Abstract
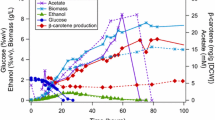
Metabolic engineering is usually focused on static control of microbial cell factories to efficient production of interested chemicals, though heterologous pathways compete with endogenous metabolism. However, products like carotenoids may cause metabolic burden on engineering strains, thus limiting product yields and influencing strain growth. Herein, a growth-phase-dependent regulation was developed to settle this matter, and its efficiency was verified using the heterogenous biosynthesis of lycopene in Saccharomyces cerevisiae as an example. Through growth-phase-dependent control of the lycopene biosynthetic pathway, limited step in MVA pathway, and competitive squalene pathway, production yield was increased by approximately 973-fold (from 0.034- to 33.1-mg/g CDW) and 1.48 g/L of production was obtained by one-stage fermentation in a 5-L bioreactor. Our study not only introduces an economically approach to the production of carotenoids, but also provides an example of dynamic regulation of biosynthetic pathways for metabolic engineering.







Similar content being viewed by others
References
Burg JM, Cooper CB, Ye Z, Reed BR, Moreb EA, Lynch MD (2016) Large-scale bioprocess competitiveness: the potential of dynamic metabolic control in two-stage fermentations. Curr Opin Chem Eng 14:121–136. https://doi.org/10.1016/j.coche.2016.09.008
Carvalho N, Coelho E, Gales L, Costa V, Teixeira JA, Moradas-Ferreira P (2016) Production of orotic acid by a Klura3Delta mutant of Kluyveromyces lactis. J Biosci Bioeng 121:625–630. https://doi.org/10.1016/j.jbiosc.2015.10.008
Chen X, Zhu P, Liu L (2016) Modular optimization of multi-gene pathways for fumarate production. Metab Eng 33:76–85. https://doi.org/10.1016/j.ymben.2015.07.007
Chen Y, Wang Y, Liu M, Qu J, Yao M, Li B, Ding M, Liu H, Xiao W, Yuan Y (2019) Primary and Secondary Metabolic Effects of a Key Gene Deletion (DeltaYPL062W) in Metabolically Engineered Terpenoid-Producing Saccharomyces cerevisiae. Appl Environ Microbiol. https://doi.org/10.1128/AEM.01990-18
Chen Y, Xiao W, Wang Y, Liu H, Li X, Yuan Y (2016) Lycopene overproduction in Saccharomyces cerevisiae through combining pathway engineering with host engineering. Microb Cell Fact 15:113. https://doi.org/10.1186/s12934-016-0509-4
Curran KA, Morse NJ, Markham KA, Wagman AM, Gupta A, Alper HS (2015) Short Synthetic Terminators for Improved Heterologous Gene Expression in Yeast. ACS Synth Biol 4:824–832. https://doi.org/10.1021/sb5003357
Dueber JE, Wu GC, Malmirchegini GR, Moon TS, Petzold CJ, Ullal AV, Prather KL, Keasling JD (2009) Synthetic protein scaffolds provide modular control over metabolic flux. Nat Biotechnol 27:753–759. https://doi.org/10.1038/nbt.1557
Gupta A, Reizman IM, Reisch CR, Prather KL (2017) Dynamic regulation of metabolic flux in engineered bacteria using a pathway-independent quorum-sensing circuit. Nat Biotechnol 35:273–279. https://doi.org/10.1038/nbt.3796
Hedl M, Tabernero L, Stauffacher CV, Rodwell VW (2004) Class II 3-hydroxy-3-methylglutaryl coenzyme A reductases. J Bacteriol 186:1927–1932. https://doi.org/10.1128/jb.186.7.1927-1932.2004
Hong J, Park SH, Kim S, Kim SW, Hahn JS (2019) Efficient production of lycopene in Saccharomyces cerevisiae by enzyme engineering and increasing membrane flexibility and NAPDH production. Appl Microbiol Biotechnol 103:211–223. https://doi.org/10.1007/s00253-018-9449-8
Jin W, Xu X, Jiang L, Zhang Z, Li S, Huang H (2015) Putative carotenoid genes expressed under the regulation of Shine-Dalgarno regions in Escherichia coli for efficient lycopene production. Biotechnol Lett 37:2303–2310. https://doi.org/10.1007/s10529-015-1922-1
