Abstract
The number of fishes exceeds that of all other vertebrates both in terms of species numbers and in their morphological and phylogenetic diversity. They are an ecologically and economically important group and play an essential role as a resource for humans. This makes the genomic exploration of fishes an important area of research, both from an applied and a basic research perspective. Fish genomes can vary greatly in complexity, which is partially due to differences in size and content of repetitive DNA, a history of genome duplication events and because fishes may be polyploid, all of which complicate the assembly and analysis of genome sequences. However, the advent of modern sequencing techniques now facilitates access to genomic data that permit genome-wide exploration of genetic information even for previously unexplored species. The development of genomic resources for fishes is spearheaded by model organisms that have been subject to genetic analysis and genome sequencing projects for a long time. These offer a great potential for the exploration of new species through the transfer of genomic information in comparative analyses. A growing number of genome sequencing projects and the increasing availability of tools to assemble and access genomic information now move boundaries between model and nonmodel species and promises progress in many interesting but unexplored species that remain to be studied.
You have full access to this open access chapter, Download protocol PDF
Similar content being viewed by others
Key words
1 Diversity of Fishes
Fishes are the most diverse group of vertebrates on earth [1]. As of 2017, a total of 33,554 species of fishes have been described [2], and many more remain to be discovered. They have colonized marine and freshwater habitats alike and display tremendous anatomical and ecological diversity. The term fishes (Pisces) includes the most basal jawless fishes (lampreys and hagfishes) that live as parasites or scavengers and the lobe-finned fishes (lungfish and coelacanths) that gave rise to the tetrapods. The exclusion of the latter renders fishes as a whole a paraphyletic group. The ray-finned fishes comprise the basal bichirs (Polypteridae) and sturgeons (Ascipenderidae) as well as the holostei [Bowfins (Amiidae) and gars (Lepisosteidae)]. The most speciose group by far is the Teleosts that have undergone a spectacular diversification since the Cretaceous period [3]. They are prevalent in many aquatic ecosystems and are of manifold importance to man. Fishes are found in the deepest ocean trenches and up to 5200 m elevation in the Himalayas [2]. They have colonized rivers, lakes, and oceans but also extreme habitats like caves where they live in constant darkness, the Arctic, or desert springs with high temperature and salt conditions. Some killifishes even survive dry periods by laying drought-resistant eggs, and these fishes are also among the most short-lived ones [4]. The oldest fishes to date may be Greenland sharks that have been estimated to be close to 400 years old [5]. The south-east Asian Paedocypris with a standard length of 7.9 mm possibly represents the world’s smallest vertebrate [6] while the largest nonmammalian vertebrate is given by the whale shark that may reach a length of up to 13 m. The diversity of reproductive modes in fishes includes egg laying and oviposition or the birth of fully developed young. Egg laying fishes have evolved numerous modes of brood care including mouthbrooding, substrate brooding, nest building, pelvic fin brooding, or ventral pouch brooding whereby the parental care may be performed through the father, the mother or both parents. Eggs of fishes may be released into the pelagial zone in mass spawning events and left to themselves, deposited into caves, mussels, gravel rudds or nests out of plant matter or air bubbles [7]. All fishes have a direct development, but may go through extended and distinct larval periods including the blind and worm-like ammocoetes larvae of lampreys or the marine leptocephalus larvae of eels and tarpon as opposed to the fully developed offspring of fishes that give birth. The feeding types of fishes are equally diverse. While some feed on microscopic algae (Silver carp, Hypophthalmichthys molitrix), scrape algae of surfaces (Nase, Chondrostoma nasus) or feed on higher plants (grass carp, Ctenoparyngodon idella) the majority of fishes feed on animals. Again, there is a range from plankton feeders (herring, Clupea harengus), to piscivorous fishes (pike, Esox lucius) and top predators like sharks that may prey upon marine mammals. There a numerous highly specialized feeding strategies in which fishes specialize on detritus, decaying wood, snails, or mussels. They eat scales, skin, eyes or parasites of other fishes or specialize on crabs, shrimp, insects, coral, fruit, sponges, and many other food items that are taken on occasion. The exploitation of these different food sources is typically facilitated by evolutionary accommodation of the feeding apparatus, which constitutes a key element that has determined the impressive adaptive radiation of fishes. Another factor that has contributed to the diversity of fishes is the many means by which they use their body or fins to move. Fishes can swim, whereby different species use fins very differently to propel themselves and for fine maneuvering. Some are constant swimmers whereas others are sit-and-wait predators or almost sessile in very confined spaces where they tend to camouflage. Some eel-shaped species, puffer fish or flatfishes are able to bury themselves into different substrates. Different fishes can use their modified mouth or fins to cling to hard substrates, enabling them to persist in strong currents or to climb steep waterfalls. Finally, mudskippers have even colonized the intertidal zone above the water level where they can move rapidly using body movements and modified fins. Fishes use the same senses to acquire signals from their environment like humans, including vision with highly developed eyes that enable them to see color, and sometimes ultraviolet or polarized light. They can hear sounds with the help of the Weberian apparatus and they have a sensory system equipped to feel pain. The smell or taste of fishes is well developed within the nose, but also through taste buds that are distributed across their body. Beyond these, fishes can detect currents or waves underwater through their lateral line and head canal system, and some are able to detect electrical fields of prey items or the earth’s magnetic field for orientation. Although fishes are mostly harmless to humans, there are species that are highly toxic or venomous, and that are of medical importance. Tissues of the Japanese pufferfish (Takifugu sp.) contain tetrodotoxin that can kill humans if consumed and tropical marine predators like moray eels or barracuda may accumulate toxins that originate from dinoflagellate blooms. Finally, there are also venomous species, such as the stonefish (Synanceia verrucosa) or the related lionfish (Pterois volitans) that possess venom and inflict painful and life-threatening injuries when touched or unintentionally stepped on.
