Abstract
Climate change, malnutrition, and food insecurity are the inevitable challenges being faced by the agriculture sector today. Plants are susceptible to extreme temperatures during the crucial phases of flowering and seed development, and elevated carbon levels also lead to yield losses. Productivity is also affected by floods and droughts. Therefore, increasing plant yield and stress tolerance are the priorities to be met through novel biotechnological interventions. The contributions of NAC genes towards enhancing plant survivability under stress is well known. Here we focus on the potential of NAC genes in the regulation of abiotic stress tolerance, secondary cell wall synthesis, lateral root development, yield potential, seed size and biomass, ROS signaling, leaf senescence, and programmed cell death. Once naturally tolerant candidate NAC genes have been identified, and the nature of their association with growth and fitness against multi-environmental stresses has been determined, they can be exploited for building inherent tolerance in future crops via transgenic technologies. An update on the latest developments is provided in this review, which summarizes the current understanding of the roles of NAC in the establishment of various stress-adaptive mechanisms in model and food crop plants.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Transcription factors (TFs) contribute to about 7% of the coding part of plant transcriptomes. Several TFs also function as immediate or early stress-responsive factors against biological triggers (Hoang et al. 2017; Lindemose et al. 2013). Transcription factors are well known to initiate the reaction cascades by binding to cis-elements upstream of target genes, which encode proteins for particular biological roles (Baillo et al. 2019). A typical TF is comprised of four parts: a conserved DNA-binding part, a variable transcription-regulation part, an oligomerization part, and a nuclear localization signal (NLS) for protein import into the nucleus. The preferential involvement of some TFs as master regulators of signaling and regulatory mechanisms in stress acclimatization is well explored (Hoang et al. 2019). For instance, members of TF sub-families (CBF/DREB, MYB, WRKY, NAC, bZIP, APETALA, C2-H2 type zinc fingers, basic helix-loop-helix etc.) are recognized to be intimately involved in transforming stress signals into alterations in gene expression and thereby triggering adaptive responses in plant cells (Kosová et al. 2015; Lata et al. 2011). There are several recent examples of stress-related TFs from the NAC (NAM, ATAF, and CUC) TF subfamily. The acronym NAC originates from three different genes (No Apical Meristem: NAM, Arabidopsis Transcription Activation Factor: ATAF, and Cup-Shaped Cotyledon: CUC), where NAC domain was first reported (Aida 1997; Sablowski et al.1998). NAC is considered one of the largest TF families in plants, with more than 100 genes reported in Arabidopsis thaliana (L.) Heynh. (Arabidopsis) as well as several other members of the plant kingdom (Table 1) (Baillo et al. 2019; Singh et al. 2019).
The NAC TFs control plant development, senescence, morphogenesis, and abiotic stress tolerances (Kosová et al. 2015; Singh et al. 2016). It is noteworthy that NAC members constitute a well conserved DNA binding (NAC) domain at the N-terminal and a varied C-terminal domain that generally has an intrinsically disordered region. The intrinsically disordered region contributes to various biological functions among sub-families, for instance, regulates transcription via an ABA-dependent or ABA-independent pathway to modulate stress-related gene expression (Baillo et al. 2019; Ernst et al. 2004). There are even more complex functions associated with NACs, for example, influencing miRNA mediated cleavage of mRNAs (Mallory et al. 2004) and ubiquitin-dependent proteolysis (Xie et al. 2002). Whether this complex TF family has roles limited to stress and plant development, is still under debate.
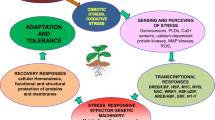
This review primarily focuses on the role this plant-specific TF subfamily plays during general developmental and adaptive regulation in food crops. Here, we summarize the evidence currently available (particularly from the past decade), concerning the range of NAC functions in controlling the growth and development of the plants, and their participation in the plant’s adaptive response against a variety of stresses (Fig. 1). We also discuss important translational aspects associated with NAC TFs in combating the latest challenges of climate change through biotechnological interventions.
NAC proteins: structure, function, regulation co-relations
As noted above, NAC proteins are represented by a conserved N-terminal DNA binding domain and a variable transcription regulatory region at the C-terminus, which plays a role in either transcriptional activation or repression of stress-induced genes and pathways (Puranik et al. 2012; Yamaguchi et al. 2010, Fig. 2). The N-terminus is separated into sub-domains dubbed A through E. Within the conserved N-terminal domain too, the sequence varies. Sub-domains C and D are highly conserved and positively charged. These sub-domains bind to DNA. The positions Lys123 and Lys126 (β4–5; sub-domain D), Val119–Ser183 (β4–6; sub-domain D–E), and Lys79, Arg85, and Arg88 (β1–2; sub-domain C) are considered essential residues for DNA binding (Chen et al. 2011; Ernst et al. 2004). Among these, Arg88 has so far been found conserved in all NAC proteins (Puranik et al. 2012). The redundancy of Lys79 and Arg85 has also been proposed as a reason for the varying DNA binding abilities of NAC proteins (Jensen et al. 2010). The nuclear localization signal rests in the sub-domain D, mediated by the lysine residues which help in nuclear shuttling (Le et al. 2011; Olsen et al. 2005a, b; Tran et al. 2009). Sub-domains B and E are relatively divergent and may be contributing to NAC protein function diversity, along with the C-terminal domains. Sub-domain A plays a role in functional dimer formation through Leu14–Thr23 and Glu26–Tyr31 residues (Ernst et al. 2004; Jensen et al. 2010; Olsen et al. 2005a; Ooka et al. 2003; Puranik et al. 2012). Model of a typical NAC protein from pearl millet (PgNAC21) showing α-helix and twirled β-sheet bound to target DNA can be seen in Fig. 3a, b.
The NAC domain has also been implicated in protein binding activities, which in turn may be crucial to various functions of the NAC proteins including stress tolerance (Olsen et al. 2005a; Tran et al. 2007; Yamaguchi et al. 2010). The C-terminal domain has a low complexity region containing serine–threonine, proline–glutamine, or acidic residue repeats (Fang et al. 2008). This causes an intrinsic disorder and renders a lack of stable three-dimensional structures (Jensen et al. 2010). However, this makes the NACs interact with diverse targets (Lipoxygenase, DEAD/DEAH box helicase, Pectin methyl esterase inhibitor, and Homeobox associated proteins coding genes, etc.) (Grover et al. 2014). These motifs are conserved within a given sub-family but vary among groups of sub-families.
To this end, there are several sub-families of the NAC proteins, classified and described in the literature (Hu et al. 2010; Lv et al. 2016). Some works reported as many as 18 sub-families for NAC TFs (Shang et al. 2013), though no definitive role has been assigned to a particular sub-family. These uncertainties emerge because of the rich diversity of NAC domains. This is also the reason for limited success in identifying the role of NAC individual genes within a given sub-family (Puranik et al. 2012). Additionally, these attributes of NAC sub-families are also indicative of redundancy for the target gene members. However, identifying the expression pattern will be the first step in understanding how these NAC members are involved in stress response.
Expression of NAC genes during development and stress response
Stress-responsive NAC genes are expressed differentially and are highly regulated at the transcriptional level. Phylogenetic analysis indicates that stress-responsive NAC TFs contain the closely homologous NAC domain (Fang et al. 2008; Tran et al. 2009). Analysis of Arabidopsis seedlings using the Agilent 22 K Oligo DNA microarray revealed that 12 ANAC genes out of 67 (reported in the genome), were induced by ABA and abiotic stresses (Fujita et al. 2004). Arabidopsis salt-stressed root transcriptome using 70mer oligomer microarray probes comprising 23,686 genes, exposed differential expression in the ANAC genes: 23 were up-regulated whereas seven genes were suppressed (Jiang and Deyholos 2006). Other work by Matsui et al. (2008) showed that, when the whole genome expression profiling (tiling array) in three-week old Arabidopsis seedlings was subjected to drought, cold, salinity, and ABA stress, 30 ANAC genes out of 108, were up-regulated under at least one of the imposed stresses. Later, Jiang et al. (2014) reported hyper-sensitivity of lines of Arabidopsis overexpressing the RhNAC3 gene from rose (Rosa hybrida) upon ABA or drought stresses during seed germination and leaf closure stages. Nevertheless, NAC family members exhibit preferential gene expression at various life cycle stages, or during the development of tissues. In this regard, NTM1-Like or “NAC with Transmembrane Motif 1”-Like (NTLs), a class of membrane-associated NAC transcription factor, which is known to be associated with transcriptional response to external stimuli, and linked to delayed flowering (NTL8). Investigations revealed that higher expression of NTLs was linked to a reduced expression of FLOWERING LOCUS T (FT). This in turn resulted in flowering delay, reduced growth and leaf curling in Arabidopsis (Kim et al. 2007).
In Medicago truncatula, out of 97 MtNAC candidate genes, 40 NACs were expressed in different tissues –roots, buds, seed pods, and flowers (Ling et al. 2017). Among the expressed NACs, nine genes were preferentially expressed in roots, 13 in seed pods, and three in buds. Moreover, RNA-seq data analysis showed that 44 MtNAC genes were found regulated by various stresses such as cold, drought, salt, freezing, and ABA-stress. Of these, 17 MtNAC genes were up-regulated, whereas only MtNAC1 was down-regulated under all stresses. Further, 33 genes were induced exclusively by cold and drought, whereas 12 genes were specifically expressed during freezing and salinity stresses. Expression of MtNAC50 was highly up-regulated during cold, and MtNAC95 was up-regulated in salt, drought, and ABA stresses. Similarly, MtNAC57 and MtNAC73 were up-regulated during all stresses apart from freezing (Ling et al. 2017).
Fang and co-authors (Fang et al. 2008) employed 70mer oligomer microarray analysis to identify 140 putative ONAC-like TFs in rice (Oryza sativa). Twenty-one of these were induced by drought or salinity and five were repressed by stress in the seedling stage. The same authors (Fang et al. 2008) validated 20 ONAC genes with elevated expression levels using rice seedlings and found that five genes were induced by dehydration, 19 by salt, and 16 by cold.
Ha et al. (2014) used phylogenetic analysis to identify 71 CaNAC genes, including eight membrane-bound NACs, from the chickpea (Cicer arietinum) genome. Nineteen of the predicted 23 dehydration-related CaNAC genes were specific to either roots or leaves. Fourteen genes were up-regulated, whereas four were down-regulated under dehydration stress in leaves. CaNAC06 and CaNAC67 were the most up-regulated genes with 200 to 300-fold. The highly down-regulated genes were CaNAC02 and CaNAC04. In root tissues, 12 genes were up-regulated, and three genes were down-regulated during dehydration. By comparison, 88 NAC genes were identified in the pigeon pea (Cajanus cajan), using homology searches, and de novo approaches based on the published pigeon pea draft genome (Satheesh et al. 2014). Of these, 36 NAC genes were identified as putatively drought-responsive, based on the phylogenetic analysis (Satheesh et al. 2014). Several stress-responsive cis-acting regulatory elements (MYB, TC rich repeats, HSE element, ABRE element) were reported from promoter regions of these NAC genes, which may contribute to enhancing the stress tolerance.
