Abstract
Extant ‘fishes’ belong to jawless, cartilaginous, actinopterygian, or sarcopterygian fish lineages. They comprise a non-monophyletic group of vertebrates from which tetrapods are excluded, and they are therefore paraphyletic. Recent advances in whole-genome sequencing have shed light on phylogenetic relationships, divergence times among major fish lineages, and the molecular basis of phenotypic diversity. This review encompasses the diversity of extant fishes and explores the variation in genomic organization and its evolutionary origins. This review begins with evaluating available genomic sequence resources with a focus on literally complete (‘telomere-to-telomere’) sequencing. Next, it discusses among-lineage variations in genomic contents, considering karyotype reports, genome sizes, and whole-genome duplication events. Additionally, we investigate three families of protein-coding genes, visual opsins, aquaporins (Aqps), and Hox genes, as they offer insights into morphological and physiological differences among major fish lineages. Overall, our technical basis of analyzing fish genomes and the knowledge of genomic organization provide valuable insights into our vertebrate-wide understanding of how molecular mechanisms specifying variable phenotypes are encoded in genomic sequence information.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Taxonomically, fishes are regarded as only one of the vertebrate classes, but phylogenetically, they almost correspond to vertebrates except that the latter also includes tetrapods (Fig. 1). Molecular synapomorphy of this paraphyletic group has been sought in relation to water-to-land transition (Falcon et al. 2023), and this transition was accompanied by molecular factors secondarily gained in the tetrapod lineage, including genes encoding receptors for airborne odors (Wang et al. 2021) as well as gene regulation enabling fin-to-limb transition (Meyer et al. 2021). The fin-to-limb transition involved the loss of the group of genes responsible for fin ray formation, actinodin, unique to the tetrapod lineage (Zhang et al. 2010; Biscotti et al. 2016).
Phylogenetic overview of different fish lineages. Phylogenetic relationships shown in this figure are based on OneZoom explorer (Wong and Rosindell 2022), and divergence times are based on the Timetree of Life project (Hedges et al. 2006). The Elopoomorpha–Osteoglossomorpha clade is based on recent literature (Parey et al. 2023). Diamonds indicate whole-genome duplication(s) (WGD), of which the ones on gray triangles indicate that each of them applies to a subset of the taxon. The timings of the WGDs before 400 million years ago, as well as those in the sturgeon/paddlefish lineage, are still controversial (see Kuraku et al. 2023; Redmond et al. 2023). Parentheses include the numbers of described species as of July 2023 based on Eschmeyer’s Catalog of Fishes (Fricke et al. 2023)
When limited to extant species, fishes consist of several distinct evolutionary lineages, cyclostomes, chondrichthyans, actinopterygians, and sarcopterygians (coelacanths and lungfishes), which diverged in more than 500 million years. Among these extant fish lineages, Actinopterygii comprises more than 27,000 described species, including sunfishes and sturgeons that exceed 3 meters in total length and the bigmouth buffalo Ictiobus cyprinellus that lives more than 120 years. Despite this deep divergence and exceptional species richness of actinopterygian fishes, only a limited number of ‘model’ species are often used in life science research as highly accessible laboratory-friendly systems (e.g., Braasch et al. 2015). Sometimes, study results from such laboratory systems are regarded as applicable widely to diverse ‘fishes’, in comparison with mammals or other vertebrates. Among typical fish models, the traditionally heavily used are zebrafish and medaka, both freshwater, short-lived, small-sized, annually reproducing species (Lleras-Forero et al. 2020). They cannot represent the whole fish diversity, since species in other fish lineages sometimes exhibit dissimilar life history characters, such as enhanced longevity, viviparity or placentation, and elongated gestation time.
The phylogenetic distinction between the different ‘fish’ lineages should also be recognized widely in life science. For this purpose, this review summarizes the current understanding of how molecular-level properties of these fish lineages differ from each other based on emerging genome sequence data.
Emerging genome sequence resources: how close to ‘T2T’?
Recent effort with massively parallel DNA sequencing methods has reached increasing fish lineages. One recent landmark study with genome assemblies of several Elopomorpha species suggested a new teleost fish phylogeny proposing a new monophyletic group Eloposteoglossocephala (Fig. 1; Parey et al. 2023). Before analyzing the genome contents, sequencing effort and subsequent quality controls of output should already encounter nonuniform characters of fish genomes, such as genome size, GC content, and repetitive element content (‘repetitiveness’). A scaffolding step such as that employing Hi-C data also instructs us about the species’ karyotype (Yamaguchi et al. 2021a).
Telomere-to-telomere (T2T) sequencing stands for complete determination of genomic DNA sequences from a telomeric end to the other for individual chromosomes of a particular species. T2T sequencing of complex eukaryotic genomes was reported initially for a human cell line (Nurk et al. 2022), followed by chicken (Huang et al. 2023). Finishing the genome assembly in the T2T grade requires a suite of sequencing and scaffolding approaches, and it would be instrumental in recognizing how close the existing resources are. Traditionally, researchers have relied on N50 lengths and ‘completeness scores’, quantified by detecting evolutionarily conserved 1-to-1 orthologs as metrics for evaluating genome assembly qualities, but the former can be excessively larger when erroneous assembly (‘overassembly’) is introduced, and the latter becomes saturated even with a genome assembly of suboptimal continuity and do not provide enough resolution for evaluation (Yamaguchi et al. 2021a). Instead, in the T2T era, the number and length of individual chromosomal sequences and the number and length of undetermined regions (‘gaps’) matter. It is also challenging to complete the sequencing of telomeric and centromeric regions, of which telomeric sequences can particularly be validated by computationally scanning the ends of chromosome-scale sequences for the canonical telomeric repeat (TTAGGG)n.
In fact, complete sequencing of fish genomes has been hindered due to various reasons. One confounding factor in cyclostome genome sequencing is somatic reorganization of the genomes termed programmed genome rearrangement (PGR) (in lampreys; Sémon et al. 2012) or chromosome elimination (in hagfishes; Nagao et al. 2023; reviewed in Smith et al. 2021). Besides, lamprey genomes exhibit a high GC-content heterogeneity with GC-rich protein-coding regions (Smith et al. 2013) as well as high interspersed repeat content derived partly from horizontal transfers of Tc1 retroelements (Kuraku et al. 2012). Another typical difficulty is in enlarged genome size as detailed later, mainly for chondrichthyans and lungfishes (see below). Among fish lineages, the highest chance of complete genome sequencing lies in the teleost fish lineage. The lineup of quality metrics for currently available genome assemblies of laboratory ‘models’ and aquaculture targets in this lineage generally shows high continuity and completeness (Fig. 2). Still, the latest genome assembly of the zebrafish Danio rerio (version GRCz11; released in 2017) contains far more than 10,000 gaps including the longest one of precisely 800 kb (Fig. 2). The currently available medaka genome assembly contains much fewer (491) gaps (that are consistently 1 kb-long), but there are nearly 900 sequences that remain to be joined into the 24 chromosomal sequences (Fig. 2; available at https://utgenome.org/medaka_v2/#!Assembly.md).
