Abstract
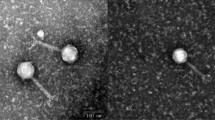
Pathogenic Escherichia coli strains cause diseases in both humans and animals. The limiting factors to prevent as well as control infections from pathogenic E. coli strains are their pathotypes, serotypes, and drug resistance. Herein, a bacteriophage (vB_EcoM-P896) has been isolated from duck sewage. Furthermore, aside from targeting intestinal pathogenic E. coli strains like enteropathogenic E. coli, Shiga toxin-producing E. coli, entero-invasive E. coli, and enteroaggregative E. coli, vB_EcoM-P896 can cause lysis in extraintestinal pathogenic E. coli strains such as avian pathogenic E. coli. Stability analysis revealed that vB_EcoM-P896 was stable under the following conditions: temperature, 4℃–50℃; pH, 3–11. The sequencing of the vB_EcoM-P896 genome was conducted utilizing an HiSeq system (Illumina, San Diego, CA) and subjected to de novo assembling with the aid of Spades 3.11.1. The characteristics of the DNA genome were as follows: size, 170,656 bp; GC content, 40.4%; the number of putative coding regions, 294. Transmission electron microscopy analysis of morphology and genome analysis revealed that the phage vB_EcoM-P896 belonged to the order Caudovirales and the family Myoviridae. The pan-genome analysis of vB_EcoM-P896 was divided into two levels. The first level involved the analysis of 91 strains of muscle tail phages, which were mainly divided into 5 groups. The second level involved the analysis of 24 strains of myophage with high homology. Of the 1480 gene clusters, 23 were shared core genes. Neighbor-joining phylogenetic trees were constructed using the Poisson model with MEGA6.0 based on the conserved sequences of phage proteins, the amino acid sequence of the terminase large subunit, and tail fibrin. Further analysis revealed that vB_EcoM-P896 was a typical T4-like potent phage with potential clinical applications.








Similar content being viewed by others
Data availability
No datasets were generated or analysed during the current study.
Abbreviations
- APEC :
-
Avian pathogenic Escherichia coli
- CDS :
-
Coding regions
- CFU :
-
Colony forming units
- DAEC :
-
Diffusely adherent Escherichia coli
- EAEC :
-
Enteroaggregative Escherichia coli
- E. coli :
-
Escherichia coli
- EC :
-
Escherichia coli
- EHEC :
-
Enterohaemorrhagic Escherichia coli
- EIEC :
-
Enteroinvasive Escherichia coli
- EPEC :
-
Enteropathogenic Escherichia coli
- ETEC :
-
Enterotoxic Escherichia coli
- ExPEC :
-
Extraintestinal Pathogenic Escherichia coli
- HGT :
-
Horizontal geng transfer
- HUS :
-
Hemolytic-uremic syndrome
- IPEC :
-
Intestinal Pathogenic Escherichia coli
- LB :
-
Luria-Bertani
- MOI :
-
Multiplicity of infection
- NA :
-
Not available
- NCBI :
-
National Center for Biotechnology Information Search database
- NJ :
-
Neighbor-joining
- NMEC :
-
Neonatal meningitis Escherichia coli
- OD :
-
Optical density 600
- PAS :
-
Phage-antibiotic synergy
- PBS :
-
Phosphate Buffered Saline
- PFU :
-
Plaque forming unit
- PHACTS :
-
Phage classification tool set
- RAST :
-
Rapid Annotation using Subsystem Technology server
- RBPs :
-
Receptor binding proteins
- rpm :
-
Revolutions per minute
- STEC :
-
Shiga toxin-producing Escherichia coli
- TerL :
-
The large terminal subunit
- UPEC :
-
Uropathogenic Escherichia coli
References
Ackermann HW (2007) 5500 phages examined in the electron microscope. Arch Virol 152:227–243
Ackermann H (2009) phage classification and characterization. Methods Mol Biol 501:127–140
Adler BA, Kazakov AE, Zhong C et al (2022) The genetic basis of phage susceptibility, cross-resistance and host-range in Salmonella. Microbiology 167(12):001126
