Abstract
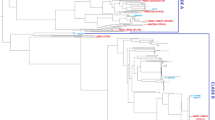
Uropathogenic Escherichia coli (UPECs) are the predominant cause of asymptomatic bacteriuria (ABU) and symptomatic UTI. In this study, multidrug-resistant (MDR) ABU-UPECs from hospitalized patients of Kolkata, India, were characterized with respect to their ESBL phenotype, acquisition of β-lactamase genes, mobile genetic elements (MGEs), phylotype property, ERIC-PCR profile, sequence types (STs), clonal complexes (CCs) and evolutionary and quantitative relationships and compared to the symptomatic ones to understand their epidemiology and evolutionary origin. Statistically significant incidence of ESBL producers, β-lactamase genes, MGEs and novel phylotype property (NPP) among ABU-UPECs similar to the symptomatic ones indicated the probable incidence of chromosomal plasticity on resistance gene acquisition through MGEs due to indiscriminate drug usage. ERIC-PCR typing and MLST analysis showed clonal heterogeneity and predominance of ST940 (CC448) among asymptomatic isolates akin to symptomatic ones along with the evidence of zoonotic transmissions. Minimum spanning tree analysis showed a close association between ABU-UPEC with known and unidentified STs having NPPs with isolates that belonged to phylogroups clade I, D, and B2. This is the first study that reported the occurrence of MGEs and NPPs among ABU-UPECs with the predominance of ESBL production which displayed the deleterious effect of MDR among this pathogen demanding alternative therapeutic interventions. Moreover, this study for the first time attempted to introduce a new approach to ascertain the phylotype property of unassigned UPECs. Withal, increased recognition, proper understanding and characterization of ABU-UPECs with the implementation of appropriate therapeutic measures against them when necessary are the need of the era which otherwise might lead to serious complications in the vulnerable population.






Similar content being viewed by others
References
Al-Guranie DR, Al-Mayahie S (2020) Prevalence of E. coli ST131 among uropathogenic E. coli isolates from Iraqi patients in Wasit Province, Iraq. Int J Microbiol. https://doi.org/10.1155/2020/8840561
Allcock S, Young EH, Holmes M, Gurdasani D, Dougan G, Sandhu MS, Solomon L, Török ME (2017) Antimicrobial resistance in human populations: challenges and opportunities. Glob Health Epidemiol Genom 10(2):e4. https://doi.org/10.1017/gheg.2017.4
Alonso CA, González-Barrio D, Ruiz-Fons F, Ruiz-Ripa L, Torres C (2017) High frequency of B2 phylogroup among non-clonally related fecal Escherichia coli isolates from wild boars, including the lineage ST131. FEMS Microbiol Ecol 93(3). https://doi.org/10.1093/femsec/fix016
Basu S, Mukherjee M (2018) Incidence and risk of co-transmission of plasmid-mediated quinolone resistance and extended-spectrum β-lactamase genes in fluoroquinolone-resistant uropathogenic Escherichia coli: a first study from Kolkata, India. J Glob Antimicrob Resist 14:217–223. https://doi.org/10.1016/j.jgar.2018.03.009
Belete MA (2020) Bacterial profile and ESBL screening of urinary tract infection among asymptomatic and symptomatic pregnant women attending antenatal care of northeastern Ethiopia region. Infect Drug Resist 13:2579–2592. https://doi.org/10.2147/IDR.S258379
Biswas P, Goel M, Negi H, Datta M (2016) An efficient greedy minimum spanning tree algorithm based on vertex associative cycle detection method. Procedia Comput Sci 92:513–519. https://doi.org/10.1016/j.procs.2016.07.376
Carone BR, Xu T, Murphy KC, Marinus MG (2014) High incidence of multiple antibiotic resistant cells in cultures of in enterohemorrhagic Escherichia coli O157:H7. Mutat Res 759:1–8. https://doi.org/10.1016/j.mrfmmm.2013.11.008
Cattoir V, Nordmann P, Silva-Sanchez J, Espinal P, Poirel L (2008) ISEcp1-mediated transposition of qnrB-like gene in Escherichia coli. Antimicrob Agents Chemother 52(8):2929–2932. https://doi.org/10.1128/AAC.00349-08
Clermont O, Bonacorsi S, Bingen E (2004) Characterization of an anonymous molecular marker strongly linked to Escherichia coli strains causing neonatal meningitis. J Clin Microbiol 42(4):1770–1772. https://doi.org/10.1128/jcm.42.4.1770-1772.2004
Clermont O, Bonacorsi S, Bingen E (2000) Rapid and simple determination of the Escherichia coli phylogenetic group. Appl Environ Microbiol 66(10):4555–4558. https://doi.org/10.1128/aem.66.10.4555-4558.2000