Statistics of selected teleost fish genome assemblies. Red bars show nuclear DNA contents estimated by Feulgen densitometry or flow cytometry analysis and karyotypes of individual species, wherever available (Gregory et al. 2007; Arai 2011). ‘Gaps’ denote those with no shorter than five undetermined bases. Gene-level completeness was evaluated by BUSCO v5 with the ortholog dataset Actinopterygii_odb10 (Seppey et al. 2019). The assemblies analyzed: medaka (Assembly version UT v2.2.4); zebrafish (GRCz11); fugu (fTakRub1.2); threespine stickleback (GAculeatus_UGA_version5); chub mackerel (fScoJap1.pri); gilthead seabream (fSpaAur1.1); channel catfish (Coco_2.0); Nile tilapia (O_niloticus_UMD_NMBU); Japanese eel (ASM2516954v1); common carp (ASM1834038v1); rainbow trout (USDA_OmykA_1.1); Atlantic salmon (Ssal_v3.1); and sterlet (ASM1064508v2)
Genome size
The ‘C-value enigma’ has attracted many researchers expecting implicit association with some phenotypic traits, but it remains elusive (Gregory 2005). Among all extant fish lineages, lungfishes have the largest known size of genomes exceeding 30 Gb (Vervoort 1980), followed by chondrichthyans whose genome sizes sometimes exceed 10 Gb (reviewed in Kuraku 2021). The difficulty in obtaining live tissues, which provide single cells as materials, prevented measuring nuclear DNA content especially of elusive shark and ray species, even though it is circumvented by applying a quantitative PCR-based method that does not require live cells (Kadota et al. 2023). It was not until long-read DNA sequencers became popular that giant (e.g., >10 Gb) vertebrate genomes were sequenced. It was initiated with the Mexican salamander Ambystoma mexicanum genome, whose resulting sequence assembly amounted to 32 Gb in length (Licht and Lowcock 1991; Nowoshilow et al. 2018). The Australian and West African lungfish genomes whose total number of nucleotides in currently available sequence assemblies amount to 40 and 35 Gb, respectively, were also sequenced to provide semi-chromosomal scaffolds, some of which exceed 1 Gb (Meyer et al. 2021; Wang et al. 2021). It should be noted that for these species with particularly enlarged genomes, the genome size estimates, independent of DNA sequence length, have not been unequivocally measured (e.g., by employing reliable reference from other species) and remain to be consolidated for controlling the genome sequencing products.
Among actinopterygians, some of the sturgeon species that underwent successive whole-genome duplications (Du et al. 2020), as well as polypterids, have genome sizes of larger than 3 Gb, while many of the remainders have genomes of smaller than 2 Gb. Although more thorough cross-species comparisons are awaited, among vertebrates, the genome sizes are correlated with intron lengths (Hara et al. 2018). Especially teleost fishes with small genome sizes have dramatically reduced intron lengths (Jakt et al. 2022). Short introns are thought to accelerate transcription and splicing (Swinburne and Silver 2008; Heyn et al. 2015; Keane and Seoighe 2016), which is implicated in rapid cellular activity, possibly leading to a short life (see below).
Whole-genome duplication
Recent accumulation of whole-genome sequences has finally encompassed all the extant vertebrate lineages, which has delivered an overview of which lineages experienced whole-genome duplication (WGD; Fig. 1). The non-teleost actinopterygian genomes have consolidated the phylogenetic position of the so-called teleost-specific genome duplication (TSGD; Thompson et al. 2021), whereas emerging chondrichthyan genomes support no additional WGD in its lineages of >450 million years (reviewed in Kuraku 2021). The most controversial is the mode of genome duplication close to the origin of vertebrates (reviewed in Kuraku et al. 2023). In the last century, hagfish and lamprey were regarded as having diverged before genome expansion occurred (Sidow 1996; Escriva et al. 2002). Later studies suggested more abundant gene repertoires in those jawless fish lineages even in the absence of whole cyclostome genome sequences (Putnam et al. 2008; Kuraku et al. 2009; Mehta et al. 2013). A series of genome analyses based on synteny conservation have suggested one round of WGD in the stem vertebrate lineage and a subsequent allotetraploidy in the stem gnathostome lineage (Simakov et al. 2020; Nakatani et al. 2021). Separately from these events, in the cyclostome lineage, additional WGD has been implicated by lamprey genome analysis (Nakatani et al. 2021), which should be ascertained by analyzing the hagfish genomes (Marlétaz et al. 2023b; Yu et al. 2023).
Karyotype
Another contrast between teleost fishes and other fish lineages is manifested in karyotypes and technical disposition for karyotyping studies. The karyotypes of teleost fishes usually harbor 44–54 chromosomes in diploid genomes, and their lengths are not dramatically variable (Arai 2011). For example, the medaka has 24 pairs of chromosomes (2n = 48) whose lengths vary only between 23 and 38 Mb, while zebrafish has 25 chromosome pairs (2n = 50) between 37 and 78 Mb. On the other hand, the karyotypes of non-teleost fishes often harbor more abundant chromosomes, including those shorter than 20 Mb (see below for those called ‘microchromosomes’) and/or longer than 100 Mb (summarized in Yamaguchi et al. 2023b). Those short chromosomes tend to be GC rich and have high gene density. As a whole, such karyotypes that exhibit a gradualism in chromosome length accommodate intragenomic heterogeneity of genomic, transcriptomic, and epigenomic properties (Hara and Kuraku 2023). Remarkably, the different fish lineages exhibit distinct modes of implementing this intragenomic heterogeneity.
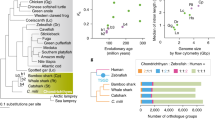
The common ancestor of teleost fishes experienced TSGD (Fig.1). Comparative genomic study between fishes including spotted gar, which is a non-teleost bony fish, and tetrapods indicates that fusions between microchromosomes and macrochromosomes resulted in 13 pairs of ancestral chromosomes that subsequently duplicated to 26 pairs (Braasch et al. 2016). After a WGD, sequence divergence between duplicated genes resulting from WGD (‘ohnologs’) gradually stops the recombination between homeologous chromosomes (Li et al. 2021). This evolutionary transition is known as a rediploidization step. The rediploidization step after the TSGD was coupled with loss of ohnologs and chromosome rearrangements that resulted in large karyotype differences between early diverged teleost lineages (Parey et al. 2022). For example, chromosome rearrangements observed between zebrafish and medaka (Fig. 3) were introduced shortly after the divergence between Otomorpha and Euteleosteomorpha (Fig. 1). Later inside the taxa Ostariophysi (including zebrafish) and Euteleostei (including medaka), the karyotypes became largely stable. Therein, not only the number of chromosomes, but also the number of visible major chromosome arms (fundamental number) are highly conserved (Yoshida and Kitano 2021). Genome-wide comparative synteny analysis within these groups confirmed minimum chromosome fusion/fission events (Kasahara et al. 2007; Star et al. 2011; Kakioka et al. 2013; Chen et al. 2014). Additional independent WGDs are observed in Salmoniformes and some Cyprinidae fishes, at least (Lien et al. 2016; Xu et al. 2019). They have exceptionally diverse karyotypes presumably due to the lineage-specific rediploidization step after the WGD (Robertson et al. 2017). Thus, teleost fishes generally have stable karyotypes, and WGD has temporally driven their karyotype evolution.
Cross-species karyotypic similarity in actinopterygian and chondrichthyan lineages. Similarity of genomic sequences for a pair of teleost fishes with a divergence time of 224 million years ago (left) is visualized with diagonal lines, together with an elasmobranch species pair with a divergence time of 270 million years ago (right). The divergence time estimates were obtained from Timetree of Life project (Hedges et al. 2006). Chromosome-scale sequences are sorted by length from top to bottom and left to right. Diagonal lines are colored in accordance with sequence divergence levels (dark green 75–100%, light green 50–75%, orange 25–50%, yellow 0–25%). Note that some regions in the zebrafish or zebra shark genome sequences used as queries are highly repetitive and are therefore causing horizontal arrays of similarity signals. In the parentheses are the total sequence lengths of the individual genome assemblies
Among non-teleost fishes, the chromosome organization in chondrichthyan genomes remained unexplored because of the lack of chromosome-scale sequence information. One recent study based on whole-genome sequencing characterized the chromosome organization of the little skate Leucoraja erinacea, which divided its chromosomes into three length categories, macro-, meso-, and microchromosomes (Marlétaz et al. 2023a). Notably, this species lacks a karyotype report, and their sequence-based findings remain to be verified. The comparison of the little skate genome with that of the zebra shark exhibited intrachromosomal breaks and few interchromosomal rearrangements (Fig. 3). The overall karyotypic organization of elasmobranchs have been maintained well since the shark–ray split which traces back to approximately 270 million years ago (Fig. 3). In comparison, much lower cross-species similarity, with a considerable number of interchromosomal rearrangements, is observed between distantly related teleost fish lineages despite a relatively short divergence time (Fig. 3).
