Abstract
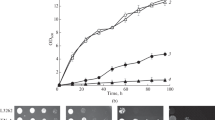
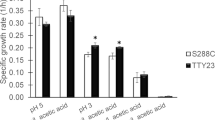
Acetic acid existing in a culture medium is one of the most limiting constraints in yeast growth and viability during ethanol fermentation. To improve acetic acid tolerance in Saccharomyces cerevisiae strains, a drug resistance marker-aided genome shuffling approach with higher screen efficiency of shuffled mutants was developed in this work. Through two rounds of genome shuffling of ultraviolet mutants derived from the original strain 308, we obtained a shuffled strain YZ2, which shows significantly faster growth and higher cell viability under acetic acid stress. Ethanol production of YZ2 (within 60 h) was 21.6% higher than that of 308 when 0.5% (v/v) acetic acid was added to fermentation medium. Membrane integrity, higher in vivo activity of the H+-ATPase, and lower oxidative damage after acetic acid treatment are the possible reasons for the acetic acid-tolerance phenotype of YZ2. These results indicated that this novel genome shuffling approach is powerful to rapidly improve the complex traits of industrial yeast strains.







Similar content being viewed by others
References
Abbott DA, Knijnenburg TA, de Poorter LMI, Reinders MJT, Pronk JT, van Maris AJA (2007) Generic and specific transcriptional responses to different weak organic acids in anaerobic chemostat cultures of Saccharomyces cerevisiae. FEMS Yeast Res 7(6):819–833
Bai FW, Anderson WA, Moo-Young M (2008) Ethanol fermentation technologies from sugar and starch feedstocks. Biotechnol Adv 26(1):89–105
Bracey D, Holyoak CD, Nebe-von Caron G, Coote PJ (1998) Determination of the intracellular pH (pH(i)) of growing cells of Saccharomyces cerevisiae: the effect of reduced-expression of the membrane H+-ATPase. J Microbiol Meth 31(3):113–125
Causton HC, Ren B, Koh SS, Harbison CT, Kanin E, Jennings EG, Lee TI, True HL, Lander ES, Young RA (2001) Remodeling of yeast genome expression in response to environmental changes. Mol Biol Cell 12(2):323–337
Chen AK, Gelling C, Rogers PL, Dawes IW, Rosche B (2009) Response of Saccharomyces cerevisiae to stress-free acidification. J Microbiol 47(1):1–8
Coyle JT, Puttfarcken P (1993) Oxidative stress, glutamate, and neurodegenerative disorders. Science 262(5134):689–695
Dai MH, Ziesman S, Ratcliffe T, Gill RT, Copley SD (2005) Visualization of protoplast fusion and quantitation of recombination in fused protoplasts of auxotrophic strains of Escherichia coli. Metab Eng 7(1):45–52
Gietz RD, Schiestl RH (2007) High-efficiency yeast transformation using the LiAc/SS carrier DNA/PEG method. Nat Protoc 2(1):31–34
Graves T, Narendranath NV, Dawson K, Power R (2006) Effect of pH and lactic or acetic acid on ethanol productivity by Saccharomyces cerevisiae in corn mash. J Ind Microbiol Biotechnol 33(6):469–474
Graves T, Narendranath NV, Dawson K, Power R (2007) Interaction effects of lactic acid and acetic acid at different temperatures on ethanol production by Saccharomyces cerevisiae in corn mash. Appl Microbiol Biotecnol 73(5):1190–1196
Guldener U, Heck S, Fiedler T, Beinhauer J, Hegemann JH (1996) A new efficient gene disruption cassette for repeated use in budding yeast. Nucleic Acids Res 24(13):2519–2524
Hida H, Yamada T, Yamada Y (2007) Genome shuffling of Streptomyces sp. U121 for improved production of hydroxycitric acid. Appl Microbiol Biotecnol 73(6):1387–1393
Hou L (2009) Novel methods of genome shuffling in Saccharomyces cerevisiae. Biotechnol Lett 31(5):671–677
Kai Z, Ping WX, Lina ZN, Jun L, Yan L, Tao J, Zhou DP (2008) Screening and breeding of high taxol producing fungi by genome shuffling. Sci China C Life Sci 51(3):222–231
Kawahata M, Masaki K, Fujii T, Iefuji H (2006) Yeast genes involved in response to lactic acid and acetic acid: acidic conditions caused by the organic acids in Saccharomyces cerevisiae cultures induce expression of intracellular metal metabolism genes regulated by Aft1p. FEMS Yeast Res 6(6):924–936