The diversity of fishes has permitted them to exploit niches in aquatic ecosystems in many specialized ways and to become dominant components in food webs. Fishes represent top predators that convert energy from lower trophic levels to biomass that is harvested by humans and other top predators. For this reason, fishes have been naturalized across the globe in hope to create prolific food resources for human consumption, including the release of carp, trout, salmon, Tilapia, Nile perch, eel, catfish, and many other species outside their native range. However, it is now clear that considerable detrimental side effects on local ecosystems are common whenever such introductions were successful. Humans also employ fishes in attempts to manipulate ecosystems as biological control agents, for example, the silver carp (Hypophthalmichthys) or grass carp (Ctenopharyngodon) to control algae, or aquatic weeds or mosquito fish (Gambusia) to control malaria vectors, again often accompanied by undesirable side effects.
Humans have long kept fishes as ornamental pets, and the history of domestication and aquaculture dates back a long time. Goldfish were already bred in China in 1000 AD [8]. Nowadays, they often are the first pets that children are acquainted with, and seed a positive image fishes have for humans. Additional species such as Koi Carp (Cyprinus carpio), Siamese fighting fish (Betta splendens), platyfish (Xiphophorus maculatus), zebrafish (Danio rerio), flowerhorn chichlids (Amphilophus hybrids), and many wild ornamental species are kept as pets. Other marine and freshwater including tuna, flatfishes, sea bass, seabream or freshwater fishes like trout, salmon, carp, Tilapia, catfishes, and sturgeon are targets of intensive aquaculture to meet the growing demands for food. Likewise, wild populations of fishes are managed and exploited as the most important food resource from aquatic environments. Together, aquaculture and fisheries provide food, income, and livelihoods of hundreds of millions of people and the world per capita fish supply reached a record high of 20 kg in 2014 [9]. Given the growing world population and the limited availability of space for agriculture, fish will play a central role in providing future generations with adequate nutrition. They not only play a direct role but are used to produce fish oil as a food complement, fish meal as food for other livestock and manure to fertilize fields. Accordingly, whole industries are built around fisheries, fish farming, and fish products. Fishes play an important socioeconomic role in recreational angling, and some can serve as flagship species to transport conservation issues into a broader public.
Due to their economic value and because of the essential role fishes play in ecosystems, they are subject to management, conservation efforts, and scientific studies. Fishes are targets of applied research that aims at improving harvests, but also out of broader interest in fish biology or because fishes can serve as model vertebrates in studies that aim at obtaining results of direct relevance to humans in fundamental medical research or ecotoxicology. Fishes are prime models in evolutionary studies. It is this prevalence of fishes and the diverse ways in which they are exploited by humans that makes them targets for genomic exploration.
2 The Genomic Makeup of Fishes
Compared to other vertebrates, fishes seem to have more plastic and variable genomes, which is associated with the fact that they display frequent polyploidization, have high speciation rates and carry a diversity of repetitive genetic elements [10, 11]. The majority of fishes that are intensively studied have relatively compact genomes, but fish genomes may vary in size between 0.35 and 133 Gb [12]. Among these, the teleosts have the most compact genomes ranging from 0.35 to 10 Gb, followed by Chondrichthyes (1.5–17.5 Gb) and finally the lobiform Dipnoi (80–132 Gb) [13]. The Japanese pufferfish Takifugu rubripes was targeted in one of the first fish genome sequencing projects because of its compact genome size of 0.39 Gb, which still marks the lower end of the spectrum of vertebrate genome sizes. The three-spined stickleback (Gasterosteus aculeatus) genome has a size of 0.46 Gb, the one of the Zebrafish Danio rerio has 1.67 Gb and the Japanese Medaka 0.70 Gb. The genome of a basal “fishes” such as the sea lamprey Petromyzon marinus has a size of 0.65 Gb and the sarcopterygian Latimeria chalumnae has a genome size of 2.86 Gb that is only a little bit smaller than our own. The largest fish genome can be found in the marbled lungfish Protopterus aethiopicus (133 Gb), which represents the largest genome known from any metazoan. Fish genome size and diversity are affected by their variable content of repetitive DNA elements [14] that make a more important relative contribution in fish genomes than in mammals [12, 15, 16]. Besides affecting genome size and structure, repetitive genetic elements have been found to be involved in functional genetic divergence among fishes, such as the rapid evolution of new sex-determining loci or the emergence of barriers to reproduction that reduce the viability of hybrid offspring [11, 17, 18]. The diversity of repetitive genetic elements in fishes exceeds that in higher vertebrates and the relative contribution of repetitive genetic elements may vary from 6% in Tetraodon to 55% in Danio. The distribution of TE families across the phylogeny demonstrates that their presence and abundance may be highly lineage-specific, and that periods of TE diversification occur independently among different lineages of fishes. The Sarcopterygii have lost TE diversity, a trend that manifested even more in the notable reduction of TE diversity in birds and mammals [15]. While the diversity of TEs in fish genomes represents an important component of their between and within lineage genomic diversity, repetitive elements pose challenges for the assembly and thus the analysis of their genomes [19].