More recently, Hussain et al. (2017) identified 139 GmNAC genes in the soybean (Glycine max), and observed genotype-based GmNAC gene expression in response to drought. Out of these 139 GmNAC genes, 28 genes were predicted to be drought-responsive, based on the phylogenetic analysis. Eight of the GmNAC genes (GmNAC004, 021, 065, 066, 073, 082, 083, 08) showed higher expression levels in drought-resistant cultivars than in drought-sensitive cultivars and were induced despite the level of dehydration sensitivity of cultivars. Earlier, Le et al. (2011) reported 50 putative stress-responsive GmNAC genes, based on the sequence alignment and phylogenetic analysis with known Arabidopsis (ANAC055, ANAC072, ANAC019) and rice (SNAC1/SNAC2) stress-responsive NAC genes. Sixteen GmNACs were tissue-specific and highly expressed in roots and flowers. Twenty-five GmNACs were induced, and six were repressed by two-fold or more under dehydration stress in roots and shoots of soybean. GmNAC085, which is identical to the widely-studied SNAC1/ONAC2, displayed induction of 390-fold in shoots and 20-fold in roots and was amongst those genes most highly expressed gene during dehydration.
The overall expression patterns observed across different plants imply the selective up-regulation of individual NAC members and indicate the contribution of NAC TFs in the stress adaptation scheme (Figs. 4, 5, 6). However, there is still debate about whether there are several NAC candidates with conserved roles across plant species in response to stress type. In the next sections, we summarize the NAC roles more specific to various stress adaptive schemes and how this information could be utilized to exploit NACs for crop improvement.
Schematic representation of NAC regulation in plant abiotic stress tolerance. Upon encountering stress, the NAC gene is induced by ABA dependent/independent pathway, which in turn binds to the promoter sequences of other genes (COR, DREB etc.), thereby regulating ROS, GSTF, FT expression. On the other hand, accumulation of unfolded/misfolded proteins triggers unfolded protein response (UPR) in the ER. Thus, membrane bound NAC domain proteins (NTLs) are activated by ER stress and undergo conformational changes (TMD cleavage) to modulate the expression of stress-induced gene. NTLs thus, plays important role in communicating ER stress signaling from PM to nucleus to mitigate the ER stress. NACRS NAC recognition site/sequence, COR Cold regulated, RD responsive to desiccation, ER endoplasmic reticulum, NTL NAC with Transmembrane Motif 1″-Like, TMD transmembrane domain, AREB abscisic acid-responsive element-binding protein, DREB drought-responsive element-binding, ROS reactive oxygen species, GSTF glutathione S-transferase; FT flowering locus T; UPR unfolded protein response, PM plasma membrane
Representation of the NAC transcriptional regulatory network associated with stress adaptation from case studies of rice (Drought: Chung et al. 2018, Heat: Fang et al. 2015) and wheat (Salinity: Zhang et al. 2016, Huang et al. 2015). a During heat, ONAC003 (SNAC3) expression is induced upon perceiving stress. Binding of ONAC003 with NAC-specific NACRS (NAC recognition sequences/site) and CDBS (Core DNA-binding sequence,) and is activated. This results in up-regulation of targets CATA (Catalase isozyme A-like), APX3 (Ascorbate peroxidase 3), APX8 (Ascorbate peroxidase 8), RbohF (NADPH oxidase), and Prx IIE2 (Peroxiredoxin), thereby reducing peroxide levels (H2O2), MDA (Malondialdehyde) and electrolytic leakage. This modulation of ROS metabolism by target gene up-regulation results in heat tolerance in rice plants. b OsNAC regulatory networks include the activation of OsNAC5, 6, 9, and 10 TFs during drought stress. Further, OsNAC activation leads to the up-regulation of various ERF (Ethylene responsive factor) domain genes. Target genes, such as OsERF1, OsERF54, OsERF57 and OsERF107, and OsERF9 are directly up-regulated by OsNAC5, OsNAC6, OsNAC9 and OsNAC10. The up-regulation of target genes (OsERF1, OsERF54, OsERF57, OsERF107, and OsERF9) leads to the increased expression of LAC26 (Laccase-22-like), TAG lipase 2 (Triacylglycerol lipase 2), Glucan endo-1,3-beta-glucosidase 6, and OsEXP7 (expansin-A7-like), which altogether resulted in alteration of the root architectures for drought tolerance and enhanced grain yield in rice. c In case of salinity stress, TaNAC47 and TaNAC29 expression is induced in response to stress signaling via the ABA dependent pathway. TaNAC47 binds to ABRE cis-elements and activates transcription. This leads to up-regulation of targets such as, RD29A (Responsive to desiccation A), RD29B (Responsive to desiccation B), GSTF6 (Glutathione S-transferase F6), RD20 (Responsive to desiccation 20), P5CS1 (Δ1-pyrroline-5-carboxylate synthetase 1), COR47 (Cold regulated 47) in Arabidopsis. This, in turn, leads to reduced water loss and increased proline and soluble sugar, further improving survival under salinity stress in plants. Similarly, increased expression of TaNAC29 during salinity stress leads to up-regulation of RD29b (Responsive to desiccation 29b) and ERD11 (Early responsive to dehydration) target genes, thereby enhancing SOD (Superoxide dismutase) and CAT (Catalase) activity, resulting in reduced water and enhanced salinity tolerance in Arabidopsis
Representation of the NAC transcriptional regulatory network associated with stress adaptation from cases studied in SAT cereals (Shinde et al. 2019) and legume (Yu et al. 2016) to provide stress tolerance in Arabidopsis and rice plants. Blue arrows indicate up-regulation; red arrows indicate down-regulation. a Chickpea CarNAC4 is induced under drought and salinity. Upon activation, CarNAC4 in Arabidopsis plants causes up-regulation of RD29, COR15A, KIN1, DREB genes, resulting in lower MDA and low water loss. b The pearl millet PgNAC21 regulatory network involves induction of NAC21 in response to salt and ABA exposure. Binding of MYB1 to upstream regulatory sequences (NACRS) of PgNAC21 in Arabidopsis plants activates its expression, which then leads to up-regulation of target gene (COR, RD20, GSTF6), resulting in salt stress adaptation. c Finger millet EcNAC67 is induced during salinity and drought stress. Activation of EcNAC67 in rice plants causes a greater abundance of gene transcripts, which result in increased plant biomass, lower yield loss and better survival of rice plants under drought and salt conditions. SAT semi-arid tropics, NACRS NAC recognition site, COR15A cold regulated, RD29 responsive to desiccation29, GSTF6 glutathione S-transferase F6, MDA malondialdehyde
Potential of NACs in crop improvement
Abiotic stress tolerance
Although NAC proteins were initially found to associate with plant development, their involvement in stress responses is now being increasingly recognized (Jensen and Skriver 2014). Classically, three closely associated NAC proteins members (ANA019, ANAC055, and ANAC072 (RD26)) respond to various abiotic stresses and hormones such as dehydration, freezing, salinity, wounding, jasmonic acid (JA), and abscisic acid (ABA). Most of the knowledge about these proteins stems from genome-wide and functional genomics studies in the most studied plants such as soybean (Hao et al. 2011), rice (Ohnishi et al. 2005), and Arabidopsis (Jiang et al. 2014), among others. Over-expression of these TFs has often provided substantial evidence supporting their activities (Table 2). For instance, transgenic rice plants overexpressing the SNAC2 gene demonstrated a significantly enhanced germination and higher growth rates than the wild type (WT) under imposed salinity (Hu et al. 2008). Additionally, these plants showed greater vigor than non-transformed controls under a freezing environment. Similarly, transgenic tobacco lines overexpressing the poplar NAC13 gene, demonstrated enhanced salt tolerance (Cheng et al. 2020).
OsSNAC1, a rice stress-responsive TF, was shown to ameliorate salinity and drought tolerance in wheat cultivars when over-expressed (Saad et al. 2013). Wheat plants overexpressing the OsSNAC1 gene had elevated sensitivity to abscisic acid (ABA), which caused higher water and chlorophyll levels in their leaf tissues with an increased fresh and dry root weight. Furthermore, a higher level of OsSNAC1 was involved in regulating the expression of abiotic stresses and ABA signaling genes such as wheat 1-phosphatidylinositol-3-phosphate-5-kinase, type 2C protein phosphatases, sucrose phosphate synthase, and the regulatory components of the ABA receptor (Saad et al. 2013). Conversely, OsSNAC1 over-expression in cotton (Gossypium hirsutum) plants improved drought and salt tolerance by facilitating vigorous root growth and lowering the transpiration frequency relative to non-transformed plants (Liu et al. 2014). OsNAC6 from rice was induced by multiple stresses such as cold, abscisic acid (ABA), drought, salt, and JA, and is thus considered a redundant candidate for signals derived from abiotic as well as biotic stresses in rice (Ohnishi et al. 2005). This gene is also induced by wounding, along with other early-responsive genes (Ohnishi et al. 2005). Ochiai et al. (2011) claimed an enhanced tolerance to Boron toxicity by inhibition of the NAC-like transcription factor BORON EXCESS TOLERANT1 (BET1) gene in transgenic rice. A recent example includes ONAC066, which is induced in response to multiple stresses: polyethylene glycol (PEG), H2O2, or salinity treatments of the ONAC066-overexpressing transgenic rice plants resulted in greater accumulation of soluble sugars and proline, reduction in reactive oxygen species (ROS), and water loss, thereby providing accelerated drought and oxidative resistance in rice plants (Yuan et al. 2019). Similarly, OsNAC14 provided increased drought resistance in over-expressed rice plants during vegetative growth by repairing the damaged DNA and defense mechanism (Shim et al. 2018). Transgenic plants also had greater panicle number and a faster grain filling rate than WT (Shim et al. 2018). OsNAC14 functions by binding to the OsRAD51A1 promoter, a constituent of DNA repair machinery. Another case reported by Fang et al. (2015) revealed the importance of the SNAC3 (ONAC003) gene in conferring heat and drought endurance through the up-regulation of high temperature responsive genes (Fig. 5a). Further, SNAC3 overexpressing rice plants exhibited lesser electrolytic leakage, Malondialdehyde (MDA) and peroxides levels than WT at high temperatures, thereby demonstrating tolerance via ROS modulation (Fang et al. 2015). Similarly, OsNAC5, 6, 9, and 10 conferred drought endurance in over-expressed rice plants by up-regulating target genes that are responsible for altering root architecture (Chung et al. 2018, Fig. 5b). Transgenic plants also had reduced grain yield loss under drought stress compared to WT.
TaNAC29, a wheat (Triticum aestivum) NAC TF, was reported to boost tolerance against salt and drought when over-expressed in Arabidopsis plants, but compromised the flowering time (Huang et al. 2015, Fig. 5c). In another study, over-expression of TaNAC47 in Arabidopsis caused ABA hypersensitivity resulting in the activation of a plethora of responses by altering gene expression and displayed enhanced resistance towards PEG, salinity, and freezing stresses in transgenic plants (Zhang et al. 2016, Fig. 5c). Another possible dehydration tolerance case occurred when TaRNAC1 was over-expressed in transgenic wheat under PEG. This generated higher aboveground biomass and yield under water-deficit conditions (Chen et al. 2018). TaNAC2L over-expression, induced by high-temperature, stimulated heat-responsive gene expression, thereby enhancing thermotolerance in transformed Arabidopsis plants (Guo et al. 2015). Earlier, Mao et al. (2012) also reported the role of the TaNAC2 allele in improving drought, salt, and freezing tolerance in TaNAC2-overexpressing Arabidopsis plants. The role of NAC family TFs in wheat is not limited to abiotic stress only. A recent report indicated that TaNACL-D1 interaction with TaFROG (Fusarium Resistance Orphan Gene) can facilitate resistance to Fusarium head blight disease. Unusually, TaNACL harbors the Triticeae-specific protein in the C-terminal region (Perochon et al. 2019). As discussed in previous sections, the virtue of complexity in the C-terminus in the NAC TF genes is fascinating. Therefore, it will be interesting to see more cases of genus-specific roles of NAC members in the future.
