Abstract
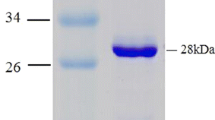
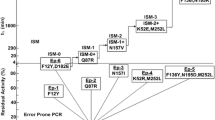
A gene encoding halohydrin dehalogenase (HHDH) from Agrobacterium tumefaciens CCTCC M 87071 was cloned and expressed in Escherichia coli. To increase activity and stability of HHDH, 14 amino acid residues around the active site and substrate-binding pocket based on the structural analysis and molecular docking were selected as targets for site-directed mutagenesis. The studies showed that the mutant HHDH (Mut-HHDH) enzyme had a more accessible substrate-binding pocket than the wild-type HHDH (Wt-HHDH). Molecular docking revealed that the distance between the substrate and active site was closer in mutant which improved the catalytic activity. The expressed Wt-HHDH and Mut-HHDH were purified and characterized using 1,3-dichloro-2-propanol (1,3-DCP) as substrates. The specific activity of the mutant was enhanced 26-fold and the value of k cat was 18.4-fold as compared to the Wt-HHDH, respectively. The Mut-HHDH showed threefold extension of half-life at 45 °C than that of Wt-HHDH. Therefore it is possible to add 1,3-DCP concentration up to 100 mM and epichlorohydrin (ECH) was produced at a relatively high conversion and yield (59.6 %) using Mut-HHDH as catalyst. This Mut-HHDH could be a potential candidate for the upscale production of ECH.










Similar content being viewed by others
References
Archelas A, Furstoss R (1997) Synthesis of enantiopure epoxides through biocatalytic approaches. Annu Rev Microbiol 51(1):491–525
Assis HMS, Sallis PJ, Bull AT, Hardman DJ (1998) Biochemical characterization of a haloalcohol dehalogenase from Arthrobacter erithii H10a. Enzyme Microb Technol 22(7):568–574
Bornscheuer UT, Hesseler M (2010) Enzymatic removal of 3-monochloro-1, 2-propanediol (3-MCPD) and its esters from oils. Eur J Lipid Sci Technol 112(5):552–556
Chung CT, Niemela SL, Miller RH (1989) One-step preparation of competent Escherichia coli: transformation and storage of bacterial cells in the same solution. Proc Natl Acad Sci USA 86(7):2172–2175
de Jong RM, Tiesinga JJW, Rozeboom HJ, Kalk KH, Tang LX, Janssen DB, Dijkstra BW (2003) Structure and mechanism of a bacterial haloalcohol dehalogenase: a new variation of the short-chain dehydrogenase/reductase fold without an NAD(P)H binding site. EMBO J 22(19):4933–4944
de Jong RM, Tiesinga JJW, Villa A, Tang LX, Janssen DB, Dijkstra BW (2005) Structural basis for the enantioselectivity of an epoxide ring opening reaction catalyzed by halo alcohol dehalogenase HheC. J Am Chem Soc 127(38):13338–13343
DeLano WL (2002) The PyMOL user’s manual. DeLano Scientific, San Carlos 452
Elenkov MM, Hauer B, Janssen DB (2006) Enantioselective ring opening of epoxides with cyanide catalysed by halohydrin dehalogenases: a new approach to non-racemic β-hydroxy nitriles. Adv Synth Catal 348(4–5):579–585
Fox RJ, Davis SC, Mundorff EC, Newman LM, Gavrilovic V, Ma SK, Chung LM, Ching C, Tam S, Muley S (2007) Improving catalytic function by ProSAR-driven enzyme evolution. Nat Biotechnol 25(3):338–344
Haak RM, Berthiol F, Jerphagnon T, Gayet AJA, Tarabiono C, Postema CP, Ritleng V, Pfeffer M, Janssen DB, Minnaard AJ (2008) Dynamic kinetic resolution of racemic β-haloalcohols: direct access to enantio-enriched epoxides. J Mol Catal B Enzym 130(41):13508–13509
Hasnaoui-Dijoux G, Majerić Elenkov M, Lutje Spelberg JH, Hauer B, Janssen DB (2008) Catalytic promiscuity of halohydrin dehalogenase and its application in enantioselective epoxide ring opening. Chem Biol Chem 9(7):1048–1051
Higgins TP, Hope SJ, Effendi AJ, Dawson S, Dancer BN (2005) Biochemical and molecular characterisation of the 2, 3-dichloro-1-propanol dehalogenase and stereospecific haloalkanoic dehalogenases from a versatile Agrobacterium sp. Biodegradation 16(5):485–492
Janssen DB, Majeric-Elenkov M, Hasnaoui G, Hauer B, Spelberg JHL (2006) Enantioselective formation and ring-opening of epoxides catalysed by halohydrin dehalogenases. Biochem Soc Trans 34(2):291–295
Jin HX, Hu ZC, Liu ZQ, Zheng YG (2012) Nitrite-mediated synthesis of chiral epichlorohydrin using halohydrin dehalogenase from Agrobacterium radiobacter AD1. Biotechnol Appl Biochem 59(3):170–177
Jin HX, Liu ZQ, Hu ZC, Zheng YG (2013) Production of (R)-epichlorohydrin from 1,3-dichloro-2-propanol by two-step biocatalysis using haloalcohol dehalogenase and epoxide hydrolase in two-phase system. Biochem Eng J 74:1–7
Kalogeris E, Antzoulatos O, Mamma D (2007) Application of different processes for the biodegradation of 1, 3-dichloro-2-propanol by the bacterium Pseudomonas putida DSM437. Chem Biochem Eng Q 21(3):297–305
Kawthekar RB, Bi W-t, Kim G-J (2008) Synthesis and application of bimetallic chiral [Co(salen)]-type complexes: a new catalytic approach to synthesis of optically pure β-blockers via kinetic resolution of epichlorohydrin. Appl Organomet Chem 22(10):583–591