Genome duplication events have been postulated to represent major evolutionary events that have facilitated the extraordinary diversification of fishes [20]. There is evidence that rounds of genome duplications have occurred in the stem lineage of the vertebrates, and that an additional round of tetraploidization followed by rediploidization has occurred early in the evolution of the ray-finned fishes (Actinopterygii). This process has generated redundant gene copies that may have vanished but also taken up new functions [11, 21, 22]. The evolution of multigene families such as the Hox cluster has been explained through ancient gene duplications and adds another level of complexity to the genetic makeup of fishes [23]. Species of fishes that deviate notably toward larger genome sizes include a range of species that have undergone lineage-specific and more recent genome duplications. Examples include the ancient tetraploid Salmonidae (trout, salmon, whitefishes), but also taxa like the sturgeons (Acipenseridae) where ploidy ranges from diploid to octaploids and that may carry several hundred chromosomes [24]. Although the genome size increases, it is common that redundant gene copies are lost in a process of rediploidization that occurs even after multiple rounds of polyploidization [25, 26]. However, it is also possible that copies of duplicated genes diverge after polyploidization to acquire new functions. Paralogy relationships within genomes can still be tracked as genomes rediploidize, as in the Atlantic salmon genome [27].
While many genome duplication events may be quite ancient [24], there is a range of polyploid species of more recent origin. The range of modes of reproduction of these fishes often leads to patterns of inheritance that deviate from a classical Mendelian pattern. While this comprises interesting phenomena in itself, it poses challenges for the exploration of their genome content as a single organism may contain more than two alleles of a given gene and because divergence between gene copies will reach levels that would otherwise be encountered in separated species. These issues are typically not considered in default parameters of data analyses tools, which can introduce massive bias in attempts to identify orthologous and paralogous sequences and in all studies on genetic variation. Examples include the Eurasian diploid–polyploid species complexes of Cobitis loaches in which polyploid hybrids can carry sets of chromosomes originating from parental species that do not co-occur with the hybrid lineages any more [28, 29]. Comparable examples belong to the cypriniform fishes such as the Iberian cyprinid Squalius alburnoides or the North American Minnow Phoxinus eos-neogaeus. All of these taxa of recent polyploidy origin are allotetraploid, that is, they have arisen as hybrids between two divergent lineages that can apparently only continue to exist when species-specific sets of chromosomes are inherited as a whole. They may use different reproductive modes to pass their genetic material on to the offspring including normal meiosis, asexual reproduction through gynogenesis, where male sperm initiates development but genetic material is excluded, and hybridogenesis [30,31,32,33]. An example from central America and the first vertebrate in which unisexuality was discovered [34] includes the Amazon molly, a species of hybrid origin, in which gynogenetic females mate with males of another species to initiate development but exclude the male genetic material from the developing zygote [35, 36]. Experimental studies [37] suggest that fishes are flexible and actively choose their mode of diploid – polyploid reproduction depending on the genotype of the parents, which explains the diversity and success of such lineages in nature. There is one species of fish, the North- and Central-American mangrove killifish Kryptolebias marmoratus that exhibits true hermaphroditism and must have existed as a self-fertilizing lineage for a long time [38, 39].
Although Teleostei are at the base of the evolution of the vertebrates, the explosive diversification that has resulted in most of today’s diversity of ray-finned fishes (Actinopterygii) has taken place between the late Mesozoic and early Cenozoic [40]. Gene sequences and the order of genes within the genome (synteny) have been conserved. This now permits a transfer of positional genomic information between fully sequenced genomes of model organisms and the wealth of emerging model systems [41, 42]. Conservation of synteny can be visualized by means of oxford grids (Fig. 1) or more elaborate circle graphs (see Fig. 2 in [27]) all of which illustrate which regions of the genome contain homologous sequences that are arranged in the same order.