Anand T, Vaid RK, Bera BC, Barua S, Riyesh T, Virmani N et al (2015) Isolation and characterization of a bacteriophage with broad host range, displaying potential in preventing bovine diarrhoea. Virus Genes 51:315–321
Atnafie B, Paulos D, Abera M, Tefera G, Hailu D, Kasaye S et al (2017) Occurrence of Escherichia Coli O157:H7 in cattle feces and contamination of carcass and various contact surfaces in abattoir and butcher shops of hawassa. Ethiopia Bmc Microbiol 17:24
Aziz RK, Bartels D, Best AA, DeJongh M, Disz T, Edwards RA et al (2008) The RAST server: rapid annotations using subsystems technology. BMC Genomics 9:75
Bachrach G, Leizerovici-Zigmond M, Zlotkin A, Naor R, Steinberg D (2003) bacteriophage isolation from human saliva. Lett Appl Microbiol 2003:50–53
Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS et al (2012) SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 19:455–477
Chan BK, Abedon ST, Loc-Carrillo C (2013) Phage cocktails and the future of phage therapy. Future Microbiol 8:769–783
Chang Y, Shin H, Lee J, Park C, Paik S, Ryu S (2015) Isolation and genome characterization of the virulent staphylococcus aureus bacteriophage SA97. Viruses 7:5225–5242
Chen X, Zhang W, Yin J, Zhang N, Geng S, Zhou X et al (2014) Escherichia Coli Isolates from sick chickens in china: changes in antimicrobial resistance between 1993 and 2013. Vet J 202:112–115
Cieplak T, Soffer N, Sulakvelidze A et al (2018) A bacteriophage cocktail targeting Escherichia coli reduces E. coli in simulated gut conditions, while preserving a non-targeted representative commensal normal microbiota. Gut Microbes 9(5):391–399
Cieplak T, Soffer N, Sulakvelidze A, Nielsen DS (2018) A bacteriophage cocktail targeting Escherichia coli reduces E. coli in simulated gut conditions, while preserving a non-targeted representative commensal normal microbiota. Gut Microbes 9(5):391–399
Coulter L, McLean R, Rohde R, Aron G (2014) Effect of bacteriophage infection in combination with tobramycin on the emergence of resistance in Escherichia Coli and pseudomonas aeruginosa biofilms. Viruses 6:3778–3786
Croxen MA, Law RJ, Scholz R, Keeney KM, Wlodarska M, Finlay BB (2013) Recent advances in understanding enteric pathogenic Escherichia Coli. Clin Microbiol Rev 26:822–880
Cui HY, Yuan L, Lin L (2017) Novel chitosan film embedded with liposome-encapsulated phage forbiocontrol of Escherichia coli O157:H7 in beef. Carbohyd Polym 177:156–164
Dissanayake U, Ukhanova M, Moye ZD, Sulakvelidze A, Mai V (2019) Bacteriophages reduce pathogenic Escherichia coli counts in mice without distorting gut microbiota. Front Microbiol 10:1984
Fratamico PM, DebRoy C, Liu Y, Needleman DS, Baranzoni GM, Feng P (2016) Advances in Molecular Serotyping and Subtyping of Escherichia Coli. Front Microbiol 7
Gill JJ, Hyman P (2010) Phage choice, isolation, and preparation for phage therapy. Curr Pharm Biotechno 11:2
Harper DR, Abedon ST, Burrowes BH, McConville ML (2019) Bacteriophages. Springer, Cham
Horiuk Y, Horiuk V, Kukhtyn M, Tsvihun A, Kernychnyi S (2020) Characterization of lytic activity of phage SAvB14 on Staphylococcus aureus variant bovis. J Adv Vet Animal Res 7(3):509–513
Huff WE, Huff GR, Rath NC, Balog JM, Donoghue AM (2002) Prevention of Escherichia coli Infection in Broiler Chickens with a Bacteriophage Aerosol Spray. Poult Sci 2002:1486–1491
Hyman P, Abedon ST (2010) Bacteriophage host range and bacterial resistance. Adv Appl Microbiol 2010:217–248