The karyotype report is also missing for some more chondrichthyan species, whose chromosome-scale genome sequences have been made available [smalltooth sawfish Pristis pectinata (Jarva et al. 2023), shortfin mako Isurus oxyrinchus (Stanhope et al. 2023), and elephant fish Callorhinchus milii (Nakatani et al. 2021)]. The lack of karyotype reports is attributed mainly to the difficulty in obtaining fresh tissue materials from which cell culture is performed for repeated experiments to consolidate reproducible results. Another critical obstacle is the unique body fluid composition of chondrichthyans, which is overcome by adding urea, NaCl, and TMAO in culturing chondrichthyan cells, to mimic the body fluid. This culture medium adaptation enabled karyotyping for four shark species (Uno et al. 2020), which serve as indispensable references for validating whole-genome sequences. The importance of karyotypic references is outstanding for chondrichthyans with high karyotypic variation for which no versatile substitute, such as genetic linkage mapping, is usually accessible, unlike teleost fishes. Despite its importance, this sort of effort has rarely been demonstrated (see Součková et al. 2023).
According to the karyotypes obtained in the above-mentioned cell culture-based study, as well as more existing reports (summarized in Stingo and Rocco 2001; Arai 2011), the chromosome numbers of sharks and rays dramatically vary from 2n = 28 (for Narcine brasiliensis; Donahue 1974) to 106 (for Chiloscyllium punctatum; Uno et al. 2020). Of those, even with a relatively deep divergence of more than 250 million years, the red stingray Hemitrygon akajei and brownbanded bamboo shark C. punctatum share similar numbers of metacentric and subtelomeric chromosomes (Asahida et al. 1987; Uno et al. 2020). Karyotypes of elasmobranchs tend to show a continuum of chromosome lengths spanning from >100 Mb to <10 Mb. Even within Chondrichthyes, holocephalans (chimaeras and ratfish) tend to have a dichotomy of chromosome length ranges between (1) nearly 100 Mb or longer (only 4–5 chromosomes) and (2) shorter than 50 Mb (Nakatani et al. 2021). Interestingly, there are closely related species whose chromosome numbers are similar, but their morphology and length differ significantly. For instance, the whale shark Rhincodon typus and the brownbanded bamboo shark have chromosome numbers of 102 and 106, respectively. The former species has eight meta- or submetacentric chromosome pairs with the largest chromosome 3.6 times longer than the smallest one, while the latter species has 26 meta- or submetacentric chromosome pairs with the largest versus smallest length ratio of approximately 7 times (Uno et al. 2020).
Many non-teleost karyotypes including those of birds and turtles exhibit a substantial variation of chromosome lengths (Burt 2002; International Chicken Genome Sequencing Consortium 2004; Waters et al. 2021). This observation prompted researchers to call their large and small components ‘macro-’ and ‘microchromosomes’, respectively (Ohno et al. 1969; Ohno 1970), but there is no consensus on their definition. For example, the smallest Australian lungfish chromosome (818 Mb) is much larger than the ‘macrochromosomes’ defined in the study of the little skate genome (>40 Mb) (Marlétaz et al. 2023a). Conversely, microchromosomes defined therein (2.5 to 20 Mb) are comparable to or sometimes larger than some of the largest chromosomes of Takifugu rubripes. In practice, the terms macrochromosome and microchromosome should thus be used to categorize chromosomes with different lengths within a karyotype, but not across different species’ karyotypes.
Sex chromosome organization
Sexes of most vertebrate species are determined genetically, and the determination mechanism, intensively explored in mammals, is triggered during embryonic development primarily by a master sex-determining (SD) gene (reviewed in Capel 2017). The SD genes are harbored on sex chromosomes that are derived from autosomal chromosome pairs and undergo various frequency of turnovers in different evolutionary lineages. In Mammalia and Aves, the XY and ZW systems, respectively, were established approximately 150 and 110 million years ago, which are the oldest among vertebrates ever investigated (Graves 2016).
These several years have witnessed elaborate studies on teleost fishes reporting a variety of SD genes (Kitano et al. 2024). Those teleost fish SD genes are often orthologous to sex determination genes identified in other vertebrates, including Dmrt1- and Sox3-related transcription factors known as the SD genes in mammals and birds or components of TGF-β signaling pathway, such as Amh, Amhr2, Bmpr1b, Gsdf, and Gdf6 (Bertho et al. 2021; Rajendiran et al. 2021). The high variety of SD genes observed even within evolutionarily young taxa shows rapid SD gene turnovers, for example, in the genera Takifugu (Kabir et al. 2022) and Oryzias (Tanaka et al. 2007; Myosho et al. 2015). In such species, sex chromosome pairs have accumulated few sequence changes, and recombination suppression, which is a hallmark of sex-determining regions, should be operating only in a small part of the genomic segment harboring the sex-determining region.
Of approximately 1300 chondrichthyan species, sex chromosomes were reported for only eight species, if limited to the solid study cases with multiple individuals for both sexes (summarized in Uno et al. 2020). They are all batoids, namely, Hypanus sabinus, Platyrhinoidis triseriata, Potamotrygon aff. motoro, Potamotrygon falkneri, Potamotrygon motoro, Potamotrygon orbignyi, Potamotrygon wallacei, and Rhinobatos productus. The other existing reports are limited to sharks, which include the large sex chromosomes of the leopard shark Triakis semifasciata, although based on a single sex (Maddock and Schwartz 1996). We recently reported a heteromorphic sex chromosome pair of the brownbanded bamboo shark Chiloscyllium punctatum and white-spotted bamboo shark Chiloscyllium plagiosum (Uno et al. 2020). Mass sequencing projects organized under the Earth BioGenome Project (Lewin et al. 2022) have released several genome assemblies of chondrichthyan species including chromosomes labeled as X (and Y, sometimes) in the NCBI Genome database, as of July 2023, which however needs to be verified by comparing between the male and female genomes.
So far, the only study that has associated a chondrichthyan sex chromosome with its DNA sequences was performed for the zebra shark Stegostoma tigrinum and the whale shark Rhincodon typus (Yamaguchi et al. 2023b). This study identified a chromosome with a conspicuously low relative sequencing depth (nearly 0.5 folds) for male versus female and designated it as the X chromosome. The zebra shark X chromosome has a pseudoautosomal region (PAR) that shows a male–female ratio of sequencing depth comparable to autosomes on one end and is homologous to the whale shark chromosome separately identified as the X chromosome. This co-occurrence suggests that this X chromosome originated before the divergence between these two shark species as early as around 50 million years ago. Further studies on species in other shark lineages are awaited.
Gene family evolution
A profound question is whether the variation of basic genomic characteristics, such as karyotype, genome size, and ploidy, is associated with the phenotypic traits. Some studies suggested the association of genome size with longevity (Griffith et al. 2003; also see Gregory 2004), body size, and/or habitat depth (Ebeling et al. 1971). Nonetheless, one solid line of examples for the association with phenotypes can be provided by studies focusing on particular gene families.