Documentation of the role of NAC members in stress adaptation is not limited to Triticeae. ZmNAC111, a maize (Zea mays) TF, was associated with increased drought tolerance of maize seedlings and water-use efficiency (WUE), along with expression of drought-responsive genes during water deficit (Mao et al. 2015). Another report claimed the involvement of ZmNAC55 in inducing drought resistance to overexpressing Arabidopsis plants (Mao et al. 2016). Further, ZmNAC55 gene had multiple cis-elements related to abiotic stress acting in the promoter region.
Soybean is a crucial legume crop that is cultivated mainly to provide cooking oil and dietary protein. GmNAC11 and GmNAC20, which are well-characterized genes in soybean, that are differentially expressed under many environmental stresses coupled with plant hormones (Hao et al. 2011). These genes encode proteins that localize to the cell nucleus and bind to a core DNA sequence CGT[G/A] (Hao et al. 2011). GmNAC11 is a transcriptional activator regulating Drought-responsive element-binding 1A (DREB1A), Early responsive to dehydration 11 (ERD11), Cold regulated 15A (COR15A), Ethylene responsive factor 5 (ERF5), Ras-related protein Rab18 (RAB18), and Potassium channel in Arabidopsis thaliana 2 (KAT2) genes, thereby building salt tolerance (Hao et al. 2011). Over-expression of GmNAC20, on the other hand, augments salt and freezing tolerance (Hao et al. 2011). In similar manner, Arabidopsis plants that over-expressed GmNAC019 showed enhanced survival rates, intense antioxidant defense, lower peroxide levels and water loss under soil drying situations (Hoang et al. 2019). The transgenic plants were also found to be hypersensitive to ABA, exhibiting lower seed germination rates with fewer green cotyledons which suggested ABA-mediated regulation. Another recent report indicated the involvement of GmSNAC49 in inducing Arabidopsis drought tolerance by up-regulating drought-responsive genes via ABA signaling period (So and Lee 2019).
Chickpea (Cicer arietinum), a vital legume crop in the semi-arid tropics (SAT), is naturally resistant to several abiotic stresses and suboptimal conditions. CarNAC6 is a chickpea nuclear protein that can bind to CGT[G/A]. Over-expression of CarNAC6 in Arabidopsis plants resulted in an enhanced drought tolerance and promoted root development under saline conditions (Liu et al. 2017). In a similar fashion, over-expression of CarNAC4 in Arabidopsis plants led to increased expression of the stress-related genes- Early Responsive to Dehydration 10 (ERD10), Cold Regulated 15A (COR15A), Responsive to desiccation 29A (RD29A), KIN1 (Stress-induced protein KIN1), Cold Regulated 47 (COR47), and Drought-responsive element-binding A (DREBA), thereby enhancing endurance to drought and saline conditions (Yu et al. 2016, Fig. 6a). CarNAC2, another chickpea gene encoding NAC protein (transcriptional activator), is a nuclear protein of 191 amino acids that showed enhanced resistance in transformed Arabidopsis plants when over-expressed (Yu et al. 2014). A few years ago, Peng et al. (2009) reported drought and hormone (indole-3-acetic acid and ABA) induction of the CarNAC3 protein with a conserved NAC domain belonging to the NAP (NAC-like, activated by APETALA3/PISTILLATA) sub-class of the NAC superfamily. Similarly, transgenic Arabidopsis plants overexpressing drought-induced AhNAC2 from peanuts (Arachis hypogaea) resulted in a greater expression of stress-related genes and a higher endurance to drought and salinity compared to the control (Liu et al. 2011). Moreover, AhNAC2 overexpressing Arabidopsis lines were ABA hypersensitive at seed germination, stomatal closure, and root growth relative to WT plants, implying the functioning of AhNAC2 in ABA signaling. Another report suggested that over-expression of AtNAC2 (ANAC092) in groundnuts provided tolerance against salinity and drought stress, and improved yield (Patil et al. 2014). Tang et al. (2017) isolated and characterized stress-responsive AhNAC4, from peanut immature seeds. Peanut AhNAC4 belonging to the ATAF subfamily was highly induced by drought. Over-expression of AhNAC4 improved drought tolerance with an increase in stomatal closure and higher WUE in transformed tobacco (Nicotiana tabacum) plants compared to WT. Interestingly, when tobacco seedlings were subjected to 15 days of drought, all WT plants displayed severe wilting because of water scarcity, whereas the transgenic plants showed delayed leaf wilting. Further, the WT plants became completely desiccated, and most of them (70%) did not recover after being watered. In contrast, a higher survival ratio was seen in AhNAC4 transgenic plants, with 90% remaining viable, indicating its importance in improving drought tolerance (Tang et al. 2017). Similarly, MuNAC4, a NAC TF from horse gram (Macrotyloma uniflorum), displayed enhanced drought tolerance in addition to proliferated lateral root growth as compared to WT when introduced into peanut plants. The imposition of long-term drought resulted in an increase in lateral roots with reduced membrane damage, increasing osmotic adjustment, and anti-oxidative enzyme regulation in transgenic peanut under stress (Pandurangaiah et al. 2014).
Pearl millet (Pennisetum glaucum) is an important cereal crop that is cultivated mainly in SAT for its high nutritional value. The plant, which is well known for its resistance to abiotic stress, has gained much attention since its whole-genome sequences became available (Varshney et al. 2017). PgNAC21, a pearl millet NAC gene, has been shown to provide salinity tolerance in transgenic Arabidopsis plant by up-regulating COR47, RD20, and GSTF6 (Glutathione S-transferase F6) target genes (Shinde et al. 2019, Fig. 6b). Another example of SAT cereal crop NAC is finger millet (Eleusine coracana). Over-expression of finger millet EcNAC67 in rice plants resulted in an increased root and shoot biomass, less reduction in grain yield and maintenance of higher water content leading to better survival against drought and salinity situations (Fig. 6c). Similarly, SbSNAC1, a member of the NAC superfamily from tropical cereal sorghum (Sorghum bicolor), is expressed during drought and salinity and at a relatively higher concentration in roots (Lu et al. 2013). Transgenic Arabidopsis plants that over-expressed SbSNAC1 showed improved survival rates under drought stress accompanied by vigorous green leaves with reduced ion leakage compared with WT plants (Lu et al. 2013).
NAC TFs are undoubtedly effective as an upstream regulator of the expression of adaptive stress by downstream genes. A schematic representation of the NAC transcriptional network along with its target genes is shown in Fig. 5 as an aid to understanding their role in stress adaption in rice and wheat crops. Fine-tuning the expression of stress-specific NAC TFs is very promising for designing plant stress tolerance. However, it is also important to identify the other parallel biological processes influenced by NACs that indirectly contribute to plant adaptive responses and yield potential under negative environmental cues.
NACs in secondary cell wall synthesis
Secondary cell walls (SCWs) are the greatest contributors to plant biomass. Secondary cell walls present in the fibers and tracheary elements of plants are comprised of cellulose, hemicelluloses and lignin. The lignocellulosic biomass represents the carbon-free raw material for generating biofuels. Thus, engineering plants with better SCW characteristics is a key approach to reducing the processing of lignocellulosic biomass. Also, plant resistance to pathogens depends on a complicated mesh of constitutive/inducible defensive barriers. In order to successfully colonize the host plant tissues, pathogens need to overcome the plant cell wall. In this context, the plant cell wall acts as a passive barrier that regulates defense processes and as a platform for signaling the molecules that activate immune responses (Miedes et al. 2014).
NAC TFs are confirmed as mediating SCW synthesis in several species (Grover et al. 2014; Valdivia et al. 2013). A sub-group of closely related NST1 (ANAC043), NST2 (ANAC066), and NST3/SND1 (ANAC012) proteins, function as master transcriptional switches in mediating SCW formation (Mitsuda and Ohme-Takagi 2008; Singh et al. 2016), (Fig. 7). Both SND2 (SECONDARY WALL NAC DOMAIN PROTEIN2) and SND3 (SECONDARY WALL NAC DOMAIN PROTEIN3) function downstream of NST1 and NST3 (Singh et al. 2016; Zhong et al. 2008). SND2 up-regulates the genes responsible for cellulose, hemicellulose, and lignin biosynthesis and polymerization. VND6 and VND7, which are Vascular NAC Domain proteins, act as regulators of SCW biosynthesis, particularly in the xylem vessels (Kubo et al. 2005; Yamaguchi et al. 2008). However, XND1, which is a Xylem NAC Domain1 protein, acts as a negative regulator of secondary cell wall formation in xylem vessels by inhibiting VND functions to activate SCW associated gene expression.
The transcriptional network regulating SCW formation by NAC domain proteins. NST1/2/3 are the master switches for controlling secondary cell wall biosynthesis in fibers. SND2/3 functions, which are downstream of NST1 and NST3, activates transcription of MYB factors and up-regulate genes associated with cellulose, xylem, hemicellulose, and lignin biosynthesis. VND6,7 and SWNs also act as regulators of SCW biosynthesis, specifically in vascular vessels. On the other hand, XND1 protein acts contrary to VND proteins by inhibiting SCW biosynthesis and PCD in xylem vessels. SCW secondary cell wall, PCD programmed cell death, NST NAC SECONDARY WALL THICKENING PROMOTING FACTOR, SND2/3 SECONDARY WALL NAC DOMAIN PROTEIN, VND VASCULAR NAC DOMAIN PROTEIN, SWNs SECONDARY WALL NAC DOMAIN PROTEINS, XND1 XYLEM NAC DOMAIN 1
Over-expression of SND2 in Arabidopsis caused up-regulation of the biosynthetic genes that encode components to initiate the formation of SCW (cellulose and lignin polymerization; Hussey et al. 2011). SND2, which occupies a central role in the transcriptional regulatory network for the SCW synthesis, also upregulates the MYB103 TF and SND1 when it is over-expressed. SND2 over-expression also negatively influences the fiber wall deposition in Arabidopsis; in contrast, its over-expression in Eucalyptus caused an increase in fiber cell area (Hussey et al. 2011). Hussey et al. (2011) also highlighted the importance of determining expression thresholds for SND2 over-expression that are optimal for an enhanced SCW deposition, since expression beyond the threshold led to co-suppression.