Oxford grids exploring synteny relationships between Cottus ssp. (European sculpin) linkage groups (x-axis) and chromosomes of model organisms such as the Stickleback (Gasterosteus aculeatus) or the zebrafish (Danio rerio) (y-axis). For this purpose, genomic fragments for which the position in Cottus was genetically mapped were BLAST searched against fully sequenced genomes of the other species. Numbers in fat squares indicate shared markers among known chromosomes and Cottus linkage groups. The synteny of Cottus and the stickleback is well conserved while the zebrafish genome is more divergent both in terms of chromosome organization and in the low number of markers that could be mapped overall (n given at bottom right of each graph). This highlights the utility of the stickleback genome as a genomic reference for the exploration of Cottus. (Modified from [42])
The exploration of fishes like alpine char (Salvelinus umbla, top) or grayling (Thymallus thymallus, middle) is facilitated through advances in genomics in the closely related Atlantic salmon (Salmo salar). The former can be referred to as satellite species of the latter as a transfer of genomic information is very promising. This in turn supports studies on their own biology in manifold ways. Less well-known fishes like loaches (Cobitis spp., bottom) have been difficult to study because of their hybrid origin and polyploid genomes. Long read sequencing and continuing development of approaches to assemble genomes now enable better access to such fascinating systems in fundamental research. (All pictures A. Hartl)
Such inference can infer homology among chromosomes, chromosome fissions and fusion and remnants of duplicated chromosomes. A related issue is that conserved synteny can support inference about homology when gene annotations are transferred between species. Finally, knowledge about syntenic relationships between two genomes lets one predict which genetic elements can be found near a given marker, even if that part of the genome of one of the species is not fully sequenced or assembled. Together with the rapidly growing number of fully sequences fish genomes that sample the fish phylogeny more and more densely, these inferences contribute greatly to the exploration of as yet unexplored species [23]. Even when genomes are not fully sequenced, the conservation of synteny can be exploited to validate newly generated genetic maps [43] or to explore the most likely gene content of QTL regions that have not been fully sequenced in the target species [42, 44]. The number of fully sequenced and annotated fish genomes that are made available through databases such as Ensembl [45] is currently rapidly growing due to the development of sophisticated assembly strategies and the rise of long read sequencing that spans genomic fragments that are difficult to assemble.
3 Genomics in Studies on the Biology of Fishes
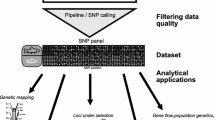
Fishes are studied genetically to infer basic biological and evolutionary processes. The details of such studies have often relied on population genetic approaches in which descriptors of population structure were inferred. This included studies on the distribution of lineages across their ranges [46] and the outcome of secondary contact when such lineages hybridize [47]. Studies often aimed to infer population structure from an evolutionary perspective [48] but also with the goal to improve stock management [49]. While such studies have been extremely successful in identifying units of biodiversity and evolutionary patterns, they have often relied on information from anonymous, neutrally evolving genetic markers and they applied the neutral evolutionary theory. However, there is a deep interest in identifying the loci that drive evolutionary processes and that determine the phenotypic properties of organisms. The latter aspects require that additional information be integrated with population genetic patterns observed at a given marker. First, anonymous markers need to be assigned to genome positions to infer whether they are associated with genetic elements and their functions. Moreover, a dense sampling of markers ordered along the genome permits powerful statistical analyses as patterns of individual markers can be combined in sliding window analyses that test for shared signals. Such data is useful to detect genetic signatures of selection that are expected when adaptive evolutionary change takes place. Moreover, genetic elements themselves have to be cataloged and functionally studied to understand their molecular functions and the higher-level phenotypes they affect. While such analyses have been conducted in the field of developmental biology and quantitative genetics the data that becomes available now permits the inference of populations genetic differentiation in conjunction with likely causative genetic variants in genome-wide studies of the association of phenotypes with genetic variation. A hallmark example in fishes is given by [50] who have studied genome-wide genetic variation in sticklebacks to identify genetic loci involved in the phenotypic and ecological diversity. Other intensively studied groups of fishes have been subject to intense genomic exploration as well as shedding light on study systems that have intrigued biologists for a long time [51]. Population genetics in fishes will doubtlessly move forward toward integrative analyses [52] that rely heavily on the interpretation of evolutionary or ecological patterns in nature in the light of detailed genomic and functional genetic information.
Fish genomics is driven by the progress that has been made in intensively studied species such as the zebrafish (Danio rerio) or medaka (Oryzias latipes). These species have long been favorite ornamental fishes and make excellent laboratory animals because of their short generation times and the ease of their care and breeding. Fundamental biological processes were uncovered in these fishes and could then be explored, generalized and extended to other species. The transfer of knowledge from model organisms has already decidedly influenced areas of applied research such as medical studies, ecotoxicology, environmental sensing systems, and sustainable aquaculture strategies [53]. The integration of knowledge from model organisms with the so-called satellite species that are related closely enough to facilitate the transfer of genomic information paves the way to study ecologically relevant taxa, and more broadly the evolution and diversity of all known fishes and other species [54]. The applicability of this approach will tremendously increase as the progress in next-generation sequencing fully includes nonmodel organisms and more and more fish genomes and biological knowledge about different species accumulates. The wave of next-generation sequencing has turned fishes into a highly informative group, a “new model army,” in which long-standing questions on the evolution of their biodiversity can be addressed [23] (Fig. 2).