In the opsin gene family, teleost fishes tend to accumulate more gene copies through tandem gene duplications as well as the above-mentioned TSGD (Lin et al. 2017; Yamaguchi et al. 2021b). One striking example is the silver spinyfin Diretmus argenteus. This deep-sea dweller has unique visual opsin gene repertoires, that is, 38 rhodopsin (RHO or Rh1) gene duplicates, which are thought to cooperatively function for sensing dim light in the deep sea (Musilova et al. 2019). In contrast, chondrichthyans generally exhibit few duplications of opsin gene repertoires and instead experience more frequent secondary gene loss of visual opsins (Hart 2020; Yamaguchi et al. 2021b). Utilizing the reduced gene repertoires, visual adaptation has been achieved by spectral tuning sometimes involving amino acid substitutions that have not been described in other fish lineages (Yamaguchi et al. 2023a).
A possible link with varying phenotypes can also be sought in gene repertoires responsible for modulating water permeability. Herein, we focus on aquaporins (AQPs) that function mainly as water channels and scrutinize its gene repertoire variation based on previous studies (Cerdà and Finn 2010; Tingaud-Sequeira et al. 2010; Finn et al. 2014). This gene family also exhibited an expansive nature of gene repertoires in the teleost fish lineage due to TSGD and a conservative nature in the chondrichthyan lineage, except a few tandem duplications (AQP3 and AQP10) in the latter lineage (Fig. 4). Also, AQP8, which is presumed to be involved in ammonia transport (Saparov et al. 2007), has not been identified in genome sequences of chondrichthyan species except the spiny dogfish Squalus acanthias (Cutler et al. 2022).
Aquaporin (AQP) gene repertoires in selected species in different fish lineages. Orthology, shown with a vertical positioning of the boxes, is supported by existing literature as well as our phylogenetic inference that will be reported elsewhere. Colored boxes with no number indicate the existence of only one ortholog. The numbers in the boxes indicate the multiplicity of the orthologs generated by lineage-specific gene duplications, while white boxes with dotted lines show the absence of possible ortholog in the currently available genome assemblies. At the top is the classification into different subfamilies including glpAQPs (aquaglyceroporins) based on existing literature (King et al. 2004; Finn et al. 2014). Red boxes indicate the lamprey genes that are potentially orthologous to multiple jawed vertebrate AQP subtypes
A more remarkable distinction is observed in the organization of Hox gene clusters, namely, the genomic regions harboring tandem copies of homeobox-containing genes responsible for regional specification of body segments along the anteroposterior axis. In general, a vertebrate Hox gene cluster harbors up to ten Hox genes tandemly duplicated before or around the time of bilaterian radiation and are found in different chromosomes as a result of whole-genome duplications. Teleost fish Hox genes are usually contained in seven or eight clusters because of the TSGD (Kuraku and Meyer 2009), and the non-teleost species that underwent additional whole-genome duplication (acipenseriforms and cyclostomes) also have more than four clusters (Mehta et al. 2013; Pascual-Anaya et al. 2018; Du et al. 2020). Chondrichthyans tend to have conservative gene repertoires with decreased molecular evolutionary rate throughout the genomes (Fig. 1), which is also reflected in the high conservation of the organization of Hox A, B, and D clusters (Hara et al. 2018). Exceptionally, the elasmobranch Hox C cluster underwent frequent gene loss and invasion of massive repeats (Hara et al. 2018), increasing the cluster length that typically fits in 100 kb (reviewed in Kuraku 2021). Interestingly, the eroding Hox C cluster is located in the PAR of the X chromosome in the zebra shark (Yamaguchi et al. 2023b). In fact, this Hox C cluster erosion is not observed in holocephalans, as indicated by as many as eleven persistent Hox C genes in a 100 kb-long cluster in the Callorhinchus milii genome (Ravi et al. 2009), and is thus confined to Elasmobranchii (Fig. 1; reviewed in Kuraku 2021). To the authors’ knowledge, shark Hox C cluster is the only documented case of a vertebrate Hox cluster residing on a sex chromosome (Yamaguchi et al. 2023b). Its localization in the PAR which should still maintain recombination between maternal and paternal chromatids likely cancels the sex-biased dosage of the expression of the Hox C genes. The possible relevance of the decreased constraint in maintaining the Hox cluster structure unique to this lineage needs to be explored in more detail.
Conclusions
These several years have witnessed the arrival of modern sequencing technologies and genome informatic analyses at some missing lineages including elasmobranchs and lungfishes, which permitted more thorough among-lineage comparisons. Fish lineages, phylogenetically divided primarily into jawless, cartilaginous, actinopterygian, and sarcopterygian fishes, have distinct genomic organization featured by size and ploidy level of the genome, and size and number of chromosomes. In particular, variable contents of repetitive sequences and functional genes have been revealed to be associated with genomic organizations, namely the genome size and ploidy levels, respectively. Phylogenetic understanding on the molecular-level variation among these different fish lineages is critical to addressing innumerable questions concerning phenotypic variations of vertebrates and their evolutionary origins, such as what in the genome of their last common ancestor permitted the advent of vertebrates and what in the sarcopterygian lineage later led to the emergence of tetrapods.
References
Arai R (2011) Fish Karyotypes: a checklist. Springer, New York
Asahida T, Ida H, Inoue S (1987) Karyotypes of three rays in the order Myliobatiformes. Japan J Ichthyol 33:426–430
Bertho S, Herpin A, Schartl M, Guiguen Y (2021) Lessons from an unusual vertebrate sex-determining gene. Philos Trans R Soc Lond B Biol Sci 376:20200092
Biscotti MA, Gerdol M, Canapa A, Forconi M, Olmo E, Pallavicini A, Barucca M, Schartl M (2016) The lungfish transcriptome: a glimpse into molecular evolution events at the transition from water to land. Sci Rep 6:21571
