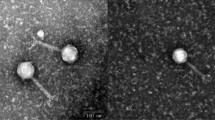
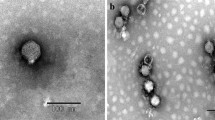
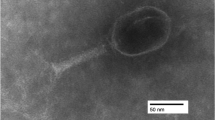
Abstract
A novel lytic bacteriophage, Escherichia phage EcS1, was isolated from sewage samples collected in Higashi-Hiroshima, Japan. The complete genome sequence of EcS1 was determined using the Illumina Miseq System. The whole genome of EcS1 was found to be 175,437 bp in length with a mean G+C content of 37.8%. A total of 295 genes were identified as structural, functional, and hypothetical genes. BLAST analysis of the EcS1 genomic sequence revealed the highest identity (79%; query cover of 73–74%) to three T4-related phages that infect Serratia sp. ATCC 39006. Host range experiments revealed that EcS1 has lytic effects on three pathogenic strains of Shigella spp. and a pathogenic strain of Salmonella enterica as well as on E. coli strains. However, two strains of Serratia marcescens showed resistance to this phage. Phylogenetic trees for phage tail fiber protein sequences revealed that EcS1 is closely related to Enterobacteriaceae-infecting phages. Thus, EcS1 is a novel phage that infects several pathogenic strains of the family Enterobacteriaceae.


Similar content being viewed by others
Change history
22 August 2018
The original version of this article unfortunately contained a mistake.
References
Starling S (2017) Shigella sonnei has the edge. Nat Rev Microbiol 15:450–451. https://doi.org/10.1038/nrmicro.2017.74
Nikfar R, Shamsizadeh A, Darbor M, Khaghani S, Moghaddam M (2017) A study of prevalence of Shigella species and antimicrobial resistance patterns in paediatric medical center, Ahvaz, Iran. Iran J Microbiol 9(5):277–282
Ansari S, Nepal HP, Gautam R, Shrestha S, Neopane P, Gurung G, Chapagain ML (2015) Community acquired multi-drug resistant clinical isolates of Escherichia coli in a tertiary care center of Nepal. Antimicrob Resist Infect Control 4:15. https://doi.org/10.1186/s13756-015-0059-2
Bao H, Zhang P, Zhang H, Zhou Y, Zhang L, Wang R (2015) Bio-control of Salmonella Enteritidis in foods using bacteriophages. Viruses 7:4836–4853. https://doi.org/10.3390/v7082847
Zhou Y, Liang Y, Lynch KH, Dennis JJ, Wishart DS (2011) PHAST: a fast phage search tool. Nucleic Acids Res 39:W347–W352
Kumar S, Stecher G, Tamura K (2016) MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol 33(7):1870–1874. https://doi.org/10.1093/molbev/msw054
Miller ES, Kutter E, Mosig G, Arisaka F, Kunisawa T, Ruger W (2003) Bacteriophage T4 genome. Microbiol Mol Biol Rev 67(1):86–156
Acknowledgements
The authors are grateful to T. Shimamoto and A. M. Soliman for host range experiments. This study was supported by the Japan Society for the Promotion of Science, KAKENHI (Grant number 15H04477). We also thank Katie Oakley, Ph.D., from Edanz Group (http://www.edanzediting.com/ac) for editing a draft of this manuscript.
Funding
This study was supported by the Japan Society for the Promotion of Science, KAKENHI (Grant number 15H04477).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflicts of interest.
Ethical approval
This article does not contain any studies with human participants or animals.
Additional information
Handling Editor: Chan-Shing Lin.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Saad, A.M., Askora, A., Kawasaki, T. et al. Full genome sequence of a polyvalent bacteriophage infecting strains of Shigella, Salmonella, and Escherichia. Arch Virol 163, 3207–3210 (2018). https://doi.org/10.1007/s00705-018-3971-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00705-018-3971-y