The zebrafish and the medaka were the first fish model systems that were intensively studied genetically and for which methods to conduct mutagenesis screens were established [55, 56]. Studies on these fishes were at the forefront of developmental biology, biomedical, and genomic studies. Moreover, they complemented each other in that they differ notably in their phylogenetic position and properties, which provided insights into the possibilities and power of comparative analyses between these models [57]. Since then, a large community of researchers has exploited the zebrafish system to pioneer many fields of fish genetics. Sophisticated methods that have been first developed in model systems [58] are now becoming applicable in other species. The wealth of knowledge that is available for the zebrafish has been collected in the form of a dedicated book [59] and is accessible online in the ZFIN database [60]. Likewise, there are comprehensive books [61, 62] and a website [63] for the medaka. Beyond the fully sequenced and annotated genomes, these resources (1) provide information on the laboratory use (protocols) of zebrafish and medaka, (2) summarize information on known mutants and transgenic strains as well as wild type strains, (3) provide access to genetic, genomic and developmental information, (4) aid in the transfer of information between species and databases, and (5) facilitate the use of fishes as a model for human focused medical research. Finally, they (6) serve as a general platform for researchers, and a collection of husbandry and laboratory protocols are provided. Only a few other model organisms parallel this rich set of genomic resources and genomic information is increasingly added to and curated in public open access databases [45].
A growing number of additional fish genomes have been fully sequenced and extend the genomic exploration of fishes. Each was initially planned with a different biological emphasis and each has distinct advantages related to the biology of the species or its use from a human perspective. Disadvantages relative to zebrafish and medaka vary and may be relatively longer generation times, more demanding husbandry and difficulties in breeding them. Pufferfish genomes such as the ones from Takifugu rubipes and Tetraodon nigroviridis were initially targeted because of their compact genome sizes [64, 65]. These studies revealed that Pufferfish, nonetheless, carries a number of genes that is comparable to the human genome and targeted a species that are of fundamental biology questions and economic importance. Other species were targeted with a more focused view on phenomena that are of medical importance. The genome of the African Killifish Nothobranchius furzeri was sequenced to gain access to a species that served as a model to study senescence [66]. This species is extremely short lived and can thus serve to genetically map traits related to ageing in relatively short experimental timescales. A species that has received interest from the field of developmental biology is the Mexican cave tetra Astyanax mexicanus [67] that lives in subterranean caves and is distinguished from its surface dwelling relatives by a number of reduced traits such as the loss of vision but also by the gain of other sensory abilities. Its genome has enabled mapping of the genetic basis of these traits and added great detail to our understanding of the genetic changes that cause phenotypic evolution. The platyfish (Xiphophorus maculatus) was sequenced as a model to study the development of skin melanoma, and to study the genetics of live-bearing and sex determination [68]. A growing number of fishes is targeted for their economic importance and with the goal to study genomic resources that may be relevant to improve aquaculture. However, studies on Atlantic salmon [27], turbot [69], the European sea bass [70], and tilapia [71] illustrate that their genomes also gave rich insights into questions related to environmental adaptation, development, and genome evolution. Other fish species have been specifically targeted with the aim to develop model systems to study evolutionary processes that have given rise to the diversity of fishes. As a prime example, the stickleback has been dubbed a supermodel that is amenable for the full integration of behavioral, developmental, ecological, and genetic data [72]. Its genome has been sequenced and served in hallmark studies in the field of ecological genomics that illustrated the power of genomics to unravel evolutionary processes and to link genotype with phenotype information [50, 73]. These studies, among many others, have carried a species that has received long-standing interest of researchers as a model in behavioral studies into the genomic era. Likewise, cichlids have been favorite study systems to understand the explosive diversification that must have occurred in the east African lakes of the rift valley where hundredth of species have evolved within each of the separated lakes. These systems have received interest to study the process of speciation, functional morphology of the feeding apparatus, and color polymorphism and its role in mate choice. Progress in these fields as well as in aspects of the molecular evolution of this group of fishes has been greatly facilitated by several cichlid genomes [51].
Clearly, the previous trend to sequence genomes only for particularly well-studied species for which a wealth of information is available will not be the only path for future research. Numerous genome sequencing projects that are not mentioned here and their number is growing exponentially. The resulting sequences and the tools to assemble and access the information make genome sequencing project feasible for more and more species that are interesting for a smaller community or single researchers. This trend clearly moves boundaries between model species and nonmodel species and promises progress in many exceptional species that remain to be studied.