Braasch I, Peterson SM, Desvignes T, McCluskey BM, Batzel P, Postlethwait JH (2015) A new model army: emerging fish models to study the genomics of vertebrate Evo-Devo. J Exp Zool B Mol Dev Evol 324:316–341
Braasch I, Gehrke AR, Smith JJ, Kawasaki K, Manousaki T, Pasquier J, Amores A, Desvignes T, Batzel P, Catchen J, Berlin AM, Campbell MS, Barrell D, Martin KJ, Mulley JF, Ravi V, Lee AP, Nakamura T, Chalopin D, Fan S, Wcisel D, Cañestro C, Sydes J, Beaudry FE, Sun Y, Hertel J, Beam MJ, Fasold M, Ishiyama M, Johnson J, Kehr S, Lara M, Letaw JH, Litman GW, Litman RT, Mikami M, Ota T, Saha NR, Williams L, Stadler PF, Wang H, Taylor JS, Fontenot Q, Ferrara A, Searle SM, Aken B, Yandell M, Schneider I, Yoder JA, Volff JN, Meyer A, Amemiya CT, Venkatesh B, Holland PW, Guiguen Y, Bobe J, Shubin NH, Di Palma F, Alföldi J, Lindblad-Toh K, Postlethwait JH (2016) The spotted gar genome illuminates vertebrate evolution and facilitates human–teleost comparisons. Nat Genet 48:427–437
Burt DW (2002) Origin and evolution of avian microchromosomes. Cytogenet Genome Res 96:97–112
Capel B (2017) Vertebrate sex determination: evolutionary plasticity of a fundamental switch. Nat Rev Genet 18:675–689
Cerdà J, Finn RN (2010) Piscine aquaporins: an overview of recent advances. J Exp Zool A Ecol Genet Physiol 313:623–650
Chen S, Zhang G, Shao C, Huang Q, Liu G, Zhang P, Song W, An N, Chalopin D, Volff JN, Hong Y, Li Q, Sha Z, Zhou H, Xie M, Yu Q, Liu Y, Xiang H, Wang N, Wu K, Yang C, Zhou Q, Liao X, Yang L, Hu Q, Zhang J, Meng L, Jin L, Tian Y, Lian J, Yang J, Miao G, Liu S, Liang Z, Yan F, Li Y, Sun B, Zhang H, Zhang J, Zhu Y, Du M, Zhao Y, Schartl M, Tang Q, Wang J (2014) Whole-genome sequence of a flatfish provides insights into ZW sex chromosome evolution and adaptation to a benthic lifestyle. Nat Genet 46:253–260
Cutler CP, Murray D, Ojo T, Harmon S, MacIver B, Cramb G, Zeidel ML (2022) Aquaporin (AQP) channels in the spiny dogfish, Squalus acanthias I: characterization of AQP3 and AQP15 function and expression, and localization of the proteins in gill and spiral valve intestine. Comp Biochem Physiol B Biochem Mol Biol 258:110702
Donahue WH (1974) A karyotypic study of three species of Rajiformes (Chondrichthyes, Pisces). Can J Genet Cytolog 16:203–211
Du K, Stöck M, Kneitz S, Klopp C, Woltering JM, Adolfi MC, Feron R, Prokopov D, Makunin A, Kichigin I, Schmidt C, Fischer P, Kuhl H, Wuertz S, Gessner J, Kloas W, Cabau C, Iampietro C, Parrinello H, Tomlinson C, Journot L, Postlethwait JH, Braasch I, Trifonov V, Warren WC, Meyer A, Guiguen Y, Schartl M (2020) The sterlet sturgeon genome sequence and the mechanisms of segmental rediploidization. Nat Ecol Evol 4:841–852
Ebeling AW, Atkin NB, Setzer PY (1971) Genome sizes of teleostean fishes: increases in some deep-sea species. Am Nat 105:549–561
Escriva H, Manzon L, Youson J, Laudet V (2002) Analysis of lamprey and hagfish genes reveals a complex history of gene duplications during early vertebrate evolution. Mol Biol Evol 19:1440–1450
Falcon F, Tanaka EM, Rodriguez-Terrones D (2023) Transposon waves at the water-to-land transition. Curr Opin Gen Dev 81:102059
Finn RN, Chauvigné F, Hlidberg JB, Cutler CP, Cerdà J (2014) The lineage-specific evolution of aquaporin gene clusters facilitated tetrapod terrestrial adaptation. PLoS One 9:e113686
Fricke R, Eschmeyer WN, Van der Laan R (2023) Eschmeyer’s Catalog of fishes: genera, species, references. http://researcharchive.calacademy.org/research/ichthyology/catalog/fishcatmain.asp. Accessed 14 July 2023
Graves J (2016) Evolution of vertebrate sex chromosomes and dosage compensation. Nat Rev Genet 17:33–46
Gregory TR (2004) Genome size is not correlated positively with longevity in fishes (or homeotherms). Exp Gerontol 39:859–860
Gregory TR (2005) Synergy between sequence and size in large-scale genomics. Nat Rev Genet 6:699–708
Gregory TR, Nicol JA, Tamm H, Kullman B, Kullman K, Leitch IJ, Murray BG, Kapraun DF, Greilhuber J, Bennett MD (2007) Eukaryotic genome size databases. Nucleic Acids Res 35:D332–338
Griffith OL, Moodie GEE, Civetta A (2003) Genome size and longevity in fish. Exp Gerontol 38:333–337
Hara Y, Yamaguchi K, Onimaru K, Kadota M, Koyanagi M, Keeley SD, Tatsumi K, Tanaka K, Motone F, Kageyama Y, Nozu R, Adachi N, Nishimura O, Nakagawa R, Tanegashima C, Kiyatake I, Matsumoto R, Murakumo K, Nishida K, Terakita A, Kuratani S, Sato K, Hyodo S, Kuraku S (2018) Shark genomes provide insights into elasmobranch evolution and the origin of vertebrates. Nat Ecol Evol 2:1761–1771
Hara Y, Kuraku S (2023) The impact of local genomic properties on the evolutionary fate of genes. eLife 12:e82290
Hart NS (2020) Vision in sharks and rays: opsin diversity and colour vision. Semin Cell Dev Biol 106:12–19
Hedges SB, Dudley J, Kumar S (2006) TimeTree: a public knowledge-base of divergence times among organisms. Bioinformatics 22:2971–2972
Heyn P, Kalinka AT, Tomancak P, Neugebauer KM (2015) Introns and gene expression: cellular constraints, transcriptional regulation, and evolutionary consequences. Bioessays 37:148–154
Huang Z, Xu Z, Bai H, Huang Y, Kang N, Ding X, Liu J, Luo H, Yang C, Chen W, Guo Q, Xue L, Zhang X, Xu L, Chen M, Fu H, Chen Y, Yue Z, Fukagawa T, Liu S, Chang G, Xu L (2023) Evolutionary analysis of a complete chicken genome. Proc Natl Acad Sci U S A 120:e2216641120
International Chicken Genome Sequencing Consortium (2004) Sequence and comparative analysis of the chicken genome provide unique perspectives on vertebrate evolution. Nature 432:695–716
Jakt LM, Dubin A, Johansen SD (2022) Intron size minimisation in teleosts. BMC Genomics 23:628
Jarva TM, Phillips NM, Von Eiff C, Poulakis GR, Naylor G, Feldheim KA, Flynt AS (2023) Gene expression and evolution in the smalltooth sawfish, Pristis pectinata. bioRxiv. https://doi.org/https://doi.org/10.1101/2023.01.12.523848
Kabir A, Ieda R, Hosoya S, Fujikawa D, Atsumi K, Tajima S, Nozawa A, Koyama T, Hirase S, Nakamura O, Kadota M, Nishimura O, Kuraku S, Nakamura Y, Kobayashi H, Toyoda A, Tasumi S, Kikuchi K (2022) Repeated translocation of a supergene underlying rapid sex chromosome turnover in Takifugu pufferfish. Proc Natl Acad Sci U S A 119:e2121469119