Rice OsSND2, a close homolog of AtSND2, was discovered to be associated with regulating SCW biosynthesis in rice and offered a scheme for engineering the green biomass production (Ye et al. 2018). OsSND2 is primarily expressed in the internodes and the panicles, and its over-expression results in up-regulation of SCW biosynthetic genes and increased cellulose content (Ye et al. 2018). Other potential candidates include SECONDARY WALL NAC DOMAIN PROTEINs (SWNs) regulating the formation of SCW in rice (OsSWNs). They are being used for engineering the cell wall in monocotyledonous forage crops applications. For instance, the OsSWN2S chimeric repressor, which has a very low ability for transcriptional activation, when driven by OsSWN1 (high transcriptional activation ability) promoter, results in a reduced cell wall thickening (sclerenchymatous cells) with low lignin and xylose contents (Yoshida et al. 2013). Similarly, Chai et al. (2015) reported that over-expression of OsSWN1 results in SCW-related gene expression with improved lignin and reduced sugar build-up in transgenic plants. Since OsSWN1-like genes are highly conserved in crops such as rice, sorghum, and switchgrass, this indicates the potential of exploiting the OsSWN1 orthologs in other crop species used as food and fodder (Chai et al. 2015).
Zhao et al. (2010) identified genes responsible for SCW biosynthesis in the model legume M. truncatula. Insertion of Tnt1 retrotransposon in a NAM-like protein MtNST1 (NAC SECONDARY WALL THICKENING PROMOTING FACTOR 1) resulted in a reduced expression of cellulose, hemicellulose, and lignin biosynthetic genes. Hence, a lack of lignin in phloem fibers, decreased vascular lignin, and reduced cell wall polysaccharide content were found associated with loss of MtNST1 function (Zhao et al. 2010). Similarly, NST1 gene in M. truncatula that has lost the function mutation (T94K) produced no lignification in interfascicular fibers (nst1-3 mutant), as in the case of tnt1 transposon insertion alleles (Wang et al. 2011). T94K mutation in SND1 (Arabidopsis homolog) caused loss of target binding with the resultant incompetence to activate secondary wall synthesis genes. Moreover, SND1 expression undergoes positive feedback control from itself, and tethers directly to a conserved motif present in its promoter region (Wang et al. 2011).
The root cap is a multilayered column consisting of parenchyma cells that lie on the top of the growing root tip and strengthen growth by taking care of the root meristem and sensing gravity in addition to rhizospheric interaction. NAC family members, viz., SMB (SOMBRERO), BRN1 (BEARSKIN1), and BRN2 (BEARSKIN2), along with VND and NST genes, were found to be involved in root cap maturation and showed similar phenotypic patterns when over-expressed (Bennett et al. 2010; Mitsuda et al. 2005, 2007; Zhong et al. 2006, 2007). Over-expression of NAC proteins (SMB, BRN1, BRN2) class IIB activated transcriptional pathways, resulting in secondary cell wall synthesis (SCW) by transcription of VND, NST, and root cap maturation genes (Bennett et al. 2010; Kamiya et al. 2016).
NACs in yield potential (grain yield), seed size and biomass
Nitrogen (N) is among the primary nutrients that influence plant productivity, and limited N supply causes major constraints on crop yield (Jones et al. 2013). Well-developed roots are required for an efficient N acquisition. Nitrogen uptake, assimilation, remobilization, and storage are regulated by complex gene networks (Masclaux-Daubresse et al. 2010). In major cereal crops, N assimilation genes often colocalize with quantitative trait loci (QTLs) influencing grain yield and nitrogen use efficiency (NUE) (He et al. 2015). Several studies have indicated that expression of various NAC members was related to nutrient seed development and deficiency stress (Agarwal et al. 2011; de Zélicourt et al. 2012; Vidal et al. 2013). These yield associated NACs hold great potential as targets for crop improvement, and their functions should be explored further. AtNAC4, a target of the AtAFB3 (AUXIN SIGNALING F-BOX3), has been shown to be a principal regulator of the nitrate-responsive network in Arabidopsis (Vidal et al. 2014). In a different finding, AtNAC4 was reported to work upstream of the OBF BINDING PROTEIN4 (AtOBP4), a zinc finger TF to induce nitrate response (Vidal et al. 2013). In rice, the PS1 (prematurely senile)/Oryza sativa NAC-like, transcriptionally activated by APETALA3/PISTILLATA (OsNAP), controls N and other nutrient stockpiling in grains. OsNAP over-expression up-regulated the genes for various amino acids and peptide transporters and significantly promoted senescence, whereas OsNAP knockdown resulted in senescence delay in rice (Liang et al. 2014). Reduced OsNAP transcript accumulation caused delay in leaf senescence including extended grain-filling duration, leading to higher yield potential (Liang et al. 2014). PS1 is a functional ortholog of AtNAP and belongs to the NAP subfamily of NAC proteins. The C-terminus of OsNAP functions as an activator, whereas the NAC sub-domains 3 and 4 function as a repressor (Liang et al. 2014). In another case, the over-expression of root specific OsNAC5 in rice plants showed an increment in grain yield of 9–23% under normal conditions (Jeong et al. 2013). However, grain yield was found to be 22–63% higher under drought condition than in WT plants. Moreover, the transgenic plants developed enlarged root diameter due to an expanded stele and aerenchyma at the flowering stage, contributing to enhanced drought tolerance (Jeong et al. 2013). In addition to this, GLP (Germin-like protein), PDX (Pyridoxin biosynthesis protein), MERI5 (Meristem protein) and O-methyltransferase were found to be up-regulated in transgenic rice. Further, OsNAC5 also plays an essential role in loading iron to the seeds via senescence signaling (Ricachenevsky et al. 2013). In contrast to the positive up-regulation of yield by NAC members, a recent study showed over-expression of miR164b and down-regulation of OsNAC2 in rice indicating improved plant architecture and increased grain yield/number than in WT plants (Jiang et al. 2018).
Rice ONAC020, ONAC026, and ONAC023 genes are highly expressed during seed development (Mathew et al. 2016). ONAC020 and ONAC026 belong to the same phylogenetic clade as CUC3, an important gene in seed development (Mathew et al. 2016). ONAC020 and ONAC026 are closely related to CUC3, and all three possess typical NAC sub-domains architecture and a NAC repression domain (NARD) (Mathew et al. 2016). It is worth mentioning that expression levels for these genes varied among rice accessions with contrast in seed size. These genes regulate downstream genes to a varying magnitude due to sequence alterations in the promoter’s regions. These ONAC020 and ONAC026 genes have a DLN stretch (ERF-associated repression motif in plants) in the B domain. Repressor ONAC026 dimerizes with trans-spliced forms of ONAC020 (ONAC020.A, ONAC020.B, ONAC020.C), or ONAC023 to form a heterodimer complex that localizes in the nucleus. It is most likely that the expression levels (repression/activation) of the complex may vary across the seed developmental stages. Further, these seed size-related NAC genes, function in seed development processes and can be utilized as possible targets for crop improvement (Mathew et al. 2016). Moreover, seed size improvement is not only an attribute of yield potential; acquiring the capability for enhanced micronutrient uptake and utilization is also very important.
Plants with stay-green phenotypes exhibit inefficient N remobilization, resulting in low harvest index, despite having the potential for higher productivity (Gregersen et al. 2008). However, plants with faster senescence exhibit an efficient N remobilization, leading to high grain protein content but with reduced grain yield (See et al. 2002). Several studies indicated that NAC genes are promising targets in breeding approaches for increased grain quality (Fig. 8) and nutritional potential. For instance, wheat Gpc-B1 (Grain protein content-B1) encodes a NAC protein NAM-B1, with accelerated senescence in flag leaves. NAM-B1 is closely related to Arabidopsis ANAC025, ANAC018, and ANAC056 proteins. At the grain filling stage, NAM-B1 (Gpc-B1) mediates nutrient redistribution from flag leaves to ears and accelerates senescence (Uauy et al. 2006). RNA interference (RNAi) mediated knockdown of NAM-B1 resulted in senescence delay, leading to lower nutritional and protein contents in the grain but increased nutritional contents and residual N in the flag leaf. Similarly, HvNAM-1 and HvNAM-2 genes, (Gpc-B1 homologs) were identified in barley (Hordeum vulgare) (Uauy et al. 2006). Spikelet initiation and growth, along with leaf senescence subsequent to floral transition, were influenced by barley GPC locus (Lacerenza et al. 2010; Parrott et al. 2012). The near-isogenic line with high GPC expression exhibited faster development and earlier flowering. (Lacerenza et al. 2010). Additionally, several genes were up-regulated in the senescing near-isogenic barley line, and hence could be involved in senescence regulation (Jukanti and Fischer 2008; Jukanti et al. 2008). These findings signify that NAM TFs are necessary for nutrient distribution not just in wheat but in other cereal crops as well.
Sorghum bicolor is extensively grown for food, forage, sugar, and biomass. The sorghum Dry Stalk (D) locus contains NAC TF SbNAC074a, and co-localizes with sugar yield-related QTL (Xia et al. 2018). SbNAC074a is associated with a premature stop codon that disrupts the NAC domain in the recessive parent (dd) and is responsible for alterations in the biomass. Thus, the near-isogenic lines (NILs) of Green midrib (dd) displayed decreased stalk lignin contents but increased soluble sugar levels and grain yields under normal field situations. (Xia et al. 2018).
An important yield related NAC gene example is Pod shattering resistance by SHAT1-5, (SHATTERING1-5). SHAT1-5 from soybean which causes fiber cap cell secondary walls thickening when they are over-expressed at 15-fold, thereby causing disruption of an upstream repressor (Dong et al. 2014). The critical cellular function of the shattering-resistant feature exists is the excess of lignification of fiber cap cells while the abscission layer is unaltered in the pod ventral suture. The subsequent loss of seed dispersal is a crucial agronomical trait that is a keystone feature in crop cultivation. The tissue development and differentiation control by NAC family TFs are therefore important factors that remain underexplored though they hold potential for improving future crops.
Lateral root development regulation by NACs
Lateral root formation is a vital root developmental feature related to the phenotypic adaptation to salinity and drought stress. NAC TFs have been found to be directly linked to lateral root initiation (Fig. 8), with AtNAC2 being perhaps the best example of this (He et al. 2005). AtNAC2 is preferentially expressed at high levels in roots and flowers. Notably, AtNAC2 expression is up-regulated in ethylene and auxin overexpressing mutants when exposed to salt stress (He et al. 2005). Under salt stress and heavy metal stress, lateral root development is promoted, while taproot growth is inhibited to reduce stress effects (Bhati et al. 2016; He et al. 2005). However, salt induction of AtNAC2 expression was not linked to the ABA signaling pathway (He et al. 2005).
AtNAC1 is also well known for its involvement in lateral root development, so its closest homolog (MtNAC1) was analyzed to determine its role in the lateral root formation in M. truncatula (D’haeseleer et al. 2011). It was found that MtNAC1 expressed a different pattern in response to auxin than did AtNAC1. Plants with MtNAC1 expression displayed no changes in the lateral root number, whereas the nodule number was reduced due to miR164 over-expression. The NAC1 regulation by miRNA is not limited to M. truncatula. Maize (Zea mays) ZmNAC1 (TC258020) expression is regulated by miR164 and thus influences the development of lateral roots in maize inbred lines, 87-1 and Zong3 (Li et al. 2012). Zong3 inbred line showed a 1.8-fold higher expression level of ZmNAC1 in its roots than did the 87-1 line. Additionally, Zong3 inbred lines showed higher lateral root density than 87-1. Over-expression of ZmNAC1 in transgenic Arabidopsis had showed enhanced lateral root growth compared to their WT (Li et al. 2012). Higher expression of mature and miR164 precursors (trans-element) was observed in 87-1 than in Zong3, which is opposite to ZmNAC1 expression patterns, thereby contributing to differences in lateral root phenotype (Li et al. 2012).
