Abstract
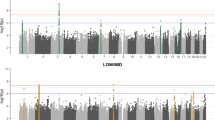
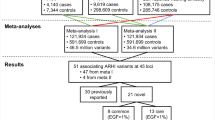
Age-related hearing loss (AHL) is characterized by a symmetric sensorineural hearing loss primarily in high frequencies and individuals have different levels of susceptibility to AHL. Heritability studies have shown that the sources of this variance are both genetic and environmental, with approximately half of the variance attributable to hereditary factors as reported by Huag and Tang (Eur Arch Otorhinolaryngol 267(8):1179–1191, 2010). Only a limited number of large-scale association studies for AHL have been undertaken in humans, to date. An alternate and complementary approach to these human studies is through the use of mouse models. Advantages of mouse models include that the environment can be more carefully controlled, measurements can be replicated in genetically identical animals, and the proportion of the variability explained by genetic variation is increased. Complex traits in mouse strains have been shown to have higher heritability and genetic loci often have stronger effects on the trait compared to humans. Motivated by these advantages, we have performed the first genome-wide association study of its kind in the mouse by combining several data sets in a meta-analysis to identify loci associated with age-related hearing loss. We identified five genome-wide significant loci (<10−6). One of these loci confirmed a previously identified locus (ahl8) on distal chromosome 11 and greatly narrowed the candidate region. Specifically, the most significant associated SNP is located 450 kb upstream of Fscn2. These data confirm the utility of this approach and provide new high-resolution mapping information about variation within the mouse genome associated with hearing loss.












Similar content being viewed by others
References
Altshuler D, Daly MJ, Lander ES (2008) Genetic mapping in human disease. Science 322(5903):881–888
Anttila V, Winsvold BS, Gormley P, Kurth T, Bettella F, McMahon G et al (2013) Genome-wide meta-analysis identifies new susceptibility loci for migraine. Nat Genet 45(8):912–917
Bennett BJ, Farber CR, Orozco L, Kang HM, Ghazalpour A, Siemers N, Neubauer M, Neuhaus I, Yordanova R, Guan B, Truong A, Yang W-PP, He A, Kayne P, Gargalovic P, Kirchgessner T, Pan C, Castellani LW, Kostem E, Furlotte N, Drake TA, Eskin E, Lusis AJ (2010) A high-resolution association mapping panel for the dissection of complex traits in mice. Genome Res 20(2):281–290
Berndt SI, Skibola CF, Joseph V, Camp NJ, Nieters A, Wang Z et al (2013) Genome-wide association study identifies multiple risk loci for chronic lymphocytic leukemia. Nat Genet 45(8):868–876
Charizopoulou N, Lelli A, Schraders M, Ray K, Hildebrand MS, Ramesh A, Srisailapathy CRS, Oostrik J, Admiraal RJC, Neely HR, Latoche JR, Smith RJH, Northup JK, Kremer H, Holt JR, Noben-Trauth K (2011) Gipc3 mutations associated with audiogenic seizures and sensorineural hearing loss in mouse and human. Nat Commun 2:201
Churchill GA, Airey DC, Allayee H, Angel JM, Attie AD, Beatty J, Beavis WD, Belknap JK, Bennett B, Berrettini W, Bleich A, Bogue M, Broman KW, Buck KJ, Buckler E, Burmeister M, Chesler EJ, Cheverud JM, Clapcote S, Cook MN, Cox RD, Crabbe JC, Crusio WE, Darvasi A, Deschepper CF, Doerge RW, Farber CR, Forejt J, Gaile D, Garlow SJ, Geiger H, Gershenfeld H, Gordon T, Gu J, Gu W, de Haan G, Hayes NL, Heller C, Himmelbauer H, Hitzemann R, Hunter K, Hsu H-CC, Iraqi FA, Ivandic B, Jacob HJ, Jansen RC, Jepsen KJ, Johnson DK, Johnson TE, Kempermann G, Kendziorski C, Kotb M, Kooy RF, Llamas B, Lammert F, Lassalle