Kadota M, Tatsumi K, Yamaguchi K, Uno Y, Kuraku S (2023) Shark and ray genome size estimation: methodological optimization for inclusive and controllable biodiversity genomics. F1000Res 12:1204. https://doi.org/10.12688/f1000research.136385.1
Kakioka R, Kokita T, Kumada H, Watanabe K, Okuda N (2013) A RAD-based linkage map and comparative genomics in the gudgeons (genus Gnathopogon, Cyprinidae). BMC Genomics 14:32
Kasahara M, Naruse K, Sasaki S, Nakatani Y, Qu W, Ahsan B, Yamada T, Nagayasu Y, Doi K, Kasai Y, Jindo T, Kobayashi D, Shimada A, Toyoda A, Kuroki Y, Fujiyama A, Sasaki T, Shimizu A, Asakawa S, Shimizu N, Hashimoto S, Yang J, Lee Y, Matsushima K, Sugano S, Sakaizumi M, Narita T, Ohishi K, Haga S, Ohta F, Nomoto H, Nogata K, Morishita T, Endo T, Shin-I T, Takeda H, Morishita S, Kohara Y (2007) The medaka draft genome and insights into vertebrate genome evolution. Nature 447:714–719
Keane PA, Seoighe C (2016) Intron length coevolution across mammalian genomes. Mol Biol Evol 33:2682–2691
King L, Kozono D, Agre P (2004) From structure to disease: the evolving tale of aquaporin biology. Nat Rev Mol Cell Biol 5:687–698
Kitano J, Ansai S, Takehana Y, Yamamoto Y (2024) Diversity and convergence of sex determination mechanisms in teleost fish. Annu Rev Anim Biosci (in press)
Kuraku S (2021) Shark and ray genomics for disentangling their morphological diversity and vertebrate evolution. Dev Biol 477:262–272
Kuraku S, Meyer A (2009) The evolution and maintenance of Hox gene in vertebrates and the teleost-specific genome duplication. Intl J Dev Biol 53:765–773
Kuraku S, Meyer A, Kuratani S (2009) Timing of genome duplications relative to the origin of the vertebrates: did cyclostomes diverge before or after? Mol Biol Evol 26:47–59
Kuraku S, Qiu H, Meyer A (2012) Horizontal transfers of Tc1 elements between teleost fishes and their vertebrate parasites, lampreys. Genome Biol Evol 4:929–936
Kuraku S, Kaiya H, Tanaka T, Hyodo S (2023) Evolution of vertebrate hormones and their receptors: insights from non-osteichthyan genomes. Annu Rev Anim Biosci 11:163–182
Lewin HA, Richards S, Lieberman Aiden E, Allende ML, Archibald JM, Bálint M, Barker KB, Baumgartner B, Belov K, Bertorelle G, Blaxter ML, Cai J, Caperello ND, Carlson K, Castilla-Rubio JC, Chaw SM, Chen L, Childers AK, Coddington JA, Conde DA, Corominas M, Crandall KA, Crawford AJ, DiPalma F, Durbin R, Ebenezer TE, Edwards SV, Fedrigo O, Flicek P, Formenti G, Gibbs RA, Gilbert MTP, Goldstein MM, Graves JM, Greely HT, Grigoriev IV, Hackett KJ, Hall N, Haussler D, Helgen KM, Hogg CJ, Isobe S, Jakobsen KS, Janke A, Jarvis ED, Johnson WE, Jones SJM, Karlsson EK, Kersey PJ, Kim JH, Kress WJ, Kuraku S, Lawniczak MKN, Leebens-Mack JH, Li X, Lindblad-Toh K, Liu X, Lopez JV, Marques-Bonet T, Mazard S, Mazet JAK, Mazzoni CJ, Myers EW, O'Neill RJ, Paez S, Park H, Robinson GE, Roquet C, Ryder OA, Sabir JSM, Shaffer HB, Shank TM, Sherkow JS, Soltis PS, Tang B, Tedersoo L, Uliano-Silva M, Wang K, Wei X, Wetzer R, Wilson JL, Xu X, Yang H, Yoder AD, Zhang G (2022) The Earth BioGenome Project 2020: starting the clock. Proc Natl Acad Sci U S A 119:e2115635118
Li Z, McKibben MTW, Finch GS, Blischak PD, Sutherland BL, Barker MS (2021) Patterns and processes of diploidization in land plants. Annu Rev Plant Biol 72:387–410
Licht LE, Lowcock LA (1991) Genome size and metabolic rate in salamanders. Comp Biochem Physiol B 100:83–92
Lien S, Koop BF, Sandve SR, Miller JR, Kent MP, Nome T, Hvidsten TR, Leong JS, Minkley DR, Zimin A, Grammes F, Grove H, Gjuvsland A, Walenz B, Hermansen RA, von Schalburg K, Rondeau EB, Di Genova A, Samy JKA, Olav Vik J, Vigeland MD, Caler L, Grimholt U, Jentoft S, Våge DI, de Jong P, Moen T, Baranski M, Palti Y, Smith DR, Yorke JA, Nederbragt AJ, Tooming-Klunderud A, Jakobsen KS, Jiang X, Fan D, Hu Y, Liberles DA, Vidal R, Iturra P, Jones SJ, Jonassen I, Maass A, Omholt SW, Davidson WS (2016) The Atlantic salmon genome provides insights into rediploidization. Nature 533:200–205
Lin JJ, Wang FY, Li WH, Wang TY (2017) The rises and falls of opsin genes in 59 ray-finned fish genomes and their implications for environmental adaptation. Sci Rep 7:15568
Lleras-Forero L, Winkler C, Schulte-Merker S (2020) Zebrafish and medaka as models for biomedical research of bone diseases. Dev Biol 457:191–205
Maddock MB, Schwartz FJ (1996) Elasmobranch cytogenetics: methods and sex chromosomes. Bull Mar Sci 58:147–155
Marlétaz F, de la Calle-Mustienes E, Acemel RD, Paliou C, Naranjo S, Martínez-García PM, Cases I, Sleight VA, Hirschberger C, Marcet-Houben M, Navon D, Andrescavage A, Skvortsova K, Duckett PE, González-Rajal Á, Bogdanovic O, Gibcus JH, Yang L, Gallardo-Fuentes L, Sospedra I, Lopez-Rios J, Darbellay F, Visel A, Dekker J, Shubin N, Gabaldón T, Nakamura T, Tena JJ, Lupiáñez DG, Rokhsar DS, Gómez-Skarmeta JL (2023a) The little skate genome and the evolutionary emergence of wing-like fins. Nature 616:495–503
Marlétaz F, Timoshevskaya N, Timoshevskiy V, Simakov O, Parey E, Gavriouchkina D, Suzuki M, Kubokawa K, Brenner S, Smith J, Rokhsar DS (2023b) The hagfish genome and the evolution of vertebrates. bioRxiv. https://doi.org/10.1101/2023.04.17.537254
Mehta TK, Ravi V, Yamasaki S, Lee AP, Lian MM, Tay B-H, Tohari S, Yanai S, Tay A, Brenner S, Venkatesh B (2013) Evidence for at least six Hox clusters in the Japanese lamprey (Lethenteron japonicum). Proc Natl Acad Sci U S A 110:16044–16049
Meyer A, Schloissnig S, Franchini P, Du K, Woltering JM, Irisarri I, Wong WY, Nowoshilow S, Kneitz S, Kawaguchi A, Fabrizius A, Xiong P, Dechaud C, Spaink HP, Volff J-N, Simakov O, Burmester T, Tanaka EM, Schartl M (2021) Giant lungfish genome elucidates the conquest of land by vertebrates. Nature 590:284–289
Musilova Z, Cortesi F, Matschiner M, Davies WIL, Patel JS, Stieb SM, de Busserolles F, Malmstrøm M, Tørresen OK, Brown CJ, Mountford JK, Hanel R, Stenkamp DL, Jakobsen KS, Carleton KL, Jentoft S, Marshall J, Salzburger W (2019) Vision using multiple distinct rod opsins in deep-sea fishes. Science 364:588–592
Myosho T, Takehana Y, Hamaguchi S, Sakaizumi M (2015) Turnover of sex chromosomes in celebensis group medaka fishes. G3 (Bethesda) 5:2685–2691