Increase in lateral root number was reported in Arabidopsis, when they are over-expressed with the soybean GmNAC004 gene. Basal expression levels in the GmNAC004 homolog (ANAC017) were induced in roots, leaves, and flowers by water deficit stress. The GmNAC004 gene works upstream of the key auxin regulators and increases the lateral root development in Arabidopsis through the auxin signaling pathway (Quach et al. 2014). GmNAC20, another soybean NAC TF, was induced to varying levels in response to plant hormones and abiotic stresses (salt and frost). Transcripts of GmNAC20 were more abundant in cotyledons and roots. Over-expression of GmNAC20 promoted the formation of lateral roots in transgenic Arabidopsis plants by altering genes related to auxin signaling (Hao et al. 2011). Similarly, over-expression of GmNAC109 (ATAF1 homolog) increased the formation of lateral roots in transgenic Arabidopsis plants by upregulating DREB1A, DREB2A, RD29A, COR15A, AREB1 (Abscisic acid-responsive element-binding protein 1) and AREB2 genes (Yang et al. 2019). The GmNAC109-overexpressing transgenic plants showed superior salt and drought tolerance than did WT Col-0 plants. ABA-responsive genes ABI1 (ABA insensitive 1) and ABI5 (ABA insensitive 5) were up-regulated in transformed Arabidopsis lines and were found hypersensitive to ABA (Yang et al. 2019). Downstream gene Auxin-induced in root cultures 3 (AIR3) expression was increased, whereas Auxin response factor 2 (ARF2) showed reduced expression in these transformed lines and helped to regulate the formation of hairy root via the auxin signaling pathway (Yang et al. 2019). In summary, these findings laid the foundation for the development of soybean lines with improved tolerance to abiotic stresses via genetic modification.
ROS signaling, leaf senescence, and programmed cell death
Reactive oxygen species (ROS) and oxides such as H2O2 serve as important signal mediators that triggers plant responses against various biotic and abiotic stresses, including heavy metal stress (Bhattacharjee 2005; Davletova et al. 2005; Petrov et al. 2015). ROS signaling is a critical component of senescence and programmed cell death (PCD). Leaf senescence is necessary for the translocation of nutrients to other plant parts such as developing tissues and storage organs (Lim et al. 2007). Some of the NAC family genes reported to be linked with leaf senescence in various plants include AtNAP, NTL4 in Arabidopsis (Guo and Gan 2006; Lee et al. 2012) and MtNAC969 (de Zélicourt et al. 2012) in M. truncatula. Additionally, several other reports have emphasized the involvement of NAC proteins in leaf senescence in crops (Fig. 9). These include SoNAP in sugarcane (Carrillo-Bermejo et al. 2020), GhNAP in cotton (Fan et al. 2015), OsNAC106 in rice (Sakuraba et al. 2015), Os07g37920 in rice, and wheat GPC (Distelfeld et al. 2012, 2014). Podzimska-Sroka et al. (2015) and Kim et al. (2016) have extensively reviewed the role of NAC proteins in leaf senescence and identified various NAC-centered senescence-related gene regulatory networks (GRNs). Pimenta et al. (2016) reported GmNAC81 mediated age-dependent senescence by endoplasmic reticulum (ER) stress-induced PCD through GmNAC81/VPE (vacuolar processing enzyme) regulatory circuit. In barley, HvNAC005 positively controls early senescence and causes stunting and delay in developmental processes. HvNAC005 binding to cis-elements of putative target genes caused up-regulation of several genes related to secondary metabolism and hormone metabolism, including those related to development, stress, and transport (Christiansen et al. 2016). Therefore, targeting HvNAC005 in future attempts, to fine-tune gene expression related to the senescence process in barley, would be an obvious strategy for improving crop yields (Christiansen et al. 2016). Chlorophyll degradation and compromised photosynthetic efficiency are typical during leaf senescence, allowing embryos to bleach, buds to break, and fruit to ripen. In Arabidopsis, ANAC046 has been identified to regulate the expression of Chlorophyll catabolic genes, namely, NON-YELLOW COLORING1, STAY-GREEN1 (SGR1), SGR2, and PHEOPHORBIDE a OXYGENASE (Oda-Yamamiz et al. 2016). ANAC046 overexpressing transgenic Arabidopsis plants showed an early-senescence phenotype and reduced chlorophyll contents compared to WT plants. This revealed that both senescence-associated genes and Chlorophyll catabolic genes were positively regulated by ANAC046 (Oda-Yamamiz et al. 2016).
Diagram showing senescence-related NAC regulation and associated phenotypic response. Blue arrows indicate up-regulation; red arrows indicate down-regulation. VPE vacuolar processing enzyme, RBCS rubisco small subunit gene, SAG senescence-associated gene, Pao pheide a oxygenase; NYC non-yellow Coloring1; NCED nine-cis-epoxycarotenoid dioxygenase3, GDH glutamate dehydrogenase, GS1 glutamine synthetase
The rice NAC gene, OsY37 (Oryza sativa Yellow37/ONAC011) accelerated heading time and promoted senescence during the terminal phase (Mannai et al. 2017). OsY37 over-expression displayed early heading and precocious senescence in rice flag leaves, whereas knockdown of OsY37 expression resulted in delayed heading time and leaf senescence, along with higher chlorophyll accumulation during the vegetative stage (Mannai et al. 2017). Similarly, OsNAC2 is involved in promoting leaf senescence via ABA biosynthesis (Mao et al. 2017). Over-expression of OsNAC2 resulted in up-regulation of OsSGR and OsNYC3 genes, which are responsible for chlorophyll degradation, and hence, in an accelerated leaf senescence. Interestingly, OsNAC2 is up-regulated by a lower ABA but down-regulated by higher ABA levels, showing that reduced expression of OsNAC2 resulted in 10% increased grain yield in knockdown lines (Mao et al. 2017). Foxtail millet (Setaria italica) is another crucial food, fodder, and potential energy crop, but little is known about the functional roles of the senescence-related NAC genes of this crop. Foxtail millet NAC1 (SiNAC1), an ortholog of Arabidopsis NAP (NAC-like, ACTIVATED BY AP3/PI), is involved in promoting natural and dark-induced leaf senescence by upregulating the NCED3 gene related to ABA biosynthesis (Ren et al. 2018).
Programmed cell death (PCD) is an inherently encoded, positively controlled cellular suicide pathway that is required for the growth and survival of life forms under a compromised environment. In plants, PCD occurs during stress responses and is also involved in regular plant development. Under environmental stress, cells, tissues, or even entire organs are sacrificed to enhance the survival probabilities of the whole plant (Gadjev et al. 2008). Two NAC genes, ANAC087 and ANAC046 have been reported to control the expression of cell death-related genes for inducing ectopic PCD in Arabidopsis columella root cap cells (Huysmans et al. 2018). ANAC087 regulates chromatin (nuclear) degradation via the nuclease BFN1 in lateral root caps. However, the genesis of cell death regulation in root caps was linked to both ANAC087 and ANAC046 in the course of its fall from the root tip (Huysmans et al. 2018). According to Yang et al. (2015), the XND1/ANAC014 gene regulated SCW biosynthesis in Arabidopsis via PCD in xylem vessels, whereas PvNAC1 and PvNAC2 caused leaf senescence in annual plant species.
The findings summarized above show that NAC TFs play crucial roles in ROS signaling, leaf senescence, and PCD (Fig. 9). These reports involving various plant species have shed light on the application of NAC genes to improving plant stress responses.
Conclusions and future prospects
Understanding the diverse mechanisms involved in stress adaptation in plants is the first step in designing suitable future-ready crops. In particular, TFs are the first line master regulators that work upstream of the gene sets responsible for the commencement of multi-omics shifts. This review highlights the NAC genes involved in the primary and secondary phases of stress adaptive responses in plants (Fig. 1). Various functional genomics approaches employed by these genes in model plants proved quite successful. Potentially, NAC genes could be great candidates for targeted engineering to develop resistance across various aspects of stress coping mechanisms, as discussed above (Table 2; Figs. 5, 6, 7, 8, 9). There remain several challenges, including the phylogenetic classification of NAC members. The C-terminus complexity and possible functional redundancy are both challenges to be overcome but both offer opportunities for exploitation. Before NAC related approaches can be integrated into breeding programs, their functional validation must be undertaken. Modern multigene/protein targeting approaches such as CRISPR-Cas9 and synthetic microProteins could help rule out the challenges of functional redundancy by identifying the NAC members with desirable functions. The growing threats of climate change-induced drought and high-temperature stresses increase the importance of identifying the specific NAC genes that can be used to construct stress-tolerant crops. It may also be possible to design and commercialize other transgenic plants that overexpress NAC genes with different stress tolerance abilities, though there have been no such reports so far. In summary, there remains great potential for the use of NAC genes as a biotechnological tool (as mentioned in Table 2) in years to come.