Liu ZL (2006) Genomic adaptation of ethanologenic yeast to biomass conversion inhibitors. Appl Microbiol Biotecnol 73(1):27–36
Ludovico P, Sousa MJ, Silva MT, Leao C, Corte-Real M (2001) Saccharomyces cerevisiae commits to a programmed cell death process in response to acetic acid. Microbiology 147:2409–2415
Mollapour M, Piper PW (2006) Hog1p mitogen-activated protein kinase determines acetic acid resistance in Saccharomyces cerevisiae. FEMS Yeast Res 6(8):1274–1280
Mollapour M, Piper PW (2007) Hog1 mitogen-activated protein kinase phosphorylation targets the yeast Fps1 aquaglyceroporin for endocytosis, thereby rendering cells resistant to acetic acid. Mol Cell Biol 27(18):6446–6456
Montooth KL, Siebenthall KT, Clark AG (2006) Membrane lipid physiology and toxin catabolism underlie ethanol and acetic acid tolerance in Drosophila melanogaster. J Exp Biol 209(Pt 19):3837–3850
Nakazawa N, Iwano K (2004) Efficient selection of hybrids by protoplast fusion using drug resistance markers and reporter genes in Saccharomyces cerevisiae. J Biosci Bioeng 98(5):353–358
Narendranath NV, Thomas KC, Ingledew WM (2001) Effects of acetic acid and lactic acid on the growth of Saccharomyces cerevisiae in a minimal medium. J Ind Microbiol Biotechnol 26(3):171–177
Otte B, Grunwaldt E, Mahmoud O, Jennewein S (2009) Genome shuffling in Clostridium diolis DSM 15410 for improved 1, 3-propanediol production. Appl Environ Microbiol 75(24):7610–7616
Pampulha ME, Loureirodias MC (1989) Combined effect of acetic-acid, pH and ethanol on intracellular pH of fermenting yeast. Appl Microbiol Biotecnol 31(5–6):547–550
Piper P, Mahe Y, Thompson S, Pandjaitan R, Holyoak C, Egner R, Muhlbauer M, Coote P, Kuchler K (1998) The Pdr12 ABC transporter is required for the development of weak organic acid resistance in yeast. EMBO J 17(15):4257–4265
Portillo F, Serrano R (1989) Growth-control strength and active-site of yeast plasma-membrane ATPase studied by site-directed mutagenesis. Eur J Biochem 186(3):501–507
Serrano R (1984) Plasma-membrane ATPase of fungi and plants as a novel type of proton pump. Curr Top Cell Regul 23:87–126
Shi DJ, Wang CL, Wang KM (2009) Genome shuffling to improve thermotolerance, ethanol tolerance and ethanol productivity of Saccharomyces cerevisiae. J Ind Microbiol Biotechnol 36(1):139–147
Viegas CA, Almeida PF, Cavaco M, Sa-Correia I (1998) The H(+)-ATPase in the plasma membrane of Saccharomyces cerevisiae is activated during growth latency in octanoic acid-supplemented medium accompanying the decrease in intracellular pH and cell viability. Appl Environ Microbiol 64(2):779–783
Wei P, Li Z, He P, Lin Y, Jiang N (2008) Genome shuffling in the ethanologenic yeast Candida krusei to improve acetic acid tolerance. Biotechnol Appl Biochem 49(2):113–120
Yenush L, Mulet JM, Arino J, Serrano R (2002) The Ppz protein phosphatases are key regulators of K+ and pH homeostasis: implications for salt tolerance, cell wall integrity and cell cycle progression. EMBO J 21(5):920–929
Zhang YX, Perry K, Vinci VA, Powell K, Stemmer WP, del Cardayre SB (2002) Genome shuffling leads to rapid phenotypic improvement in bacteria. Nature 415(6872):644–646
Acknowledgments
We thank Dr. Johannes H. Hegemann and Thomas D. Petes for supply of plasmids pSH65 and pFA6a-kanMX4. Thanks also to Henan Tianguan Fuel Ethanol Co. Ltd. for yeast strains providing.
Author information
Authors and Affiliations
Corresponding author
Additional information
D.-Q. Zheng and X.-C. Wu contributed equally to this work.
Rights and permissions
About this article
Cite this article
Zheng, DQ., Wu, XC., Wang, PM. et al. Drug resistance marker-aided genome shuffling to improve acetic acid tolerance in Saccharomyces cerevisiae . J Ind Microbiol Biotechnol 38, 415–422 (2011). https://doi.org/10.1007/s10295-010-0784-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10295-010-0784-8