References
Helfman G, Collette BB, Facey DE, Bowen BW (2009) The diversity of fishes: biology, evolution, and ecology, 2nd edn. Wiley-Blackwell, Hoboken. 736 pages, ISBN: 978-1-405-12494-2
Froese R, Pauly D (2018) FishBase (version Jun 2017). In: Roskov Y, Abucay L, Orrell T, Nicolson D, Bailly N, Kirk PM, Bourgoin T, DeWalt RE, Decock W, De Wever A, van Nieukerken E, Zarucchi J, Penev L (eds) Species 2000 & ITIS catalogue of life. Species 2000, Naturalis, Leiden. ISSN 2405-8858. , 30th January 2018. Digital resource at www.catalogueoflife.org/col.
Betancur R, Broughton RE, Wiley EO et al (2013) The tree of life and a new classification of bony fishes. PLOS Curr Tree Life. [last modified: 2013 Apr 23]
Valenzano DR, Benayoun BA, Singh PP et al (2015) The African turquoise killifish genome provides insights into evolution and genetic architecture of lifespan. Cell 163(6):1539–1554
Nielsen J, Hedeholm RB, Heinemeier J, Bushnell PG, Christiansen JS, Olsen J, Ramsey CB, Brill RW, Simon M, Steffensen KF, Steffensen JF (2016) Eye lens radiocarbon reveals centuries of longevity in the Greenland shark (Somniosus microcephalus). Science 353(6300):702–704
Kottelat M, Britz R, Tan HH, Witte KE (2005) Paedocypris, a new genus of Southeast Asian cyprinid fish with a remarkable sexual dimorphism, comprises the world's smallest vertebrate. Proc R Soc B 273:895–899
Breder DM, Rosen DE (1966) Modes of reproduction in fishes, American Museum of Natural History. Natural History Press, Garden City, NY. 941 pp
Komiyama T, Kobayashi H, Tateno Y, Inoko H, Gojobori T, Ikeo K (2009) An evolutionary origin and selection process of goldfish. Gene 430(1–2):5–11
FAO (2016) The State of World Fisheries and Aquaculture 2016. Contributing to food security and nutrition for all. FAO, Rome. 200 pp. isbn:978-92-5-109185-2
Venkatesh B (2003) Evolution and diversity of fish genomes. Curr Opin Genet Dev 13:588–592
Volff JN (2005) Genome evolution and biodiversity in teleost fish. Heredity 94:280–294
Kapusta A, Suh A, Feschotte C (2017) Dynamics of genome size evolution in birds and mammals. Proc Natl Acad Sci U S A 114:E1460–E1469
Gregory, T.R. (2005). Animal genome size database. http://www.genomesize.com
Shao F, Wang J, Xu H, Peng Z (2018) FishTEDB: a collective database of transposable elements identified in the complete genomes of fish. Database 2018
Chalopin D, Naville M, Plard F, Galiana D, Volff JN (2015) Comparative analysis of transposable elements highlights mobilome diversity and evolution in vertebrates. Genome Biol Evol 7:567–580
Gao B et al (2016) The contribution of transposable elements to size variations between four teleost genomes. Mob DNA 7:4
Cioffi MB, Bertollo LAC (2012) Chromosomal distribution and evolution of repetitive DNAs in fish. In: Garrido-Ramos MA (ed) Repetitive DNA, vol 7. Genome Dyn, Basel, Karger, pp 197–221
Schartl M et al (2018) Sox5 is involved in germ-cell regulation and sex determination in medaka following co-option of nested transposable elements. BMC Biol 16:16
Sotero-Caio CG, Platt RN, Suh A, Ray DA (2017) Evolution and diversity of transposable elements in vertebrate genomes. Genome Biol Evol 9:161–177
Taylor JS, Van de Peer Y, Braasch I, Meyer A (2001) Comparative genomics provides evidence for an ancient genome duplication event in fish. Philos Trans R Soc Lond Ser B Biol Sci 356:1661–1679
Jaillon O et al (2004) Genome duplication in the teleost fish Tetraodon nigroviridis reveals the early vertebrate proto-karyotype. Nature 431:946–957
Meyer A, Van de Peer Y (2005) From 2R to 3R: evidence for a fish-specific genome duplication (FSGD). BioEssays 27:937–945
Braasch I, Peterson SM, Desvignes T, McCluskey BM, Batzel P, Postlethwait JH (2015) A new model army: emerging fish models to study the genomics of vertebrate Evo-Devo. J Exp Zool Mol Dev Evol 324:316–341
Leggatt RA, Iwama GK (2003) Occurrence of polyploidy in the fishes. Rev Fish Biol Fish 13:237–246
Ludwig A, Belfiore NM, Pitra C, Svirsky V, Jenneckens I (2001) Genome duplication events and functional reduction of ploidy levels in sturgeon (Acipenser, Huso and Scaphirhynchus). Genetics 158:1203–1215