Change history
09 April 2021
A Correction to this paper has been published: https://doi.org/10.1007/s10265-021-01281-9
References
Agarwal P, Kapoor S, Tyagi AK (2011) Transcription factors regulating the progression of monocot and dicot seed development. BioEssays 33:189–202
Aida M (1997) Genes involved in organ separation in Arabidopsis: an analysis of the cup-shaped cotyledon mutant. Plant Cell Online 9:841–857
Al Abdallat AM, Ayad JY, Abu Elenein JM et al (2014) Over-expression of the transcription factor HvSNAC1 improves drought tolerance in barley (Hordeum vulgare L.). Mol Breed 33:401–414
Baillo EH, Kimotho RN, Zhang Z, Xu P (2019) Transcription factors associated with abiotic and biotic stress tolerance and their potential for crops improvement. Genes 10:771
Bennett T, van den Toom A, Sanchez-Perez GF, Campilho A, Willemsen V, Snel B, Scheres B (2010) SOMBERO, BEARSKIN1, and BEARSKIN2 regulate root cap maturation in Arabidopsis. Plant Cell 22:640–654
Bhati KK, Alok A, Kumar A, Kaur J, Tiwari S, Pandey AK (2016) Silencing of ABCC13 transporter in wheat reveals its involvement in grain development, phytic acid accumulation and lateral root formation. J Exp Bot 67:4379–4389
Bhattacharjee S (2005) Reactive oxygen species and oxidative burst: roles in stress, senescence and signal transduction in plants. Curr Sci 89:1113–1121
Carrillo-Bermejo EA, Gamboa-Tuz SD, Pereira-Santana A et al (2020) The SoNAP gene from sugarcane (Saccharum officinarum) encodes a senescence-associated NAC transcription factor involved in response to osmotic and salt stress. J Plant Res 133:897–909
Chai M, Bellizzi M, Wan C, Cui Z, Li Y, Wang G-L (2015) The NAC transcription factor OsSWN1 regulates secondary cell wall development in Oryza sativa. J Plant Biol 58:44–51
Chen Q, Wang Q, Xiong L, Lou Z (2011) A structural view of the conserved domain of rice stress-responsive NAC1. Protein Cell 2:55–63
Chen D, Chai S, McIntyre CL, Xue GP (2018) Over-expression of a predominantly root-expressed NAC transcription factor in wheat roots enhances root length, biomass and drought tolerance. Plant Cell Rep 37:225–237
Cheng Z, Zhang X, Zhao K et al (2020) Ectopic expression of a poplar gene NAC13 confers enhanced tolerance to salinity stress in transgenic Nicotiana tabacum. J Plant Res 133:727–737
Christiansen MW, Matthewman C, Podzimska-Sroka D, O’Shea C, Lindemose S, Møllegaard NE, Holme IB, Hebelstrup K, Skriver K, Gregersen PL (2016) Barley plants overexpressing the NAC transcription factor gene HvNAC005 show stunting and delay in development combined with early senescence. J Exp Bot 67:5259–5273
Chung PJ, Jung H, Choi YD, Kim JK (2018) Genome-wide analyses of direct target genes of four rice NAC-domain transcription factors involved in drought tolerance. BMC Genom 19:40
D’haeseleer K, Herder GD, Laffont C, Plet J, Mortier V, Lelandais-Briere C, De Bodt S, Keyser AD, Crespi M, Holsters M, Frugier F, Goormachtig S (2011) Transcriptional and post-transcriptional regulation of a NAC1 transcription factor in Medicago truncatula roots. New Phytol 191:647–661
Davletova S, Rizhsky L, Liang HJ, Zhong SQ, Oliver DJ, Coutu J, Shualev V, Schlauch K, Mittler R (2005) Cytosolic ascorbate peroxidase 1 is a central component of the reactive oxygen gene network of Arabidopsis. Plant Cell 17:268–281
de Zélicourt A, Diet A, Marion J, Laffont C, Ariel F, Moison M, Zahaf O, Crespi M, Gruber V, Frugier F (2012) Dual involvement of a Medicago truncatula NAC transcription factor in root abiotic stress response and symbiotic nodule senescence. Plant J 70:220–230
Distelfeld A, Pearce SP, Avni R, Scherer B, Uauy C, Piston F, Slade A, Zhao R, Dubcovsky J (2012) Divergent functions of orthologous NAC transcription factors in wheat and rice. Plant Mol Biol 78:515–524
Distelfeld A, Avni R, Fischer AM (2014) Senescence, nutrient remobilization, and yield in wheat and barley. J Exp Bot 65:3783–3798
Dong Y, Yang X, Liu J, Wang B-H, Liu B-L, Wang Y-Z (2014) Pod shattering resistance associated with domestication is mediated by a NAC gene in soybean. Nat Commun 5:3352
Du X, He F, Zhu B et al (2020) NAC transcription factors from Aegilops markgrafii reduce cadmium concentration in transgenic wheat. Plant Soil 449:39–50
Ernst HA, Olsen AN, Larsen S, Lo Leggio L (2004) Structure of the conserved domain of ANAC, a member of the NAC family of transcription factors. EMBO Rep 5:297–303
Fan K, Bibi N, Gan S, Li F, Yuan S, Ni M, Wang M, Shen H, Wang X (2015) A novel NAP member GhNAP is involved in leaf senescence in Gossypium hirsutum. J Exp Bot 66:4669–4682
Fang Y, You J, Xie K, Xie W, Xiong L (2008) Systematic sequence analysis and identification of tissue-specific or stress-responsive genes of NAC transcription factor family in rice. Mol Genet Genom 280:535–546
Fang Y, Liao K, Du H et al (2015) A stress-responsive NAC transcription factor SNAC3 confers heat and drought tolerance through modulation of reactive oxygen species in rice. J Exp Bot 66:6803–6817
Fujita M, Fujita Y, Maruyama K, Seki M, Hiratsu K, Ohme-Takagi M, Tran LS, Yamaguchi-Shinozaki K, Shinozaki K (2004) A dehydration-induced NAC protein, RD26, is involved in a novel ABA-dependent stress-signaling pathway. Plant J 39:863–876
Gadjev I, Stone JM, Gechev TS (2008) Programmed cell death in plants: new insights into redox regulation and the role of hydrogen peroxide. Int Rev Cell Mol Biol 270:87–144
Gregersen PL, Holm PB, Krupinska K (2008) Leaf senescence and nutrient remobilisation in barley and wheat. Plant Bio Stuttg 10:S37–S49
Grover A, Singh S, Pandey P et al (2014) Over-expression of NAC gene from Lepidium latifolium L. enhances biomass, shortens life cycle and induces cold stress tolerance in tobacco: potential for engineering fourth generation biofuel crops. Mol Biol Rep 41:7479–7489
Guo Y, Gan S (2006) AtNAP, a NAC family transcription factor, has an important role in leaf senescence. Plant J 46:601–612
Guo W, Zhang J, Zhang N, Xin M, Peng H, Hu Z, Ni Z, Du J (2015) The Wheat NAC Transcription Factor TaNAC2L is regulated at the transcriptional and post-translational levels and promotes heat stress tolerance in transgenic Arabidopsis. PLoS One 10:e0135667
Ha CV, Nasr Esfahani M, Watanabe Y, Tran UT, Sulieman S et al (2014) Genome-wide identification and expression analysis of the CaNAC family members in chickpea during development. Dehydration and ABA treatments. PLoS One 9:e114107
Hao YJ, Wei W, Song QX, Chen HW, Zhang YQ, Wang F, Zou HF, Lei G, Tian AG, Zhang WK, Ma B, Zhang JS, Chen SY (2011) Soybean NAC transcription factors promote abiotic stress tolerance and lateral root formation in transgenic plants. Plant J 68:302–313
He XJ, Mu RL, Cao WH, Zhang ZG, Zhang JS, Chen SY (2005) AtNAC2, a transcription factor downstream of ethylene and auxin signaling pathways, is involved in salt stress response and lateral root development. Plant J 44:903–916
He X, Qu B, Li W, Zhao Xueqiang, Teng Wan, Ma Wenying, Ren Yongzhe, Li Bin, Li Zhensheng, Tong Yiping (2015) The nitrate-inducible NAC transcription factor TaNAC2-5A controls nitrate response and increases wheat yield. Plant Physiol 169:1991–2005
Hoang XLT, Nhi DNH, Thu NBA, Thao NP, Tran LP (2017) Transcription factors and their roles in signal transduction in plants under abiotic stresses. Curr Genom 18:483–497
Hoang XLT, Nguyen NC, Nguyen YH, Watanabe Y, Tran LP, Thao NP (2019) The soybean GmNAC019 transcription factor mediates drought tolerance in Arabidopsis. Int J Mol Sci 21:286
Hong Y, Zhang H, Huang L, Li D, Song F (2016) Over-expression of a stress-responsive NAC transcription factor gene ONAC022 improves drought and salt tolerance in rice. Front Plant Sci 7:4
Hu H, You J, Fang Y, Zhu X, Qi Z, Xiong L (2008) Characterization of transcription factor gene SNAC2 conferring cold and salt tolerance in rice. Plant Mol Biol 67:169–181
Hu R, Qi G, Kong Y, Kong D, Gao Q, Zhou G (2010) Comprehensive analysis of NAC domain transcription factor gene family in Populus trichocarpa. BMC Plant Biol 10:145
Huang Q, Wang Y, Li B et al (2015) TaNAC29, a NAC transcription factor from wheat, enhances salt and drought tolerance in transgenic Arabidopsis. BMC Plant Biol 15:268
Huang L, Hong Y, Zhang H, Li D, Song F (2016) Rice NAC transcription factor ONAC095 plays opposite roles in drought and cold stress tolerance. BMC Plant Biol 16:203
Hussain et al (2017) The essence of NAC gene family to the cultivation of drought-resistant soybean (Glycine max L. Merr.) cultivars. BMC Plant Biol 17:55
Hussey SG, Mizrachi E, Spokevicius AV, Bossinger G, Berger DK, Myburg AA (2011) SND2, a NAC transcription factor gene regulates genes involved in secondary cell wall development in Arabidopsis fibres and increases fibre cell area in Eucalyptus. BMC Plant Biol 11:173
Huysmans M, Buono RA, Skorzinski N, Radio MC, Winter FD, Parizot B, Mertens J, Karimi M, Fendrych M, Nowacka MK (2018) NAC transcription factors ANAC087 and ANAC046 control distinct aspects of programmed cell death in the Arabidopsis Columella and lateral root cap. Plant Cell 30:2197–2213
Jensen MK, Skriver K (2014) NAC transcription factor gene regulatory and protein–protein interaction networks in plant stress responses and senescence. IUBMB Life 66:156–166
Jensen MK, Kjaersgaard T, Nielsen NM, Galberg P, Petersen K, O’Shea C, Skriver K (2010) The Arabidopsis thaliana NAC transcription factor family: structure-function relationships and determinants of ANAC019 stress signalling. Biochem J 426:183–196
Jeong JS, Kim YS, Redillas MCFR, Jang G, Jung H, Bang SW, Choi YD, Ha S-H, Reuzeau C, Kim J-K (2013) OsNAC5 over-expression enlarges root diameter in rice plants leading to enhanced drought tolerance and increased grain yield in the field. Plant Biotechnol J 11:101–114
Jiang Y, Deyholos MK (2006) Comprehensive transcriptional profiling of NaCl-stressed Arabidopsis roots reveals novel classes of responsive genes. BMC Plant Biol 6:25
Jiang G, Jiang X, Lü P, Liu J, Gao J, Zhang C (2014) The rose (Rosa hybrida) NAC transcription factor 3 gene, RhNAC3, involved in ABA signaling pathway both in rose and Arabidopsis. PLoS One 9:e109415
Jiang D, Chen W, Dong J, Li J, Yang F, Wu Z, Zhou H, Wang W, Zhuang C (2018) Over-expression of miR164b-resistant OsNAC2 improves plant architecture and grain yield in rice. J Exp Bot 69:1533–1543