Nagao K, Tanaka Y, Kajitani R, Toyoda A, Itoh T, Kubota S, Goto Y (2023) Bioinformatic and fine-scale chromosomal mapping uncover the essence and evolution of eliminated chromosomes in the Japanese hagfish, Eptatretus burgeri, through repetitive DNA family analysis. bioRxiv. https://doi.org/10.1101/2023.05.28.542657
Nakatani Y, Shingate P, Ravi V, Pillai NE, Prasad A, McLysaght A, Venkatesh B (2021) Reconstruction of proto-vertebrate, proto-cyclostome and proto-gnathostome genomes provides new insights into early vertebrate evolution. Nat Commun 12:4489
Nowoshilow S, Schloissnig S, Fei J-F, Dahl A, Pang AWC, Pippel M, Winkler S, Hastie AR, Young G, Roscito JG, Falcon F, Knapp D, Powell S, Cruz A, Cao H, Habermann B, Hiller M, Tanaka EM, Myers EW (2018) The axolotl genome and the evolution of key tissue formation regulators. Nature 554:50–55
Nurk S, Koren S, Rhie A, Rautiainen M, Bzikadze AV, Mikheenko A, Vollger MR, Altemose N, Uralsky L, Gershman A, Aganezov S, Hoyt SJ, Diekhans M, Logsdon GA, Alonge M, Antonarakis SE, Borchers M, Bouffard GG, Brooks SY, Caldas GV, Chen NC, Cheng H, Chin CS, Chow W, de Lima LG, Dishuck PC, Durbin R, Dvorkina T, Fiddes IT, Formenti G, Fulton RS, Fungtammasan A, Garrison E, Grady PGS, Graves-Lindsay TA, Hall IM, Hansen NF, Hartley GA, Haukness M, Howe K, Hunkapiller MW, Jain C, Jain M, Jarvis ED, Kerpedjiev P, Kirsche M, Kolmogorov M, Korlach J, Kremitzki M, Li H, Maduro VV, Marschall T, McCartney AM, McDaniel J, Miller DE, Mullikin JC, Myers EW, Olson ND, Paten B, Peluso P, Pevzner PA, Porubsky D, Potapova T, Rogaev EI, Rosenfeld JA, Salzberg SL, Schneider VA, Sedlazeck FJ, Shafin K, Shew CJ, Shumate A, Sims Y, Smit AFA, Soto DC, Sović I, Storer JM, Streets A, Sullivan BA, Thibaud-Nissen F, Torrance J, Wagner J, Walenz BP, Wenger A, Wood JMD, Xiao C, Yan SM, Young AC, Zarate S, Surti U, McCoy RC, Dennis MY, Alexandrov IA, Gerton JL, O'Neill RJ, Timp W, Zook JM, Schatz MC, Eichler EE, Miga KH, Phillippy AM (2022) The complete sequence of a human genome. Science 376:44–53
Ohno S (1970) Evolution by gene duplication. Springer, New York
Ohno S, Muramoto J, Stenius C, Christian L, Kittrell WA, Atkin NB (1969) Microchromosomes in holocephalian, chondrostean and holostean fishes. Chromosoma 26:35–40
Parey E, Louis A, Montfort J, Guiguen Y, Roest Crollius H, Berthelot C (2022) An atlas of fish genome evolution reveals delayed rediploidization following the teleost whole-genome duplication. Genome Res 32:1685–1697
Parey E, Louis A, Montfort J, Bouchez O, Roques C, Iampietro C, Lluch J, Castinel A, Donnadieu C, Desvignes T, Floi Bucao C, Jouanno E, Wen M, Mejri S, Dirks R, Jansen H, Henkel C, Chen W-J, Zahm M, Cabau C, Klopp C, Thompson AW, Robinson-Rechavi M, Braasch I, Lecointre G, Bobe J, Postlethwait JH, Berthelot C, Roest Crollius H, Guiguen Y (2023) Genome structures resolve the early diversification of teleost fishes. Science 379:572–575
Pascual-Anaya J, Sato I, Sugahara F, Higuchi S, Paps J, Ren Y, Takagi W, Ruiz-Villalba A, Ota KG, Wang W, Kuratani S (2018) Hagfish and lamprey Hox genes reveal conservation of temporal colinearity in vertebrates. Nat Ecol Evol 2:859–866
Putnam NH, Butts T, Ferrier DEK, Furlong RF, Hellsten U, Kawashima T, Robinson-Rechavi M, Shoguchi E, Terry A, Yu J-K, Benito-Gutiérrez EL, Dubchak I, Garcia-Fernàndez J, Gibson-Brown JJ, Grigoriev IV, Horton A C, de Jong PJ, Jurka J, Kapitonov VV, Kohara Y, Kuroki Y, Lindquist E, Lucas S, Osoegawa K, Pennacchio LA, Salamov AA, Satou Y, Sauka-Spengler T, Schmutz J, Shin-I T, Toyoda A, Bronner-Fraser M, Fujiyama A, Holland LZ, Holland PW, Satoh N, Rokhsar DS (2008) The amphioxus genome and the evolution of the chordate karyotype. Nature 453:1064–1071
Rajendiran P, Jaafar F, Kar S, Sudhakumari C, Senthilkumaran B, Parhar IS (2021) Sex determination and differentiation in teleost: roles of genetics, environment, and brain. Biology 10:973
Ravi V, Lam K, Tay B-H, Tay A, Brenner S, Venkatesh B (2009) Elephant shark (Callorhinchus milii) provides insights into the evolution of Hox gene clusters in gnathostomes. Proc Natl Acad Sci U S A 106:16327–16332
Redmond AK, Casey D, Gundappa MK, Macqueen DJ, McLysaght A (2023) Independent rediploidization masks shared whole genome duplication in the sturgeon-paddlefish ancestor. Nat Commun 14:2879
Robertson FM, Gundappa MK, Grammes F, Hvidsten TR, Redmond AK, Lien S, Martin SAM, Holland PWH, Sandve SR, Macqueen DJ (2017) Lineage-specific rediploidization is a mechanism to explain time-lags between genome duplication and evolutionary diversification. Genome Biol 18:111
Saparov SM, Liu K, Agre P, Pohl P (2007) Fast and selective ammonia transport by aquaporin-8. J Biol Chem 282:5296–5301
Sémon M, Schubert M, Laudet V (2012) Programmed genome rearrangements: in lampreys, all cells are not equal. Curr Biol 22:R641–R643
Seppey M, Manni M, Zdobnov EM (2019) BUSCO: assessing genome assembly and annotation completeness. Methods Mol Biol 1962:227–245
Sidow A (1996) Gen(om)e duplications in the evolution of early vertebrates. Curr Opin Genet Dev 6:715–722
Simakov O, Marlétaz F, Yue JX, O'Connell B, Jenkins J, Brandt A, Calef R, Tung CH, Huang TK, Schmutz J, Satoh N, Yu JK, Putnam NH, Green RE, Rokhsar DS (2020) Deeply conserved synteny resolves early events in vertebrate evolution. Nat Ecol Evol 4:820–830
Smith JJ, Kuraku S, Holt C, Sauka-Spengler T, Jiang N, Campbell MS, Yandell MD, Manousaki T, Meyer A, Bloom OE, Morgan JR, Buxbaum J D, Sachidanandam R, Sims C, Garruss AS, Cook M, Krumlauf R, Wiedemann LM, Sower SA, Decatur WA, Hall JA, Amemiya CT, Saha NR, Buckley KM, Rast JP, Das S, Hirano M, McCurley N, Guo P, Rohner N, Tabin CJ, Piccinelli P, Elgar G, Ruffier M, Aken BL, Searle SM, Muffato M, Pignatelli M, Herrero J, Jones M, Brown CT, Chung-Davidson YW, Nanlohy KG, Libants SV, Yeh CY, McCauley DW, Langeland JA, Pancer Z, Fritzsch B, de Jong PJ, Zhu B, Fulton LL, Theising B, Flicek P, Bronner ME, Warren WC, Clifton SW, Wilson RK, Li W (2013) Sequencing of the sea lamprey (Petromyzon marinus) genome provides insights into vertebrate evolution. Nat Genet 45:415–421
Smith JJ, Timoshevskiy VA, Saraceno C (2021) Programmed DNA elimination in vertebrates. Annu Rev Anim Biosci 9:173–201
Součková K, Jasík M, Sovadinová I, Sember A, Sychrová E, Konieczna A, Bystrý V, Dyková I, Blažek R, Lukšíková K, Pavlica T, Jankásek M, Altmanová M, Žák J, Zbončáková A, Reichard M, Slabý O (2023) From fish to cells: establishment of continuous cell lines from embryos of annual killifish Nothobranchius furzeri and N. kadleci. Aquat Toxicol 259:106517