J-MM, Lowenstein PR, Lu L, Lusis A, Manly KF, Marcucio R, Matthews D, Medrano JF, Miller DR, Mittleman G, Mock BA, Mogil JS, Montagutelli X, Morahan G, Morris DG, Mott R, Nadeau JH, Nagase H, Nowakowski RS, O’Hara BF, Osadchuk AV, Page GP, Paigen B, Paigen K, Palmer AA, Pan H-JJ, Peltonen-Palotie L, Peirce J, Pomp D, Pravenec M, Prows DR, Qi Z, Reeves RH, Roder J, Rosen GD, Schadt EE, Schalkwyk LC, Seltzer Z, Shimomura K, Shou S, Sillanpää MJ, Siracusa LD, Snoeck H-WW, Spearow JL, Svenson K, Tarantino LM, Threadgill D, Toth LA, Valdar W, de Villena FP-M, Warden C, Whatley S, Williams RW, Wiltshire T, Yi N, Zhang D, Zhang M, Zou F, Consortium CT (2004) The collaborative cross, a community resource for the genetic analysis of complex traits. Nat Genet 36(11):1133–1137
Davis RC, van Nas A, Bennett B, Orozco L, Pan C, Rau CD, Eskin E, Lusis AJ (2013) Genome-wide association mapping of blood cell traits in mice. Mamm Genome 24(3–4):105–118
DerSimonian R, Laird N (1986) Meta-analysis in clinical trials. Control Clin Trials 7(3):177–188
Devlin B, Roeder K, Bacanu S-A (2001) Unbiased methods for population-based association studies. Genet Epidemiol 21(4):273–284
Drayton M, Noben-Trauth K (2006) Mapping quantitative trait loci for hearing loss in black swiss mice. Hear Res 212(1–2):128–139
Flint J, Eskin E (2012) Genome-wide association studies in mice. Nat Rev Genet 13(11):807
Friedman RA, Van Laer L, Huentelman MJ, Sheth SS, Van Eyken E, Corneveaux JJ, Tembe WD, Halperin RF, Thorburn AQ, Thys S, Bonneux S, Fransen E, Huyghe J, Pyykkö I, Cremers CWRJ, Kremer H, Dhooge I, Stephens D, Orzan E, Pfister M, Bille M, Parving A, Sorri M, Van de Heyning PH, Makmura L, Ohmen JD, Linthicum FH, Fayad JN, Pearson JV, Craig DW, Stephan DA, Van Camp G (2009) Grm7 variants confer susceptibility to age-related hearing impairment. Hum Mol Genet 18(4):785–796
Furlotte NA, Kang EY, Van Nas A, Farber CR, Lusis AJ, Eskin E (2012) Increasing association mapping power and resolution in mouse genetic studies through the use of meta-analysis for structured populations. Genetics 191(3):959–967
Gates GA, Mills JH (2005) Presbycusis. Lancet 366(9491):1111–1120
Ghazalpour A, Rau CD, Farber CR, Bennett BJ, Orozco LD, van Nas A, Pan C, Allayee H, Beaven SW, Civelek M, Davis RC, Drake TA, Friedman RA, Furlotte N, Hui ST, Jentsch JD, Kostem E, Kang HM, Kang EY, Joo JW, Korshunov VA, Laughlin RE, Martin LJ, Ohmen JD, Parks BW, Pellegrini M, Reue K, Smith DJ, Tetradis S, Wang J, Wang Y, Weiss JN, Kirchgessner T, Gargalovic PS, Eskin E, Lusis AJ, LeBoeuf RC (2012) Hybrid mouse diversity panel: a panel of inbred mouse strains suitable for analysis of complex genetic traits. Mamm Genome 23(9–10):680–692
Han B, Eskin E (2011) Random-effects model aimed at discovering associations in meta-analysis of genome-wide association studies. Am J Hum Genet 88(5):586–598
Han B, Eskin E (2012) Interpreting meta-analyses of genome-wide association studies. PLoS Genet 8(3):e1002555
Hardy RJ, Thompson SG (1996) A likelihood approach to meta-analysis with random effects. Stat Med 15(6):619–629
Hinds DA, McMahon G, Kiefer AK, Do CB, Eriksson N, Evans DM, St Pourcain B, Ring SM, Mountain JL, Francke U, Davey-Smith G, Timpson NJ, Tung JY (2013). A genome-wide association meta-analysis of self-reported allergy identifies shared and allergy-specific susceptibility loci. Nat Genet
Huang Q, Tang J (2010) Age-related hearing loss or presbycusis. Eur Arch Otorhinolaryngol 267(8):1179–1191
Johnson KR, Erway LC, Cook SA, Willott JF, Zheng QY (1997) A major gene affecting age-related hearing loss in c57bl/6j mice. Hear Res 114(1–2):83–92