Jensen EC, Schrader HS, Rieland B, Thompson TL, Lee KW, Nickerson KW et al (1998) Prevalence of broad-host-range lytic bacteriophages of Sphaerotilus Natans, Escherichia Coli, and Pseudomonas Aeruginosa. Appl Environ Microbiol 64:575–580
Johnson JR, Russo TA (2002) Uropathogenic Escherichia coli as agents of diverse non-urinary tract extraintestinal infections. J Infect Dis 186:859–864
Kaper BJ, Nataro PJ, Mobley LH (2004) Pathogenic Escherichia Coli. Nat Rev Microbiol 2:123–140
Khan Mirzaei M, Nilsson AS (2015) Isolation of phages for phage therapy: a comparison of spot tests and efficiency of plating analyses for determination of host range and efficacy. PLoS ONE 10:e118557
Kim KS (2012) Current concepts on the pathogenesis of Escherichia Coli Meningitis: implications for therapy and prevention. Curr Opin Infect Dis 25:273–278
Knezevic P, Kostanjsek R, Obreht D, Petrovic O (2009) Isolation of pseudomonas aeruginosa specific phages with broad activity spectra. Curr Microbiol 59:173–180
Lavigne R, Darius P, Summer EJ, Seto D, Mahadevan P, Nilsson AS et al (2009) Classification of myoviridae bacteriophages using protein sequence similarity. Bmc Microbiol 9:224
Legrand P, Collins B, Blangy S, Murphy J, Spinelli S, Gutierrez C et al (2016) The atomic structure of the phage tuc 2009 baseplate tripod suggests that host recognition involves two different carbohydrate binding modules. Mbio 7:e1715–e1781
Liu M, Bischoff KM, Gill JJ, Mire-Criscione MD, Berry JD, Young R, Summer EJ (2015) Bacteriophage application restores ethanol fermentation characteristicsdisrupted by Lactobacillus fermentum. Biotechnol Biofuels 8:132
Machuca P, Daille L, Vinés E, Berrocal L, Bittner M (2010) Isolation of a Novel Bacteriophage Specific for the Periodontal PathogenFusobacterium nucleatum. Appl Environ Microb 76:7243–50
Mavrich TN, Hatfull GF (2017) Bacteriophage evolution differs by host, Lifestyle and Genome. Nat Microbiol 2:17112
McNair K, Bailey BA, Edwards RA (2012) PHACTS, a computational approach to classifying the lifestyle of phages. Bioinformatics 28:614–618
Mukherjee M (2013) molecular characterization of uropathogenic Escherichia coli: nalidixic acid and ciprofloxacin resistance, virulent factors and phylogenetic background. J Clin Diagn Res 7:2727–2731
Nobrega FL, Costa AR, Kluskens LD, Azeredo J (2015) Revisiting phage therapy: new applications for old resources. Trends Microbiol 23:185–191
North OI, Davidson AR, O’Toole G (2021) Phage Proteins required for tail fiber assembly also bind specifically to the surface of host bacterial strains. J Bacteriol 203:e406–e420
Orskov I, Orskov F, Jann B, Jann K (1977) Serology, chemistry, and genetics of o and k antigens of Escherichia coli. Bacteriol Rev 41:667–710
Page AJ, Cummins CA, Hunt M, Wong VK, Reuter S, Holden MTG et al (2015) Roary: rapid large-scale prokaryote pan genome analysis. Bioinformatics 31:3691–3693
Parasion S, Kwiatek M, Mizak L, Gryko R, Bartoszcze M, Kocik J (2012) Isolation and characterization of a novel bacteriophage φ4D lytic against enterococcus faecalis strains. Curr Microbiol 65:284–289
Park DW, Park JH (2021) Characterization of a novel phage depolymerase specific to Escherichia coli O157:H7 and biofilm control on abiotic surfaces. J Microbiol 17(26):30–64
Pires DP, Cleto S, Sillankorva S, Azeredo J, Lu TK (2016) Genetically engineered phages: a review of advances over the last decade. Microbiol Mol Biol R 80:523–543
Quagliarello V, Scheld WM (1992) Bacterial meningitis: pathogenesis, pathophysiology, and progress. N Engl J Med 327:864–872
Ross A, Ward S, Hyman P (2016) More is Better: Selecting for Broad Host Range Bacteriophages. Front Microbiol 7.