Stanhope MJ, Ceres KM, Sun Q, Wang M, Zehr JD, Marra NJ, Wilder AP, Zou C, Bernard AM, Pavinski-Bitar P, Lokey MG, Shivji MS (2023) Genomes of endangered great hammerhead and shortfin mako sharks reveal historic population declines and high levels of inbreeding in great hammerhead. iScience 26:105815
Star B, Nederbragt AJ, Jentoft S, Grimholt U, Malmstrøm M, Gregers TF, Rounge TB, Paulsen J, Solbakken MH, Sharma A, Wetten OF, Lanzén A, Winer R, Knight J, Vogel J-H, Aken B, Andersen O, Lagesen K, Tooming-Klunderud A, Edvardsen RB, Tina KG, Espelund M, Nepal C, Previti C, Karlsen BO, Moum T, Skage M, Berg PR, Gjøen T, Kuhl H, Thorsen J, Malde K, Reinhardt R, Du L, Johansen SD, Searle S, Lien S, Nilsen F, Jonassen I, Omholt SW, Stenseth NC, Jakobsen KS (2011) The genome sequence of Atlantic cod reveals a unique immune system. Nature 477:207–210
Stingo V, Rocco L (2001) Selachian cytogenetics: a review. Genetica 111:329–347
Swinburne IA, Silver PA (2008) Intron delays and transcriptional timing during development. Dev Cell 14:324–330
Tanaka K, Takehana Y, Naruse K, Hamaguchi S, Sakaizumi M (2007) Evidence for different origins of sex chromosomes in closely related Oryzias fishes: substitution of the master sex-determining gene. Genetics 177:2075–2081
Thompson AW, Hawkins MB, Parey E, Wcisel DJ, Ota T, Kawasaki K, Funk E, Losilla M, Fitch OE, Pan Q, Feron R, Louis A, Montfort J, Milhes M, Racicot BL, Childs KL, Fontenot Q, Ferrara A, David SR, McCune AR, Dornburg A, Yoder JA, Guiguen Y, Roest Crollius H, Berthelot C, Harris MP, Braasch I (2021) The bowfin genome illuminates the developmental evolution of ray-finned fishes. Nat Genet 53:1373–1384
Tingaud-Sequeira A, Calusinska M, Finn RN, Chauvigné F, Lozano J, Cerdà J (2010) The zebrafish genome encodes the largest vertebrate repertoire of functional aquaporins with dual paralogy and substrate specificities similar to mammals. BMC Evol Biol 10:38
Uno Y, Nozu R, Kiyatake I, Higashiguchi N, Sodeyama S, Murakumo K, Sato K, Kuraku S (2020) Cell culture-based karyotyping of orectolobiform sharks for chromosome-scale genome analysis. Commun Biol 3:652
Vervoort A (1980) Tetraploidy inProtopterus (Dipnoi). Experientia 36:294–296
Wang K, Wang J, Zhu C, Yang L, Ren Y, Ruan J, Fan G, Hu J, Xu W, Bi X, Zhu Y, Song Y, Chen H, Ma T, Zhao R, Jiang H, Zhang B, Feng C, Yuan Y, Gan X, Li Y, Zeng H, Liu Q, Zhang Y, Shao F, Hao S, Zhang H, Xu X, Liu X, Wang D, Zhu M, Zhang G, Zhao W, Qiu Q, He S, Wang W (2021) African lungfish genome sheds light on the vertebrate water-to-land transition. Cell 184:1362–1376
Waters PD, Patel HR, Ruiz-Herrera A, Álvarez-González L, Lister NC, Simakov O, Ezaz T, Kaur P, Frere C, Grützner F, Georges A, Graves JAM (2021) Microchromosomes are building blocks of bird, reptile, and mammal chromosomes. Proc Natl Acad Sci U S A 118:e2112494118
Wong Y, Rosindell J (2022) Dynamic visualisation of million-tip trees: the OneZoom project. Methods Ecol Evol 13:303–313
Xu P, Xu J, Liu G, Chen L, Zhou Z, Peng W, Jiang Y, Zhao Z, Jia Z, Sun Y, Wu Y, Chen B, Pu F, Feng J, Luo J, Chai J, Zhang H, Wang H, Dong C, Jiang W, Sun X (2019) The allotetraploid origin and asymmetrical genome evolution of the common carp Cyprinus carpio. Nat Commun 10:4625
Yamaguchi K, Kadota M, Nishimura O, Ohishi Y, Naito Y, Kuraku S (2021a) Technical considerations in Hi-C scaffolding and evaluation of chromosome‐scale genome assemblies. Mol Ecol 30:5923–5934
Yamaguchi K, Koyanagi M, Kuraku S (2021b) Visual and nonvisual opsin genes of sharks and other nonosteichthyan vertebrates: Genomic exploration of underwater photoreception. J Evol Biol 34:968–976
Yamaguchi K, Koyanagi M, Sato K, Terakita A, Kuraku S (2023a) Whale shark rhodopsin adapted to deep-sea lifestyle by a substitution associated with human disease. Proc Natl Acad Sci U S A 120:e2220728120
Yamaguchi K, Uno Y, Kadota M, Nishimura O, Murakumo K, Nozu R, Matsumoto R, Sato K, Kuraku S (2023b) Elasmobranch genome sequencing reveals evolutionary trends of vertebrate karyotypic organization. Genome Res 33:1527–1540
Yoshida K, Kitano J (2021) Tempo and mode in karyotype evolution revealed by a probabilistic model incorporating both chromosome number and morphology. PLoS Genet 17:e1009502
Yu D, Ren Y, Uesaka M, Beavan AJS, Muffato M, Shen J, Li Y, Sato I, Wan W, Clark JW, Keating JN, Carlisle EM, Dearden RP, Giles S, Randle E, Sansom RS, Feuda R, Fleming JF, Sugahara F, Cummins C, Patricio M, Akanni W, D’Aniello S, Bertolucci C, Irie N, Alev C, Sheng G, de Mendoza A, Maeso I, Irimia M, Fromm B, Peterson KJ, Das S, Hirano M, Rast JP, Cooper MD, Paps J, Pisani D, Kuratani S, Martin FJ, Wang W, Donoghue PCJ, Zhang YE, Pascual-Anaya J (2023) Hagfish genome illuminates vertebrate whole genome duplications and their evolutionary consequences. bioRxiv. https://doi.org/10.1101/2023.04.08.536076
Zhang J, Wagh P, Guay D, Sanchez-Pulido L, Padhi BK, Korzh V, Andrade-Navarro MA, Akimenko M-A (2010) Loss of fish actinotrichia proteins and the fin-to-limb transition. Nature 466:234–237
Acknowledgements
We thank Jun Kitano, Kazuaki Yamaguchi, Susumu Hyodo, Keiichi Sato, Yasuhisa Kobayashi, Taiki Niwa, and Akane Kawaguchi for the insightful discussion. This study was funded by intramural budgets granted by the National Institute of Genetics, as well as JSPS KAKENHI Grant number 20H03269.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethics approval
Not applicable.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
This article is published under an open access license. Please check the 'Copyright Information' section either on this page or in the PDF for details of this license and what re-use is permitted. If your intended use exceeds what is permitted by the license or if you are unable to locate the licence and re-use information, please contact the Rights and Permissions team.
About this article
Cite this article
Kuraku, S., Sato, M., Yoshida, K. et al. Genomic reconsideration of fish non-monophyly: why cannot we simply call them all ‘fish’?. Ichthyol Res 71, 1–12 (2024). https://doi.org/10.1007/s10228-023-00939-9
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10228-023-00939-9