Kang W, Ma T, Liu M, Qu J, Liu Z, Zhang H, Shi B, Fu S, Ma J, Lai LTF, He S, Qu J, Wing-Ngor AuS, Ho Kang B, Yu Lau WC, Deng Z, Xia J, Liu T (2019) Modular enzyme assembly for enhanced cascade biocatalysis and metabolic flux. Nat commun 10:4248. https://doi.org/10.1038/s41467-019-12247-w
Keung AJ, Joung JK, Khalil AS, Collins JJ (2015) Chromatin regulation at the frontier of synthetic biology. Nat Rev Genet 16:159–171. https://doi.org/10.1038/nrg3900
Krivoruchko A, Zhang Y, Siewers V, Chen Y, Nielsen J (2015) Microbial acetyl-CoA metabolism and metabolic engineering. Metab Eng 28:28–42. https://doi.org/10.1016/j.ymben.2014.11.009
Lalwani MA, Zhao EM, Avalos JL (2018) Current and future modalities of dynamic control in metabolic engineering. Curr Opin Biotechnol 52:56–65. https://doi.org/10.1016/j.copbio.2018.02.007
Lian J, Mishra S, Zhao H (2018) Recent advances in metabolic engineering of Saccharomyces cerevisiae: New tools and their applications. Metab Eng 50:85–108. https://doi.org/10.1016/j.ymben.2018.04.011
Liu D, Mannan AA, Han Y, Oyarzun DA, Zhang F (2018) Dynamic metabolic control: towards precision engineering of metabolism. J Ind Microbiol Biotechnol 45:535–543. https://doi.org/10.1007/s10295-018-2013-9
Ma SM, Garcia DE, Redding-Johanson AM, Friedland GD, Chan R, Batth TS, Haliburton JR, Chivian D, Keasling JD, Petzold CJ, Lee TS, Chhabra SR (2011) Optimization of a heterologous mevalonate pathway through the use of variant HMG-CoA reductases. Metab Eng 13:588–597. https://doi.org/10.1016/j.ymben.2011.07.001
Ma T, Shi B, Ye Z, Li X, Liu M, Chen Y, Xia J, Nielsen J, Deng Z, Liu T (2019) Lipid engineering combined with systematic metabolic engineering of Saccharomyces cerevisiae for high-yield production of lycopene. Metab Eng 52:134–142. https://doi.org/10.1016/j.ymben.2018.11.009
Moon TS, Dueber JE, Shiue E, Prather KL (2010) Use of modular, synthetic scaffolds for improved production of glucaric acid in engineered E. coli. Metab Eng 12:298–305. https://doi.org/10.1016/j.ymben.2010.01.003
Naito Y, Hino K, Bono H, Ui-Tei K (2015) CRISPRdirect: software for designing CRISPR/Cas guide RNA with reduced off-target sites. Bioinformatics 31:1120–1123. https://doi.org/10.1093/bioinformatics/btu743
Niu FX, Lu Q, Bu YF, Liu JZ (2017) Metabolic engineering for the microbial production of isoprenoids: Carotenoids and isoprenoid-based biofuels. Synth Syst Biotechnol 2:167–175. https://doi.org/10.1016/j.synbio.2017.08.001
Papapetridis I, van Dijk M, Dobbe AP, Metz B, Pronk JT, van Maris AJ (2016) Improving ethanol yield in acetate-reducing Saccharomyces cerevisiae by cofactor engineering of 6-phosphogluconate dehydrogenase and deletion of ALD6. Microb Cell Fact 15:67. https://doi.org/10.1186/s12934-016-0465-z
Partow S, Siewers V, Bjorn S, Nielsen J, Maury J (2010) Characterization of different promoters for designing a new expression vector in Saccharomyces cerevisiae. Yeast 27:955–964. https://doi.org/10.1002/yea.1806
Reider Apel A, d'Espaux L, Wehrs M, Sachs D, Li RA, Tong GJ, Garber M, Nnadi O, Zhuang W, Hillson NJ, Keasling JD, Mukhopadhyay A (2017) A Cas9-based toolkit to program gene expression in Saccharomyces cerevisiae. Nucleic Acids Res 45:496–508. https://doi.org/10.1093/nar/gkw1023
Scalcinati G, Knuf C, Partow S, Chen Y, Maury J, Schalk M, Daviet L, Nielsen J, Siewers V (2012) Dynamic control of gene expression in Saccharomyces cerevisiae engineered for the production of plant sesquitepene alpha-santalene in a fed-batch mode. Metab Eng 14:91–103. https://doi.org/10.1016/j.ymben.2012.01.007