Ryan EM, Alkawareek MY, Donnelly RF, Gilmore BF (2012) Synergistic phage-antibiotic combinations for the control of Escherichia coli biofilms in vitro. FEMS Immunol Med Microbiol 65(2)395–398
Sabouri GM, Mohammadi A (2012) Bacteriophage: time to re-evaluate the potential of phage therapy as a promising agent to control multidrug-resistant bacteria. Iran J Basic Med Sci 15:693–701
Safwat MD, Farouk AE, Mohamed MA et al (2018) Isolation and evaluation of cocktail phages for the control of multidrug-resistant Escherichia coli serotype O104:H4 and E. coli O157:H7 isolates causing diarrhea. FEMS Microbiol Lett 365(2):33–45
Seemann T (2014) Prokka: rapid prokaryotic genome annotation. Bioinformatics 30:2068–2069
Smith HW, Huggins MB (1982) Successful treatment of experimental Escherichia Coli infections in mice using phage: its general superiority over antibiotics. J Gen Microbiol 2:307–318
Smith HW, Huggins MB (1983) Effectiveness of phages in treating experimental Escherichia Coli diarrhoea in calves, piglets and lambs. J Gen Microbiol 8:2659–2675
Smith HW, Huggins MB, Shaw KM (1987) The control of experimental Escherichia Coli diarrhoea in calves by means of bacteriophages. J Gen Microbiol 5:1111–1126
Sulakvelidze A, Alavidze Z, Morris JG Jr (2001) Bacteriophage Therapy. Antimicrob Agents Chemother 2001:649–59
Tagliaferri TL, Jansen M, Horz H (2019) Fighting Pathogenic Bacteria on Two Fronts: Phages and Antibiotics as Combined Strategy. Front Cell Infect Mi 9
Torres-Barceló C (2018) The disparate effects of bacteriophages on antibiotic-resistant bacteria. Emerg Microbes Infec 7:1–12
Tremblay DM, Tegoni M, Spinelli S, Campanacci VR, Blangy SP, Huyghe CL et al (2006) Receptor-binding protein of lactococcus lactis phages: identification and characterization of the saccharide receptor-binding site. J Bacteriol 7:2400–2410
Whitfield C (2006) Biosynthesis and assembly of capsular polysaccharides in Escherichia Coli. Annu Rev Biochem 75:39–68
Yehl K, Lemire S, Yang AC, Ando H, Mimee M, Torres MDT et al (2019) Engineering phage host-range and suppressing bacterial resistance through phage tail fiber mutagenesis. Cell 179:459–469
Yi Y, Abdelhamid AG, Xu YM, Yousef AE (2021) Characterization of broad-host lytic Salmonella phages isolated from livestock farms and application against Salmonella enteritidis in liquid whole egg. LWT 144:111269
Zhang C, Li W, Liu W, Zou L, Yan C, Lu K et al (2013) T4-like phage Bp7, a potential antimicrobial agent for controlling drug-resistant Escherichia coli in chickens. Appl Environ Microb 79:5559–5565
Funding
This study was supported by the following funds: Natural Science Foundation of Anhui Province (2022AH052192). NHC Key laboratory of Enteric Pathogenic Microbiology (Jiangsu Provincial Center for Disease Control and Prevention, EM202303). "National Visiting Scholar Program for Young Backbone Teachers of Higher Education Institutions in Central and Western China" by the Ministry of Education. The Natural Science Research Project of Anhui Province (KJ2021ZD0153). The Project of Cultivating Outstanding Talents in Colleges (gxgnfx2022135). Wuhu Institute of Technology level science and technology team (wzykjtd202002).Academic and technical leader of the "Talent Project" at Wuhu Institute of Technology (rc2022dtr03). Guizhou Province Science and Technology Plan Project (Grant No. QKH [2023]008).
Author information
Authors and Affiliations
Contributions
Zhang H.Y. and Zhang S. wrote the main manuscript text. Su X.Z. and Zheng X.K. performed the data analyses. Liu M.H., Zhao C.X., Liu X. and Ma Z.X. prepared the visualization and data presentation. Zhang W. contributed to the conception of the study. All authors reviewed the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Zhang, H., Su, X., Zheng, X. et al. vB_EcoM-P896 coliphage isolated from duck sewage can lyse both intestinal pathogenic Escherichia coli and extraintestinal pathogenic E. coli. Int Microbiol (2024). https://doi.org/10.1007/s10123-024-00519-5
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s10123-024-00519-5