is efficient combination for proper derivatization and identification of metabolites. This protocol resulted in identifying a total of 87 metabolites including various amino acids, carboxylic acids, fatty acids, and polysaccharides. The optimized method provides an efficient, robust, and reproducible analytical method for metabolomic analysis in human plasma.
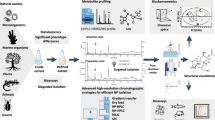
Graphical Abstract
Schematic representation of metabolite extraction and analysis using gas chromatography mass spectrometry








Similar content being viewed by others
Availability of Data and Materials
The detailed protocols, data, and material are available upon request.
References
Wang JH, Byun J, Pennathur S (2010) Analytical approaches to metabolomics and applications to systems biology. Semin Nephrol 30(5):500–511. https://doi.org/10.1016/j.semnephrol.2010.07.007
Zhang A, Sun H, Wang P, Han Y, Wang X-J (2011) Modern analytical techniques in metabolomics analysis. Analyst 137:293–300. https://doi.org/10.1039/c1an15605e
Liu X, Locasale JW (2017) Metabolomics: a primer. Trends Biochem Sci 42(4):274–284. https://doi.org/10.1016/j.tibs.2017.01.004
Chen Y, Xu J, Zhang R, Abliz Z (2016) Methods used to increase the comprehensive coverage of urinary and plasma metabolomes by MS. Bioanalysis 8(9):981–997. https://doi.org/10.4155/bio-2015-0010
Moros G, Chatziioannou AC, Gika HG, Raikos N, Theodoridis G (2016) Investigation of the derivatization conditions for GC–MS metabolomics of biological samples. Bioanalysis 9(1):53–65. https://doi.org/10.4155/bio-2016-0224
Kvitvang H, Andreassen T, Adam T, Villas-Bôas S, Bruheim P (2011) Highly sensitive GC/MS/MS method for quantitation of amino and nonamino organic acids. Anal Chem 83:2705–2711. https://doi.org/10.1021/ac103245b
Wishart DS, Jewison T, Guo AC, Wilson M, Knox C, Liu Y, Djoumbou Y, Mandal R, Aziat F, Dong E, Bouatra S, Sinelnikov I, Arndt D, Xia J, Liu P, Yallou F, Bjorndahl T, Perez-Pineiro R, Eisner R, Allen F, Neveu V, Greiner R, Scalbert A (2013) HMDB 3.0–the human metabolome database in 2013. Nucleic Acids Res 41:D801-807. https://doi.org/10.1093/nar/gks1065
Klupczyńska A, Dereziński P, Kokot ZJ (2015) Metabolomics in medical sciences–trends challenges and perspectives. Acta Poloniae Pharm 72(4):629–641
Dunn WB, Ellis DI (2005) Metabolomics: current analytical platforms and methodologies. TrAC Trends Anal Chem 24(4):285–294. https://doi.org/10.1016/j.trac.2004.11.021
Yin P, Lehmann R, Xu G (2015) Effects of pre-analytical processes on blood samples used in metabolomics studies. Anal Bioanal Chem 407(17):4879–4892. https://doi.org/10.1007/s00216-015-8565-x
Paiva M, Menezes H, Cardeal Z (2014) Sampling and analysis of metabolomes in biological fluids. Analyst 139:3683. https://doi.org/10.1039/c4an00583j
Shriver LP (2016) Mass spectrometry-based metabolomics: a practical guide. J Am Soc Mass Spectrom 27(1):1–2. https://doi.org/10.1007/s13361-015-1246-3
Khamis MM, Adamko DJ, El-Aneed A (2017) Mass spectrometric based approaches in urine metabolomics and biomarker discovery. Mass Spectrom Rev 36(2):115–134. https://doi.org/10.1002/mas.21455
Zhou B, Xiao JF, Tuli L, Ressom HW (2012) LC-MS-based metabolomics. Mol BioSyst 8(2):470–481. https://doi.org/10.1039/c1mb05350g
Lopes AS, Cruz EC, Sussulini A, Klassen A (2017) Metabolomic strategies involving mass spectrometry combined with liquid and gas chromatography. Adv Exp Med Biol 965:77–98. https://doi.org/10.1007/978-3-319-47656-8_4
Gowda GAN, Djukovic D (2014) Overview of mass spectrometry-based metabolomics: opportunities and challenges. Methods Mol Biol 1198:3–12. https://doi.org/10.1007/978-1-4939-1258-2_1
Alonso A, Marsal S, Julià A (2015) Analytical methods in untargeted metabolomics: state of the art in 2015. Front Bioeng Biotechnol 3:23–23. https://doi.org/10.3389/fbioe.2015.00023
Rodríguez-Morató J, Pozo ÓJ, Marcos J (2018) Targeting human urinary metabolome by LC-MS/MS: a review. Bioanalysis 10(7):489–516. https://doi.org/10.4155/bio-2017-0285
NIST20: Updates to the NIST Tandem and Electron Ionization Spectral Libraries. National Institute of Standards and Technology ( June 11, 2020, Updated June 12, 2020)
Dettmer K, Aronov PA, Hammock BD (2007) Mass spectrometry-based metabolomics. Mass Spectrom Rev 26(1):51–78. https://doi.org/10.1002/mas.20108
Nezami Ranjbar MR, Luo Y, Di Poto C, Varghese RS, Ferrarini A, Zhang C, Sarhan NI, Soliman H, Tadesse MG, Ziada DH, Roy R, Ressom HW (2015) GC-MS based plasma metabolomics for identification of candidate biomarkers for hepatocellular Carcinoma in Egyptian Cohort. PLoS ONE 10(6):e0127299–e0127299. https://doi.org/10.1371/journal.pone.0127299