Jones DL, Clode PL, Kilburn MR, Stockdale EA, Murphy DV (2013) Competition between plant and bacterial cells at the microscale regulates the dynamics of nitrogen acquisition in wheat (Triticum aestivum). New Phytol 200:796–807
Jukanti AK, Fischer AM (2008) A high-grain protein content locus on barley (Hordeum vulgare) chromosome 6 is associated with increased flag leaf proteolysis and nitrogen remobilization. Physiol Plant 132:426–439
Jukanti AK, Heidlebaugh NM, Parrott DL, Fischer IA, McInnerney K, Fischer AM (2008) Comparative transcriptome profiling of near-isogenic barley (Hordeum vulgare) lines differing in the allelic state of a major grain protein content locus identifies genes with possible roles in leaf senescence and nitrogen reallocation. New Phytol 177:333–349
Kamiya M, Higashio SY, Isomoto A et al (2016) Control of root cap maturation and cell detachment by BEARSKIN transcription factors in Arabidopsis. Development 143:4063–4072
Kim SG, Kim SY, Park CM (2007) A membrane-associated NAC transcription factor regulates salt-responsive flowering via FLOWERING LOCUS T in Arabidopsis. Planta 226:647–654
Kim JH, Nam HG, Lim PO (2016) Regulatory network of NAC transcription factors in leaf senescence. Curr Opin Plant Biol 33:48–56
Kosová K, Vítámvás P, Urban MO, Klíma M, Roy A, Tom Prášil I (2015) Biological networks underlying abiotic stress tolerance in temperate crops-a proteomic perspective. Int J Mol Sci 16:20913–20942
Kubo M, Udagawa M, Nishikubo N, Horiguchi G, Yamaguchi M, Ito J, Mimura T, Fukuda H, Demura T (2005) Transcription switches for protoxylem and metaxylem vessel formation. Genes Dev 19:1855–1860
Lacerenza JA, Parrott DL, Fischer AM (2010) A major grain protein content locus on barley (Hordeum vulgare L.) chromosome 6 influences flowering time and sequential leaf senescence. J Exp Bot 61:3137–3149
Lata C, Yadav A, Prasad M (2011) Role of plant transcription factors in abiotic stress tolerance. In: Shanker A (Ed) Abiotic stress response in plants—physiological, biochemical and genetic perspectives. InTech, New Delhi, India. http://www.intechopen.com/books/abiotic-stress-response-in-plants-physiological-biochemical-and-geneticperspectives/role-of-plant-transcription-factors-in-abiotic-stress-tolerance (online only)
Le DT, Nishiyama R, Watanabe Y, Mochida K, Yamaguchi-Shinozaki K, Shinozaki K, Tran LS (2011) Genome-wide survey and expression analysis of the plant-specific NAC transcription factor family in soybean during development and dehydration stress. DNA Res 18:263–276
Lee S, Seo PJ, Lee HJ, Park CM (2012) A NAC transcription factor NTL4 promotes reactive oxygen species production during drought-induced leaf senescence in Arabidopsis. Plant J 70:831–844
Li J, Guo G, Guo W, Guo G, Tong D, Ni Z, Sun Q, Yao Y (2012) miRNA 164-directed cleavage of ZmNAC1 confers lateral root development in maize (Zea mays L.). BMC Plant Biol 12:220
Liang C, Wang Y, Zhu Y, Tang J, Hu B, Liu L, Ou S, Wu H, Sun X, Chu J et al (2014) OsNAP connects abscisic acid and leaf senescence by fine tuning abscisic acid biosynthesis and directly targeting senescence associated genes in rice. Proc Natl Acad Sci USA 111:10013–10018
Lim PO, Kim HJ, Nam HG (2007) Leaf senescence. Annu Rev Plant Biol 58:115–136
Lindemose S, O’Shea C, Jensen MK, Skriver K (2013) Structure, function and networks of transcription factors involved in abiotic stress responses. Int J Mol Sci 14:5842–5878
Ling L, Song L, Wang Y, Guo C (2017) Genome-wide analysis and expression patterns of the NAC transcription factor family in Medicago truncatula. Physiol Mol Biol Plants 23:343–356
Liu X, Hong L, Li XY, Yao Y, Hu B, Li L (2011) Improved drought and salt tolerance in transgenic Arabidopsis overexpressing a NAC transcriptional factor from Arachis hypogaea. Biosci Biotechnol Biochem 75:443–450
Liu X, Liu S, Wu J et al (2013) Over-expression of Arachis hypogaea NAC3 in tobacco enhances dehydration and drought tolerance by increasing superoxide scavenging. Plant Physiol Biochem 70:354–359
Liu G, Li X, Jin S, Liu X, Zhu L et al (2014) Over-expression of rice NAC gene SNAC1 improves drought and salt tolerance by enhancing root development and reducing transpiration rate in transgenic cotton. PLoS One 9:e86895
Liu Y, Yu X, Liu S et al (2017) A chickpea NAC-type transcription factor, CarNAC6, confers enhanced dehydration tolerance in Arabidopsis. Plant Mol Biol Rep 35:83–96
Lu M, Ying S, Zhang DF et al (2012) A maize stress-responsive NAC transcription factor, ZmSNAC1, confers enhanced tolerance to dehydration in transgenic Arabidopsis. Plant Cell Rep 31:1701–1711
Lu M, Zhang D-F, Shi Y-S, Song Y-C, Wang T-Y, Li Y (2013) Expression of SbSNAC1, a NAC transcription factor from sorghum, confers drought tolerance to transgenic Arabidopsis. Plant Cell Tiss Organ Cult 115:443–455
Lv X et al (2016) Global expressions landscape of NAC transcription factor family and their responses to abiotic stresses in Citrullus lanatus. Sci Rep 6:30574
Mallory AC, Dugas DV, Bartel DP, Bartel B (2004) MicroRNA regulation of NAC-domain targets is required for proper formation and separation of adjacent embryonic, vegetative, and floral organs. Curr Biol 14:1035–1046
Mannai YE, Akabane K, Hiratsu K, Satoh-Nagasawa N, Wabiko H (2017) The NAC transcription factor gene OsY37 (ONAC011) promotes leaf senescence and accelerates heading time in rice. Int J Mol Sci 18:2165
Mao X, Zhang H, Qian X, Li A, Zhao G, Jing R (2012) TaNAC2, a NAC-type wheat transcription factor conferring enhanced multiple abiotic stress tolerances in Arabidopsis. J Exp Bot 63:2933–2946
Mao H, Wang H, Liu S, Li Z, Yang X, Yan J, Li J, Tran L-SP, Qin Feng (2015) A transposable element in a NAC gene is associated with drought tolerance in maize seedlings. Nat Commun 6:8326
Mao H, Yu L, Han R, Li Z, Liu H (2016) ZmNAC55, a maize stress-responsive NAC transcription factor, confers drought resistance in transgenic Arabidopsis. Plant Physiol Biochem 105:55–66
Mao C, Lu S, Lv B, Zhang B, Shen J, He J, Luo L, Xi D, Chen X, Ming F (2017) A rice NAC transcription factor promotes leaf senescence via ABA biosynthesis. Plant Physiol 174:1747–1763
Masclaux-Daubresse C, Daniel-Vedele F, Dechorgnat J, Chardon F, Gaufichon L, Suzuki A (2010) Nitrogen uptake, assimilation and remobilization in plants: challenges for sustainable and productive agriculture. Ann Bot 105:1141–1157
Mathew IE, Das S, Mahto A, Agarwal P (2016) Three rice NAC transcription factors heteromerize and are associated with seed size. Front Plant Sci 7:1638
Matsui A, Ishida J, Morosawa T, Mochizuki Y, Kaminuma E, Endo TA et al (2008) Arabidopsis transcriptome analysis under drought, cold, high-salinity and ABA treatment conditions using a tiling array. Plant Cell Physiol 49:1135–1149
Miedes E, Vanholme R, Boerjan W, Molina A (2014) The role of the secondary cell wall in plant resistance to pathogens. Front Plant Sci 5:358
Mitsuda N, Ohme-Takagi M (2008) NAC transcription factors NST1 and NST3 regulate pod shattering in a partially redundant manner by promoting secondary wall formation after the establishment of tissue identity. Plant J 56:768–778
Mitsuda N, Seki M, Shinozaki K, Ohme-Takagi M (2005) The NAC transcription factors NST1 and NST2 of Arabidopsis regulate secondary wall thickenings and are required for anther dehiscence. Plant Cell 17:2993–3006
Mitsuda N, Iwase A, Yamamoto H, Yoshida M, Seki M, Shinozaki K, Ohme-Takagi M (2007) NAC transcription factors, NST1 and NST3, are key regulators of the formation of secondary walls in woody tissues of Arabidopsis. Plant Cell 19:270–280
Ochiai K, Shimizu A, Okumoto Y, Fujiwara T, Matoh T (2011) Suppression of a NAC-like transcription factor gene improves boron-toxicity tolerance in rice. Plant Physiol 156:1457–1463
Ohnishi T, Sugahara S, Yamada T, Kikuchi K, Yoshiba Y, Hirano HY, Tsutsumi N (2005) OsNAC6, a member of the NAC gene family, is induced byvarious stresses in rice. Genes Genet Syst 80:135–139
Oda-Yamamizo C, Mitsuda N, Sakamoto S, Ogawa D, Ohme-Takagi M, Ohmiya A (2016) The NAC transcription factor ANAC046 is a positive regulator of chlorophyll degradation and senescence in Arabidopsis leaves. Sci Rep 6:23609
Olsen AN, Ernst HA, Leggio LL, Skriver K (2005a) DNA-binding specificity and molecular functions of NAC transcription factors. Plant Sci 169:785–797
Olsen AN, Ernst HA, Leggio LL, Skriver K (2005b) NAC transcription factors: structurally distinct, functionally diverse. Trends Plant Sci 10:79–87
Ooka H, Satoh K, Dosssi K, Nagata T, Otomo Y, Murakami K, Matsubara K, Osato N, Kawai J, Carninci P, Hayashizaki Y, Suzuki K, Kojima K, Takahara Y, Yamamoto K, Kikuchi S (2003) Comprehensive analysis of NAC family genes in Oryza sativa and Arabidopsis thaliana. DNA Res 10:239–247
Pandurangaiah M, Rao GL, Sudhakarbabu O, Nareshkumar A, Kiranmai K, Lokesh U, Thapa G, Sudhakar C (2014) Over-expression of Horsegram (Macrotyloma uniflorum Lam. Verdc.) NAC transcriptional factor (MuNAC4) in groundnut confers enhanced drought tolerance. Mol Biotechnol 56:758–769
Parrott DL, Downs EP, Fischer AM (2012) Control of barley (Hordeum vulgare L.) development and senescence by the interaction between a chromosome six grain protein content locus, day length, and vernalization. J Exp Bot 63:1329–1339
Patil M, Ramu SV, Jathish P, Sreevathsa R, Reddy PC, Prasad TG, Udayakumar M (2014) Over-expression of AtNAC2 (ANAC092) in groundnut (Arachis hypogaea L.) improves abiotic stress tolerance. Plant Biotechnol Rep 8:161–169
Peng H, Cheng HY, Chen C, Yu XW, Yang JN, Gao WR, Shi QH, Zhang H, Li JG, Ma H (2009) A NAC transcription factor gene of Chickpea (Cicer arietinum), CarNAC3, is involved in drought stress response and various developmental processes. J Plant Physiol 166:1934–1945
Perochon A, Kahla A, Vranic M, Jia J, Malla KB, Craze M, Wallington E, Doohan FM (2019) A wheat NAC interacts with an orphan protein and enhances resistance to Fusarium head blight disease. Plant Biotechnol J 17:1892–1904
Petrov V, Hille J, Mueller-Roeber B, Gechev TS (2015) ROS-mediated abiotic stress-induced programmed cell death in plants. Front Plant Sci 6:69
Pimenta MR, Silva PA, Mendes GC, Alves JR, Caetano HDN, Machado JPB, Brustolini OJB, Carpinetti PA, Melo BP, Silva JCF, Rosado GL, Ferreira MFS, Dal-Bianco M, Picoli EADT, Aragao FJL, Ramos HJO, Fontes EPB (2016) The stress-induced soybean NAC transcription factor GmNAC81 plays a positive role in developmentally programmed leaf senescence. Plant Cell Physiol 57:1098–1114