Rajkov J, Zhaojun S, Berrebi P (2014) Evolution of polyploidy and functional diploidization in sturgeons: microsatellite analysis in 10 sturgeon species. J Hered 105:521–531. https://doi.org/10.1093/jhered/esu027
Lien S, Ben F, Koop BF et al (2016) The Atlantic salmon genome provides insights into rediploidization. Nature 533:200–205
Saitoh K, Chen W-J, Mayden RL (2010) Extensive hybridization and tetrapolyploidy in spined loach fish. Mol Phylogenet Evol 56:1001–1010
Vasil’ev VP, Lebedeva EB, Vasil’eva ED (2011) Evolutionary ecology of clonal-bisexual complexes in spined loaches from genus Cobitis (Pisces, Cobitidae). J Ichthyol 51:932–940
Bohlen J, Ritterbusch D (2000) Which factors affect sex ratio of spined loach (genus Cobitis) in lake Müggelsee? Environ Biol Fish 59:347–352
Goddard KA, Megwinoff O, Wessner LL, Giaimo F (1998) Confirmation of gynogenesis in Phoxinus eos-neogaeus (Pisces: Cyprinidae). J Hered 89:151–157
Collares-Pereira MJ, Coelho MM (2010) Reconfirming the hybrid origin and generic status of the Iberian cyprinid complex Squalius alburnoides. J Fish Biol 76(3):707–715. https://doi.org/10.1111/j.1095-8649.2009.02460.x
Cunha C et al (2011) The evolutionary history of the allopolyploid Squalius alburnoides (Cyprinidae) complex in the northern Iberian Peninsula. Heredity 106:100–112. https://doi.org/10.1038/hdy.2010.7
Hubbs CL, Hubbs LC (1932) Apparent parthenogenesis in nature, in a form of fish of hybrid origin. Science 76(1983):628–630
Schlupp I (2005) The evolutionary ecology of gynogenesis. Annu Rev Ecol Evol Syst 36:399–417
Lampert KP, Schartl M (2008) The origin and evolution of a unisexual hybrid: Poecilia formosa. Philos T R Soc B 363:2901–2909
Zhang J, Sun M, Zhou L, Li Z, Liu Z, Li XY, Liu XL, Wei Liu W, Gui JF (2015) Meiosis completion and various sperm responses lead to unisexual and sexual reproduction modes in one clone of polyploid Carassius gibelio. Sci Report 5:10898
Harrington RW (1961) Oviparous hermaphroditic fish with internal self-fertilization. Science 134:1749–1750
Tatarenkov A, Lima SMQ, Taylor DS, Avise JC (2009) Long-term retention of self-fertilization in a fish clade. PNAS 106(34):14456–14459
Near TJ, Eytan RI, Dornburg A, Kuhn KL, Moore JA, Davis MP, Wainwright PC, Friedman M, Smith WL (2012) Resolution of ray-finned fish phylogeny and timing of diversification. PNAS 109:13698–13703
Sarropoulou E, Fernandes JMO (2011) Comparative genomics in teleost species: knowledge transfer by linking the genomes of model and non-model fish species. Compar Biochem Physiol 6:92–102
Cheng J, Czypionka T, Nolte AW (2013) The genomics of incompatibility factors and sex determination in hybridizing species of Cottus (Pisces). Heredity 111:520–529
Rexroad CE, Palti Y, Gahr SA, Vallejo RL (2008) A second generation genetic map for rainbow trout (Oncorhynchus mykiss). BMC Genet 9:74
Cheng J, Sedlazeck F, Altmüller J, Nolte AW (2015) Ectodysplasin signalling genes and phenotypic evolution in sculpins (Cottus). Proc Royal Soc Ser B 282(1815):20150746
Zerbino DR, Achuthan P, Wasiu Akanni W et al (2018) Ensembl 2018. Nucleic Acids Res 46(D1):D754–D761
Bernatchez L, Wilson CC (1998) Comparative phylogeography of nearctic and palearctic fishes. Mol Ecol 7:431–452
Nolte AW, Freyhof J, Tautz D (2006) When invaders meet locally adapted types: rapid moulding of hybrid zones between two species of sculpins (Cottus, pisces). Mol Ecol 15:1983–1993
Barluenga M, Stölting KN, Salzburger W, Muschick M, Axel Meyer A (2006) Sympatric speciation in Nicaraguan crater lake cichlid fish. Nature 439:719–723
Bradbury IR, Hamilton LC, Sheehan TF, Chaput G, Robertson MJ, Dempson JB, Reddin D, Morris V, King T, Louis Bernatchez L (2016) Genetic mixed-stock analysis disentangles spatial and temporal variation in composition of the West Greenland Atlantic Salmon fishery. ICES J Marine Sci 73(9):2311–2321
Jones FC, Grabherr MG et al (2012) The genomic basis of adaptive evolution in threespine sticklebacks. Nature 484:55–61
Brawand D, Wagner CE et al (2014) The genomic substrate for adaptive radiation in African cichlid fish. Nature 513:375–381
Bernatchez L, Renaut S, Whiteley AR, Campbell D, Derome N, Jeukens J, Landry L, Lu G, Nolte AW, Østbye K, Rogers SM, St-Cyr J (2010) On the origins of species: insights from the ecological genomics of whitefish. Philos Trans R Soc Lond 365:1783–1800