Sogin EM, Puskás E, Dubilier N, Liebeke M (2019) Marine metabolomics: a method for nontargeted measurement of metabolites in seawater by gas chromatography-mass spectrometry. mSystems 4(6):e00638-e1619. https://doi.org/10.1128/mSystems.00638-19
Naz S, Moreira dos Santos DC, García A, Barbas C (2014) Analytical protocols based on LC-MS, GC-MS and CE-MS for nontargeted metabolomics of biological tissues. Bioanalysis 6(12):1657–1677. https://doi.org/10.4155/bio.14.119
Shareef A, Angove MJ, Wells JD (2006) Optimization of silylation using N-methyl-N-(trimethylsilyl)-trifluoroacetamide, N, O-bis-(trimethylsilyl)-trifluoroacetamide and N-(tert-butyldimethylsilyl)-N-methyltrifluoroacetamide for the determination of the estrogens estrone and 17alpha-ethinylestradiol by gas chromatography-mass spectrometry. J Chromatogr A 1108(1):121–128. https://doi.org/10.1016/j.chroma.2005.12.098
Pasikanti KK, Ho PC, Chan ECY (2008) Gas chromatography/mass spectrometry in metabolic profiling of biological fluids. J Chromatogr B 871(2):202–211. https://doi.org/10.1016/j.jchromb.2008.04.033
Shareef A, Parnis CJ, Angove MJ, Wells JD, Johnson BB (2004) Suitability of N, O-bis(trimethylsilyl)trifluoroacetamide and N-(tert-butyldimethylsilyl)-N-methyltrifluoroacetamide as derivatization reagents for the determination of the estrogens estrone and 17alpha-ethinylestradiol by gas chromatography-mass spectrometry. J Chromatogr A 1026(1–2):295–300. https://doi.org/10.1016/j.chroma.2003.10.110
Salvatore M, Salvatore F (2015) A strategy for GC/MS quantification of polar compounds via their silylated surrogates: silylation and quantification of biological amino acids. J Analy Bioanal Techn 6:1–16. https://doi.org/10.4172/2155-9872.1000263
Villas-Bôas SG, Smart KF, Sivakumaran S, Lane GA (2011) Alkylation or silylation for analysis of amino and non-amino organic acids by GC-MS? Metabolites 1(1):3–20. https://doi.org/10.3390/metabo1010003
Abdel-Khalik J, Björklund E, Hansen M (2013) Simultaneous determination of endogenous steroid hormones in human and animal plasma and serum by liquid or gas chromatography coupled to tandem mass spectrometry. J Chromatogr B 928:58–77. https://doi.org/10.1016/j.jchromb.2013.03.013
Fiehn O, Robertson D, Griffin J, van der Werf M, Nikolau B, Morrison N, Sumner LW, Goodacre R, Hardy NW, Taylor C, Fostel J, Kristal B, Kaddurah-Daouk R, Mendes P, van Ommen B, Lindon JC, Sansone S-A (2007) The metabolomics standards initiative (MSI). Metabolomics 3(3):175–178. https://doi.org/10.1007/s11306-007-0070-6
Pang Z, Chong J, Zhou G, de Lima Morais DA, Chang L, Barrette M, Gauthier C, Jacques P-É, Li S, Xia J (2021) MetaboAnalyst 5.0: narrowing the gap between raw spectra and functional insights. Nucl Acids Res 49(1):388–396. https://doi.org/10.1093/nar/gkab382
Moros G, Chatziioannou AC, Gika HG, Raikos N, Theodoridis G (2017) Investigation of the derivatization conditions for GC-MS metabolomics of biological samples. Bioanalysis 9(1):53–65. https://doi.org/10.4155/bio-2016-0224
Fang M, Ivanisevic J, Benton HP, Johnson CH, Patti GJ, Hoang LT, Uritboonthai W, Kurczy ME, Siuzdak G (2015) Thermal degradation of small molecules: a global metabolomic investigation. Anal Chem 87(21):10935–10941. https://doi.org/10.1021/acs.analchem.5b03003
Fritsche-Guenther R, Gloaguen Y, Bauer A, Opialla T, Kempa S, Fleming CA, Redmond HP, Kirwan JA (2021) Optimized workflow for on-line derivatization for targeted metabolomics approach by gas chromatography-mass spectrometry. Metabolites 11(12):888. https://doi.org/10.3390/metabo11120888
Funding
The laboratory of Dr. Ashutosh Shrivastava is supported by the King George’s Medical University Intramural Grant Award and a research grant from CCRH, Ministry of AYUSH, Government of India (Z.28015/01/2018-HPC-EMR-AYUSH-D). Alok Mishra is the recipient of senior research fellowship from the University Grant Commission, New Delhi, India.
Author information
Authors and Affiliations
Contributions
AS and AS conceptualized and planned the study. AS and AM conducted the experiments. AS, AS, and AM analyzed the data and wrote the manuscript. All authors agree with the final version of the manuscript.
Corresponding author
Ethics declarations
Conflict of Interest
The authors have No competing interests to declare.
Ethical Approval
The study was approved by the KGMU institutional ethics committee (Ref. Code: 107. ECM II B-Ph.D./P2). Informed consent was obtained from the subjects before collection of the samples.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Srivastava, A., Mishra, A. & Shrivastava, A. Optimizing Solvents and Derivatizing Agents for Metabolomic Profiling of Human Plasma Using GC–MS. Chromatographia 86, 523–534 (2023). https://doi.org/10.1007/s10337-023-04266-z
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10337-023-04266-z