Podzimska-Sroka D, O’Shea C, Gregersen PL, Skriver K (2015) NAC transcription factors in senescence: from molecular structure to function in crops. Plants 4:412–448
Puranik S, Sahu PP, Srivastava PS, Prasad M (2012) NAC proteins: regulation 13 and role in stress tolerance. Trends Plant Sci 17:369–381
Quach TN, Tran L-SP, Valliyodan B, Nguyen HT, Kumar R et al (2014) Functional analysis of water stress-responsive soybean GmNAC003 and GmNAC004 transcription factors in lateral root development in Arabidopsis. PLoS One 9:e84886
Rahman H, Ramanathan V, Nallathambi J, Duraialagaraja S, Muthurajan R (2016) Over-expression of a NAC67 transcription factor from finger millet (Eleusinecoracana L.) confers tolerance against salinity and drought stress in rice. BMC Biotechnol 16:35
Ramegowda V, Senthil-Kumar M, Nataraja KN, Reddy MK, Mysore KS, Udayakumar M (2012) Expression of a finger millet transcription factor, EcNAC1, in tobacco confers abiotic stress-tolerance. PLoS One 7:e40397
Ren T, Wang J, Zhao M, Gong X, Wang S, Wang G, Zhou Chunjiang (2018) Involvement of NAC transcription factor SiNAC1 in a positive feedback loop via ABA biosynthesis and leaf senescence in foxtail millet. Planta 247:53–68
Ricachenevsky FK, Menguer PK, Sperotto RA (2013) kNACking on heaven’s door: how important are NAC transcription factors for leaf senescence and Fe/Zn remobilization to seeds? Front Plant Sci 4:226
Saad ASI, Li X, Li H-P, Huang T, Gao C-S, Guo M-W, Cheng W, Zhao G-Y, Yu-Cai Liao (2013) A rice stress-responsive NAC gene enhances tolerance of transgenic wheat to drought and salt stresses. Plant Sci 203–204:33–40
Sablowski RW, Meyerowitz EM, Souer E, van Houwelingen A, Kloos D, Mol J, Koes R (1998) A homolog of NO APICAL MERISTEM is an immediate target of the floral homeotic genes APETALA3/PISTILLATA. Cell 92:93–103
Sakuraba Y, Piao W, Lim JH, Han SH, Kim YS, An G, Paek NC (2015) Rice ONAC106 Inhibits leaf senescence and increases salt tolerance and tiller angle. Plant Cell Physiol 56:2325–2339
Satheesh V, Jagannadham PTK, Chidambaranathan P et al (2014) NAC transcription factor genes: genome-wide identification, phylogenetic, motif and cis-regulatory element analysis in pigeonpea (Cajanus cajan (L.) Millsp.). Mol Biol Rep 41:7763–7773
See D, Kanazin V, Kephart K, Blake T (2002) Mapping genes controlling variation in barley grain protein concentration. Crop Sci 42:680–685
Shang H, Wei L, Zou C, Yuan Y (2013) Analyses of the NAC transcription factor gene family in Gossypium raimondii Ulbr.: chromosomal location, structure, phylogeny, and expression patterns. J Integr Plant Biol 55:663–676
Shen J, Lv B, Luo L, He J, Mao C, Xi D, Ming F (2017) The NAC-type transcription factor OsNAC2 regulates ABA-dependent genes and abiotic stress tolerance in rice. Sci Rep 7:40641
Shim JS, Oh N, Chung PJ, Kim YS, Choi YD, Kim J (2018) Over-expression of OsNAC14 improves drought tolerance in rice. Front Plant Sci 9:1–14
Shinde H, Dudhate A, Tsugama D, Gupta SK, Liu S, Takano Tetsuo (2019) Pearl millet stress-responsive NAC transcription factor PgNAC21 enhances salinity stress tolerance in Arabidopsis. Plant Physiol Biochem 135:546–553
Singh S, Grover A, Nasim M (2016) Biofuel Potential of plants transformed genetically with NAC family genes. Front Plant Sci 7:22
Singh S, Khalid H, Grover A et al (2019) Altered physiological responses of LlaNAC gene of Lepidium latifolium overexpressing tobacco plants. Acta Physiol Plant 41:139
So H, Lee J (2019) NAC transcription factors from soybean (Glycine max L.) differentially regulated by abiotic stress. J Plant Biol 62:147–160
Tang Y, Liu M, Gao S, Zhang Z, Zhao X, Zhao C et al (2012) Molecular characterization of novel TaNAC genes in wheat and over-expression of TaNAC2a confers drought tolerance in tobacco. Physiol Plant 144:210–224
Tang GY, Shao FX, Xu PL, Shan L, Liu ZJ (2017) Over-expression of a peanut NAC Gene, AhNAC4, confers enhanced drought tolerance in tobacco. Rus J Plant Physiol 64:525–535
Tran LSP, Nakashima K, Sakuma Y, Osakabe Y, Qin F, Simpson SD, Maruyama K, Fujita Y, Shinozaki K, Yamaguchi-Shinozaki K (2007) Co-expression of the stress inducible zinc finger homeodomain ZFHD1 and NAC transcription factors enhances expression of the ERD1 gene in Arabidopsis. Plant J 49:46–63
Tran LSP, Quach TN, Guttikonda SK, Aldrich DL, Kumar R, Neelakandan A, Valliyod B, Nguyen HT (2009) Molecular characterization of stress-inducible GmNAC genes in soybean. Mol Genet Genomics 281:647–664
Uauy C, Brevis JC, Dubcovsky J (2006) The high grain protein content gene Gpc-B1 accelerates senescence and has pleiotropic effects on protein content in wheat. J Exp Bot 57:2785–2794
Valdivia ER, Herrera MT, Gianzo C, Fidalgo J, Revilla G, Zarra I, Sampedro J (2013) Regulation of secondary wall synthesis and cell death by NAC transcription factors in the monocot Brachypodium distachyon. J Exp Bot 64:1333–1343
Varshney R, Shi C, Thudi M et al (2017) Pearl millet genome sequence provides a resource to improve agronomic traits in arid environments. Nat Biotechnol 35:969–976
Vidal EA, Moyano TC, Riveras E, Contreras-López O, Gutiérrez RA (2013) Systems approaches map regulatory networks downstream of the auxin receptor AFB3 in the nitrate response of Arabidopsis thaliana roots. Proc Natl Acad Sci USA 110:12840–12845
Vidal EA, Álvarez JM, Gutiérrez RA (2014) Nitrate regulation of AFB3 and NAC4 gene expression in Arabidopsis roots depends on NRT1.1 nitrate transport function. Plant Signal Behav 9:e28501
Wang H, Zhao Q, Chen F, Wang M, Dixon RA (2011) NAC domain function and transcriptional control of a secondary cell wall master switch. Plant J 68:1104–1114
Xia J, Zhao Y, Burks P, Pauly M, Brown PJ (2018) A sorghum NAC gene is associated with variation in biomass properties and yield potential. Plant Direct 1–11
Xie Q, Guo HS, Dallman G, Fang S, Weissman AM, Chua NH (2002) SINAT5 promotes ubiquitin-related degradation of NAC1 to attenuate auxin signals. Nature 419:167–170
Xu Z, Gongbuzhaxi Wang C, Xue F, Zhang H, Ji W (2015) Wheat NAC transcription factor TaNAC29 is involved in response to salt stress. Plant Physiol Biochem 96:356–363
Xue GP, Way HM, Richardson T, Drenth J, Joyce PA, McIntyre CL (2011) Over-expression of TaNAC69 leads to enhanced transcript levels of stress up-regulated genes and dehydration tolerance in bread wheat. Mol Plant 4:697–712
Yamaguchi M, Kubo M, Fukuda H, Demura T (2008) Vascular-related NAC-DOMAIN7 is involved in the differentiation of all types of xylem vessels in Arabidopsis roots and shoots. Plant J 55:652–664
Yamaguchi M, Ohtani M, Mitsuda N, Kubo M, Ohme-Takagi M, Fukuda H, Demura T (2010) VND-INTERACTING2, a NAC domain transcription factor, negatively regulates xylem vessel formation in Arabidopsis. Plant Cell 22:1249–1263
Yang J, Worley E, Torres-Jerez I, Miller R, Wang M, Fu C, Wang Z-Y, Tang Y, Udvardi M (2015) PvNAC1 and PvNAC2 are associated with leaf senescence and nitrogen use efficiency in switchgrass. Bioenerg Res 8:868–880
Yang X, Kim MY, Ha J, Lee S-H (2019) Over-expression of the Soybean NAC gene GmNAC109 increases lateral root formation and abiotic stress tolerance in transgenic Arabidopsis Plants. Front Plant Sci 10:1036
Ye Y, Wu K, Chen J, Liu Q, Wu Y, Liu B, Fu X (2018) OsSND2, a NAC family transcription factor, is involved in secondary cell wall biosynthesis through regulating MYBs expression in rice. Rice 11:36
Yoshida K, Sakamoto S, Kawai T, Kobayashi Y, Sato K, Ichinose Y, Yaoi K, Akiyoshi-Endo M, Sato H, Takamizo T, Ohme-Takagi M, Mitsuda N (2013) Engineering the Oryza sativa cell wall with rice NAC transcription factors regulating secondary wall formation. Front Plant Sci 4:383
Yu X, Peng H, Liu Y et al (2014) CarNAC2, a novel NAC transcription factor in chickpea (Cicer arietinum L.), is associated with drought-response and various developmental processes in transgenic Arabidopsis. J Plant Biol 57:55–66
Yu X, Liu Y, Wang S et al (2016) CarNAC4, a NAC-type chickpea transcription factor conferring enhanced drought and salt stress tolerances in Arabidopsis. Plant Cell Rep 35:613–627
Yuan X, Wang H, Cai J, Bi Y, Li D, Song F (2019) Rice NAC transcription factor ONAC066 functions as a positive regulator of drought and oxidative stress response. BMC Plant Biol 19:278
Zhang L, Zhang L, Xia C, Zhao G, Jia J, Kong X (2016) The novel wheat transcription factor TaNAC47 enhances multiple abiotic stress tolerances in transgenic plants. Front Plant Sci 6:1–12
Zhao Q, Gallego-Giraldo L, Wang H, Zeng Y, Ding SY, Chen F, Dixon RA (2010) A NAC transcription factor orchestrates multiple features of cell wall development in Medicago truncatula. Plant J 63:100–114
Zhong R, Demura T, Ye ZH (2006) SND1, a NAC domain transcription factor, is a key regulator of secondary wall synthesis in fibers of Arabidopsis. Plant Cell 18:3158–3170
Zhong R, Richardson EA, Ye ZH (2007) Two NAC domain transcription factors, SND1 and NST1, function redundantly in regulation of secondary wall synthesis in fibers of Arabidopsis. Planta 225:1603–1611
Zhong R, Lee C, Zhou J, McCarthy RL, Ye ZH (2008) A battery of transcription factors involved in the regulation of secondary cell wall biosynthesis in Arabidopsis. Plant Cell 20:2763–2782
Acknowledgements
Sadhana Singh acknowledges the grant received from Science and Engineering Research Board-Department of Science and Technology (SERB-DST), Government of India.
Funding
This work was supported by the SERB-DST scheme for National post-doctoral fellowship (File no. PDF/2016/001586).
Author information
Authors and Affiliations
Contributions
SS conceived the idea and wrote the article. SS, HK and KKB performed critical document review. SS, HK, KKB, and AA were involved in editing of the MS. All authors read and approved the final MS.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
The original online version of this article was revised due to retrospective open access order.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Singh, S., Koyama, H., Bhati, K.K. et al. The biotechnological importance of the plant-specific NAC transcription factor family in crop improvement. J Plant Res 134, 475–495 (2021). https://doi.org/10.1007/s10265-021-01270-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10265-021-01270-y