Spaink HP, Jansen HJ, Dirks RP (2013) Advances in genomics of bony fish. Brief Funct Genomics 13:144–156
Tagu D, Colbourne JK, Nègre N (2014) Genomic data integration for ecological and evolutionary traits in non-model organisms. BMC Genomics 15(1):490
Shima A, Shimada A (1991) Development of a possible nonmammalian test system for radiation-induced germ-cell mutagenesis using a fish, the Japanese medaka (Oryzias latipes). Proc Natl Acad Sci U S A 88:2545–2549
Mullins MC, Hammerschmidt M, Haffter P, Nüsslein-Volhard C (1994) Large-scale mutagenesis in the zebrafish: in search of genes controlling development in a vertebrate. Curr Biol 4(3):189–202
Wittbrodt J, Shima A, Schartl M (2002) Medaka—a model organism from the far east. Nat Rev Genet 3(1):53–64
Kirchmaier S, Naruse K, Wittbrodt J, Felix Loosli F (2015) The genomic and genetic toolbox of the teleost medaka (Oryzias latipes). Genetics 199(4):905–918
Westerfield M (2007) The zebrafish book, 5th ed; a guide for the laboratory use of zebrafish (Danio rerio). University of Oregon Press, Eugene
Howe DG, Bradford YM, Conlin T, Eagle AE, Fashena D, Frazer K, Knight J, Mani P, Martin R, Moxon SA, Paddock H, Pich C, Ramachandran S, Ruef BJ, Ruzicka L, Schaper K, Shao X, Singer A, Sprunger B, Van Slyke CE, Westerfield M (2013) ZFIN, the Zebrafish Model Organism Database: increased support for mutants and transgenics. Nucleic Acids Res 41(Database issue):D854–D860
Kinoshita M, Murata K, Naruse K, Tanaka M (2012) Medaka: biology, management, and experimental protocols. Wiley, New York. 9780813808710
Naruse K, Tanaka M, Takeda H (eds) (2011) Medaka: a model for organogenesis, human disease, and evolution. Springer, New York. ISBN 978-4-431-92690-0
NBRP Medaka. https://shigen.nig.ac.jp/medaka/
Aparicio S, Chapman J, Stupka E et al (2002) Whole-genome shotgun assembly and analysis of the genome of Fugu rubripes. Science 297:1301–1310
Jaillon O, Aury JM et al (2004) Genome duplication in the teleost fish Tetraodon nigroviridis reveals the early vertebrate proto-karyotype. Nature 431:946–957
Valenzano DR, Benayoun BA et al (2015) The African turquoise killifish genome provides insights into evolution and genetic architecture of lifespan. Cell 163:1539–1554
McGaugh SE, Gross JB et al (2014) The cavefish genome reveals candidate genes for eye loss. Nat Commun 5:5307
Schartl M, Walter RB, Shen Y et al (2013) The genome of the platyfish, Xiphophorus maculatus, provides insights into evolutionary adaptation and several complex traits. Nat Genet 45:567–572
Figueras A, Robledo D et al (2016) Whole genome sequencing of turbot (Scophthalmus maximus; Pleuronectiformes): a fish adapted to demersal life. DNA Res 23:181–192
Tine M, Kuhl H et al (2014) European sea bass genome and its variation provide insights into adaptation to euryhalinity and speciation. Nat Commun 5:5770
Conte MA, Gammerdinger WJ et al (2017) A high quality assembly of the Nile Tilapia (Oreochromis niloticus) genome reveals the structure of two sex determination regions. BMC Genomics 18:341
Gibson G (2005) The synthesis and evolution of a super-model. Science 307:1890–1891
Colosimo PF, Hosemann KE et al (2005) Widespread parallel evolution in sticklebacks by repeated fixation of Ectodysplasin alleles. Science 307(5717):1928–1933
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Open Access This chapter is licensed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license and indicate if changes were made.
The images or other third party material in this chapter are included in the chapter's Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the chapter's Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder.
Copyright information
© 2020 The Author(s)
About this protocol
Cite this protocol
Nolte, A.W. (2020). Genomic Access to the Diversity of Fishes. In: Dutheil, J.Y. (eds) Statistical Population Genomics. Methods in Molecular Biology, vol 2090. Humana, New York, NY. https://doi.org/10.1007/978-1-0716-0199-0_16
Download citation
DOI: https://doi.org/10.1007/978-1-0716-0199-0_16
Published:
Publisher Name: Humana, New York, NY
Print ISBN: 978-1-0716-0198-3
Online ISBN: 978-1-0716-0199-0
eBook Packages: Springer Protocols