Johnson KR, Zheng QY, Erway LC (2000) A major gene affecting age-related hearing loss is common to at least ten inbred strains of mice. Genomics 70(2):171–180
Johnson KR, Zheng QY, Bykhovskaya Y, Spirina O, Fischel-Ghodsian N (2001) A nuclear-mitochondrial dna interaction affecting hearing impairment in mice. Nat Genet 27(2):191–194
Johnson KR, Zheng QY, Weston MD, Ptacek LJ, Noben-Trauth K (2005) The mass1frings mutation underlies early onset hearing impairment in bub/bnj mice, a model for the auditory pathology of usher syndrome iic. Genomics 85(5):582–590
Johnson KR, Longo-Guess C, Gagnon LH, Yu H, Zheng QY (2008) A locus on distal chromosome 11 (ahl8) and its interaction with cdh23 ahl underlie the early onset, age-related hearing loss of dba/2j mice. Genomics 92(4):219–225
Johnson KR, Gagnon LH, Longo-Guess C, Kane KL (2012) Association of a citrate synthase missense mutation with age-related hearing loss in a/j mice. Neurobiol Aging 33(8):1720–1729
Kang HM, Zaitlen NA, Wade CM, Kirby A, Heckerman D, Daly MJ, Eskin E (2008) Efficient control of population structure in model organism association mapping. Genetics 178(3):1709
Kang EY, Han B, Furlotte N, Joo JW, Shih D, Davis CR, Lusis JA, Eskin E (2014) Meta-analysis identifies gene-by-environment interactions as demonstrated in a study of 4,965 mice. PLoS Genet 10(1):e1004022
Keller JM, Noben-Trauth K (2012) Genome-wide linkage analyses identify hfhl1 and hfhl3 with frequency-specific effects on the hearing spectrum of nih swiss mice. BMC Genet 13:32
Keller JM, Neely HR, Latoche JR, Noben-Trauth K (2011) High-frequency sensorineural hearing loss and its underlying genetics (hfhl1 and hfhl2) in nih swiss mice. J Assoc Res Otolaryngol 12(5):617–631
Kirby A, Kang HM, Wade CM, Cotsapas C, Kostem E, Han B, Furlotte N, Kang EY, Rivas M, Bogue MA, Frazer KA, Johnson FM, Beilharz EJ, Cox DR, Eskin E, Daly MJ (2010) Fine mapping in 94 inbred mouse strains using a high-density haplotype resource. Genetics 185(3):1081–1095
Lange K (2002) Mathematical and statistical methods for genetic analysis. Springer Verlag
Latoche JR, Neely HR, Noben-Trauth K (2011) Polygenic inheritance of sensorineural hearing loss (snhl2, -3, and -4) and organ of corti patterning defect in the alr/ltj mouse strain. Hear Res 275(1-2):150–159
Lindblad-Toh K, Winchester E, Daly MJ, Wang DG, Hirschhorn JN, Laviolette JP, Ardlie K, Reich DE, Robinson E, Sklar P, Shah N, Thomas D, Fan JB, Gingeras T, Warrington J, Patil N, Hudson TJ, Lander ES (2000) Large-scale discovery and genotyping of single-nucleotide polymorphisms in the mouse. Nat Genet 24(4):381–386
Lippert C, Quon G, Kang EY, Kadie CM, Listgarten J, Heckerman D (2013) The benefits of selecting phenotype-specific variants for applications of mixed models in genomics. Scientific Reports, 3
Listgarten J, Lippert C, Kang EY, Xiang J, Kadie CM, Heckerman D (2013) A powerful and efficient set test for genetic markers that handles confounders. Bioinformatics 29(12):1526–1533
Manolio TA, Collins FS, Cox NJ, Goldstein DB, Hindorff LA, Hunter DJ, McCarthy MI, Ramos EM, Cardon LR, Chakravarti A, Cho JH, Guttmacher AE, Kong A, Kruglyak L, Mardis E, Rotimi CN, Slatkin M, Valle D, Whittemore AS, Boehnke M, Clark AG, Eichler EE, Gibson G, Haines JL, Mackay TFC, McCarroll SA, Visscher PM (2009) Finding the missing heritability of complex diseases. Nature 461(7265):747–753
Mott R, Flint J (2013) Dissecting quantitative traits in mice. Annu Rev Genomics Hum Genet
Newman DL, Fisher LM, Ohmen J, Parody R, Fong C-TT, Frisina ST, Mapes F, Eddins DA, Robert Frisina D, Frisina RD, Friedman RA (2012) Grm7 variants associated with age-related hearing loss based on auditory perception. Hear Res 294(1–2):125–132