Scott KJ (2001) Detection and measurement of carotenoids by UV/VIS spectrophotometry. Curr Protoc Food Anal Chem F2.2.1–F2.2.10
Shi B, Ma T, Ye Z, Li X, Huang Y, Zhou Z, Ding Y, Deng Z, Liu T (2019) Systematic Metabolic Engineering of Saccharomyces cerevisiae for Lycopene Overproduction. J Agric Food Chem 67:11148–11157. https://doi.org/10.1021/acs.jafc.9b04519
Sun J, Shao Z, Zhao H, Nair N, Wen F, Xu JH, Zhao H (2012) Cloning and characterization of a panel of constitutive promoters for applications in pathway engineering in Saccharomyces cerevisiae. Biotechnol Bioeng 109:2082–2092. https://doi.org/10.1002/bit.24481
Tan SZ, Prather KL (2017) Dynamic pathway regulation: recent advances and methods of construction. Curr Opin Chem Biol 41:28–35. https://doi.org/10.1016/j.cbpa.2017.10.004
Verwaal R, Wang J, Meijnen JP, Visser H, Sandmann G, van den Berg JA, van Ooyen AJ (2007) High-level production of beta-carotene in Saccharomyces cerevisiae by successive transformation with carotenogenic genes from Xanthophyllomyces dendrorhous. Appl Environ Microbiol 73:4342–4350. https://doi.org/10.1128/AEM.02759-06
Wu S, Letchworth GJ (2004) High efficiency transformation by electroporation of Pichia pastoris pretreated with lithium acetate and dithiothreitol. Biotechniques 36:152–154. https://doi.org/10.2144/04361DD02
Wu XL, Li BZ, Zhang WZ, Song K, Qi H, Dai JB, Yuan YJ (2017) Genome-wide landscape of position effects on heterogeneous gene expression in Saccharomyces cerevisiae. Biotechnol Biofuels 10:189. https://doi.org/10.1186/s13068-017-0872-3
Xie W, Liu M, Lv X, Lu W, Gu J, Yu H (2014) Construction of a controllable β-carotene biosynthetic pathway by decentralized assembly strategy in Saccharomyces cerevisiae. Biotechnol Bioeng 111:125–133
Xie W, Lv X, Ye L, Zhou P, Yu H (2015) Construction of lycopene-overproducing Saccharomyces cerevisiae by combining directed evolution and metabolic engineering. Metab Eng 30:69–78. https://doi.org/10.1016/j.ymben.2015.04.009
Xie W, Ye L, Lv X, Xu H, Yu H (2015) Sequential control of biosynthetic pathways for balanced utilization of metabolic intermediates in Saccharomyces cerevisiae. Metab Eng 28:8–18. https://doi.org/10.1016/j.ymben.2014.11.007
Xiong L, Zeng Y, Tang RQ, Alper HS, Bai FW, Zhao XQ (2018) Condition-specific promoter activities in Saccharomyces cerevisiae. Microb Cell Fact 17:58. https://doi.org/10.1186/s12934-018-0899-6
Yu T, Dabirian Y, Liu Q, Siewers V, Nielsen J (2019) Strategies and challenges for metabolic rewiring. Curr Opin Syste Biol 15:30–38. https://doi.org/10.1016/j.coisb.2019.03.004
Yuan J, Ching CB (2015) Dynamic control of ERG9 expression for improved amorpha-4,11-diene production in Saccharomyces cerevisiae. Microb Cell Fact 14:38. https://doi.org/10.1186/s12934-015-0220-x
Zhou P, Xie W, Yao Z, Zhu Y, Ye L, Yu H (2018) Development of a temperature-responsive yeast cell factory using engineered Gal4 as a protein switch. Biotechnol Bioeng 115:1321–1330. https://doi.org/10.1002/bit.26544
Acknowledgements
We would like to thank Guangdong Province Science and Technology Innovation Strategy Special Fund (Grant No. 2018B020206001), GDAS' Special Project of Science and Technology Development (Grant No. 2020GDASYL-20200302002), and the Science and Technology Plan Project of Guangdong Province (2016A010105013, 2019B030316017) for financial support.
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Su, B., Song, D., Yang, F. et al. Engineering a growth-phase-dependent biosynthetic pathway for carotenoid production in Saccharomyces cerevisiae. J Ind Microbiol Biotechnol 47, 383–393 (2020). https://doi.org/10.1007/s10295-020-02271-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10295-020-02271-x