Laemmli UK (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227(5259):680–685
Liu ZQ, Zhang LP, Cheng F, Ruan LT, Hu ZC, Zheng YG, Shen YC (2011) Characterization of a newly synthesized epoxide hydrolase and its application in racemic resolution of (R, S)-epichlorohydrin. Catal Commun 16(1):133–139
Lutje Spelberg JH, Tang LX, van Gelder M, Kellogg RM, Janssen DB (2002) Exploration of the biocatalytic potential of a halohydrin dehalogenase using chromogenic substrates. Tetrahedron Asymmetry 13(10):1083–1089
Majeric Elenkov M, Tang LX, Hauer B, Janssen DB (2006) Sequential kinetic resolution catalyzed by halohydrin dehalogenase. Org Lett 8(19):4227–4229
Nagasawa T, Nakamura T, Yu F, Watanabe I, Yamada H (1992) Purification and characterization of halohydrin hydrogen-halide lyase from a recombinant Escherichia coli containing the gene from a Corynebacterium sp. Appl Microbiol Biotechnol 36(4):478–482
Nouailhas H, Aouf C, Le Guerneve C, Caillol S, Boutevin B, Fulcrand H (2011) Synthesis and properties of biobased epoxy resins. Part 1. Glycidylation of flavonoids by epichlorohydrin. J Polym Sci A 49(10):2261–2270
Sambrook J, Russell DW, Russell DW (2001) Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, New York
Smith PK, Krohn RI, Hermanson GT, Mallia AK, Gartner FH, Provenzano MD, Fujimoto EK, Goeke NM, Olson BJ, Klenk DC (1985) Measurement of protein using bicinchoninic acid. Anal Biochem 150(1):76–85
Tang LX, Torres Pazmiño DE, Fraaije MW, de Jong RM, Dijkstra BW, Janssen DB (2005) Improved catalytic properties of halohydrin dehalogenase by modification of the halide-binding site. Biochemistry 44(17):6609–6618
Tang LX, van Hylckama Vlieg JET, Lutje Spelberg JH, Fraaije MW, Janssen DB (2002) Improved stability of halohydrin dehalogenase from Agrobacterium radiobacter AD1 by replacement of cysteine residues. Enzyme Microb Technol 30(2):251–258
Tang LX, van Merode AEJ, Lutje Spelberg JH, Fraaije MW, Janssen DB (2003) Steady-state kinetics and tryptophan fluorescence properties of halohydrin dehalogenase from Agrobacterium radiobacter. Roles of W139 and W249 in the active site and halide-induced conformational change. Biochemistry 42(47):14057–14065
Van Den Wijngaard AJ, Janssen DB, Witholt B (1989) Degradation of epichlorohydrin and halohydrins by bacterial cultures isolated from freshwater sediment. J Gen Microbiol 135(8):2199–2208
van den Wijngaard AJ, Reuvekamp PT, Janssen DB (1991) Purification and characterization of haloalcohol dehalogenase from Arthrobacter sp. strain AD2. J Bacteriol 173(1):124–129
van Hylckama Vlieg JET, Tang LX, Spelberg JHL, Smilda T, Poelarends GJ, Bosma T, van Merode AEJ, Fraaije MW, Janssen DB (2001) Halohydrin dehalogenases are structurally and mechanistically related to short-chain dehydrogenases/reductases. J Bacteriol 183(17):5058–5066
Yonetani R, Ikatsu H, Miyake-Nakayama C, Fujiwara E, Maehara Y, Miyoshi S, Matsuoka H, Shinoda S (2004) Isolation and characterization of a 1, 3-dichloro-2-propanol-degrading bacterium. J Health Sci 50(6):605–612
You ZY, Liu ZQ, Zheng YG, Shen YC (2013) Characterization and application of a newly synthesized 2-deoxyribose-5-phosphate aldolase. J Ind Microbiol Biotechnol 40(1):29–39
Yu CL, Parales RE, Gibson DT (2001) Multiple mutations at the active site of naphthalene dioxygenase affect regioselectivity and enantioselectivity. J Ind Microbiol Biotechnol 27(2):94–103
Yu F, Nakamura T, Mizunashi W, Watanabe I (1994) Cloning of two halohydrin hydrogen-halide-lyase genes of Corynebacterium sp. strain N-1074 and structural comparison of the genes and gene products. Biosci Biotechnol Biochem 58(8):1451–1457
Zheng HC, Liu YH, Sun MZ, Han Y, Wang JL, Sun JS, Lu FP (2013) Improvement of alkali stability and thermostability of Paenibacillus campinasensis family-11 xylanase by directed evolution and site-directed mutagenesis. J Ind Microbiol Biotechnol 41(1):153–162
Zou SP, Du EH, Hu ZC, Zheng YG (2013) Enhanced biotransformation of 1,3-dichloro-2-propanol to epichlorohydrin via resin-based in situ product removal process. Biotechnol Lett 35(6):937–942
Acknowledgments
This work was financially supported by the National Natural Science Foundation of China (No. 21176224), National Basic Research Program of China (973 Program) (No. 2011CB710806), National Major Project of Scientific Instruments Development of China (No. 2012YQ150087) and the Natural Science Foundation of Zhejiang Province of China (No. Z4080032 and R3110155).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Liu, ZQ., Gao, AC., Wang, YJ. et al. Expression, characterization, and improvement of a newly cloned halohydrin dehalogenase from Agrobacterium tumefaciens and its application in production of epichlorohydrin. J Ind Microbiol Biotechnol 41, 1145–1158 (2014). https://doi.org/10.1007/s10295-014-1443-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10295-014-1443-2