Noben-Trauth K, Zheng QY, Johnson KR (2003) Association of cadherin 23 with polygenic inheritance and genetic modification of sensorineural hearing loss. Nat Genet 35(1):21–23
Shin J-BB, Longo-Guess CM, Gagnon LH, Saylor KW, Dumont RA, Spinelli KJ, Pagana JM, Wilmarth PA, David LL, Gillespie PG, Johnson KR (2010) The r109h variant of fascin-2, a developmentally regulated actin crosslinker in hair-cell stereocilia, underlies early-onset hearing loss of dba/2j mice. J Neurosci 30(29):9683–9694
Valdar W, Solberg LC, Gauguier D, Burnett S, Klenerman P, Cookson WO, Taylor MS, Rawlins JNP, Mott R, Flint J (2006) Genome-wide genetic association of complex traits in heterogeneous stock mice. Nat Genet 38(8):879–887
Van Laer L, Huyghe JR, Hannula S, Van Eyken E, Stephan DA, Mäki-Torkko E, Aikio P, Fransen E, Lysholm-Bernacchi A, Sorri M, Huentelman MJ, Van Camp G (2010) A genome-wide association study for age-related hearing impairment in the saami. Eur J Hum Genet 18(6):685–693
Voight BF, Pritchard JK (2005) Confounding from cryptic relatedness in case-control association studies. PLoS Genet 1(3):e32
Wiltshire T, Pletcher MT, Batalov S, Barnes SW, Tarantino LM, Cooke MP, Wu H, Smylie K, Santrosyan A, Copeland NG, Jenkins NA, Kalush F, Mural RJ, Glynne RJ, Kay SA, Adams MD, Fletcher CF (2003) Genome-wide single-nucleotide polymorphism analysis defines haplotype patterns in mouse. Proc Natl Acad Sci U S A 100(6):3380–3385
Yalcin B, Willis-Owen SAG, Fullerton J, Meesaq A, Deacon RM, Rawlins JNP, Copley RR, Morris AP, Flint J, Mott R (2004) Genetic dissection of a behavioral quantitative trait locus shows that rgs2 modulates anxiety in mice. Nat Genet 36(11):1197–1202
Yalcin B, Nicod J, Bhomra A, Davidson S, Cleak J, Farinelli L, Osterras M, Whitley A, Yuan W, Gan X, Goodson M, Klenerman P, Satpathy A, Mathis D, Benoist C, Adams DJ, Mott R, Flint J (2010) Commercially available outbred mice for genome-wide association studies. PLoS Genet 6(9):e1001085
Yu J, Pressoir G, Briggs WH, Vroh Bi I, Yamasaki M, Doebley JF, McMullen MD, Gaut BS, Nielsen DM, Holland JB, Kresovich S, Buckler ES (2006) A unified mixed-model method for association mapping that accounts for multiple levels of relatedness. Nat Genet 38(2):203–208
Zheng QY, Johnson KR, Erway LC (1999) Assessment of hearing in 80 inbred strains of mice by abr threshold analyses. Hear Res 130(1–2):94–107
Zheng QY, Ding D, Yu H, Salvi RJ, Johnson KR (2009) A locus on distal chromosome 10 (ahl4) affecting age-related hearing loss in a/j mice. Neurobiol Aging 30(10):1693–1705
Acknowledgments
J.O., X.L., and R.F. are supported by the National Institutes of Health grants NIDCD R01DC010856-01. E.Y.K., J.W.J, F.H., and E.E. are supported by the National Science Foundation grants 0513612, 0731455, 0729049, 0916676, and 1065276, and the National Institutes of Health grants K25-HL080079, U01-DA024417, P01-HL30568, and PO1-HL28481. Q.Y.Z is supported by the US National Institutes of Health grants NIDCD R01DC009246.
Conflict of Interest
The authors declare that they have no conflict of interest.
Author information
Authors and Affiliations
Corresponding author
Additional information
Jeffrey Ohmen and Eun Yong Kang contributed equally to this work.
Rights and permissions
About this article
Cite this article
Ohmen, J., Kang, E.Y., Li, X. et al. Genome-Wide Association Study for Age-Related Hearing Loss (AHL) in the Mouse: A Meta-Analysis. JARO 15, 335–352 (2014). https://doi.org/10.1007/s10162-014-0443-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10162-014-0443-2