Clermont O, Christenson JK, Denamur E, Gordon DM (2013) The Clermont Escherichia coli phylo-typing method revisited: improvement of specificity and detection of new phylo-groups. Environ Microbiol Rep 5(1):58–65. https://doi.org/10.1111/1758-2229.12019
Clermont O, Olier M, Hoede C, Diancourt L, Brisse S, Keroudean M, Glodt J, Picard B, Oswald E, Denamur E (2011) Animal and human pathogenic Escherichia coli strains share common genetic backgrounds. Infect Genet Evol 11(3):654–662. https://doi.org/10.1016/j.meegid.2011.02.005
Cohen KA, Manson AL, Abeel T, Desjardins CA, Chapman SB, Hoffner S, Birren BW, Earl AM (2019) Extensive global movement of multidrug-resistant M. tuberculosis strains revealed by whole-genome analysis. Thorax 74(9):882–889. https://doi.org/10.1136/thoraxjnl-2018-211616
Cortes-Penfield NW, Trautner BW, Jump RLP (2017) Urinary tract infection and asymptomatic bacteriuria in older adults. Infect Dis Clin North Am 31(4):673–688. https://doi.org/10.1016/j.idc.2017.07.002
da Cruz Campos AC, Cavallo FM, Andrade NL, van Dijl JM, Couto N, Zrimec J, Lo Ten Foe JR, Rosa ACP, Damasco PV, Friedrich AW, Chlebowicz-Flissikowska MA, Rossen JWA (2019) Determining the virulence properties of Escherichia coli ST131 containing bacteriocin-encoding plasmids using short- and long-read sequencing and comparing them with those of other E. coli lineages. Microorganisms 7(11):534. https://doi.org/10.3390/microorganisms7110534
Dagher C, Salloum T, Alousi S, Arabaghian H, Araj GF, Tokajian S (2018) Molecular characterization of carbapenem resistant Escherichia coli recovered from a tertiary hospital in Lebanon. PLoS ONE 13(9):e0203323. https://doi.org/10.1371/journal.pone.0203323
Durmaz S, Bal EBB, Gunaydin M, ErkanYula E, Percin D (2015) Detection of β-lactamase genes, ERIC-PCR typing and phylogenetic groups of ESBL producing quinolone resistant clinical Escherichia coli isolates. Biomed Res-India 26(1):43–50
El Tawab AA, Mahmoud HB, Fatma I, El-khayat ME (2016) Prevalence of blaTEM and blaSHV genes in genomic and plasmid DNA of ESBL producing Escherichia coli clinical isolates from chicken. Benha Vet Med J 31:167–177
Gauthier L, Dortet L, Cotellon G, Creton E, Cuzon G, Ponties V, Bonnin RA, Naas T (2018) Diversity of carbapenemase-producing Escherichia coli isolates in France in 2012–2013. Antimicrob Agents Chemother 62(8):e00266-e318. https://doi.org/10.1128/AAC.00266-18
Giufrè M, Graziani C, Accogli M, Luzzi I, Busani L, Cerquetti M (2012) Escherichia coli Study Group. Escherichia coli of human and avian origin: detection of clonal groups associated with fluoroquinolone and multidrug resistance in Italy. J Antimicrob Chemother 67(4):860–867. https://doi.org/10.1093/jac/dkr565
Ghosh A, Bandyopadhyay D, Koley S, Mukherjee M (2021) Uropathogenic Escherichia coli in India-an overview on recent research advancements and trends. Appl Biochem Biotechnol. https://doi.org/10.1007/s12010-021-03521-z
Ghosh A, Mukherjee M (2019) Incidence of multidrug resistance, pathogenicity island markers, and pathoadaptive FimH mutations in uropathogenic Escherichia coli isolated from asymptomatic hospitalized patients. Folia Microbiol 64(4):587–600. https://doi.org/10.1007/s12223-019-00685-4
Gibreel T (2011) Molecular epidemiology of uropathogenic Escherichia coli in North West England and characterisation of the ST131 clone in the region. Thesis. Retrieved from http://www.manchester.ac.uk/escholar/uk-ac-man-scw:120702
Harmer CJ, Hall RM (2019) An analysis of the IS6/IS26 family of insertion sequences: is it a single family? Microb Genom 5(9):e000291. https://doi.org/10.1099/mgen.0.000291
Hassuna NA, Khairalla AS, Farahat EM, Hammad AM, Abdel-Fattah M (2020) Molecular characterization of extended-spectrum β lactamase-producing E. coli recovered from community-acquired urinary tract infections in Upper Egypt. Sci Rep 10(1):2772. https://doi.org/10.1038/s41598-020-59772-z
He K, Hu Y, Shi JC, Zhu YQ, Mao XM (2018) Prevalence, risk factors and microorganisms of urinary tract infections in patients with type 2 diabetes mellitus: a retrospective study in China. Ther Clin Risk Manag 14:403–408. https://doi.org/10.2147/TCRM.S147078
Hellmuth JE, Hitzeroth AC, Bragg RR, Boucher CE (2017) Evaluation of the ERIC-PCR as a probable method to differentiate Avibacterium paragallinarum serovars. Avian Pathol 46(3):272–277. https://doi.org/10.1080/03079457.2016.1259610
Hoeksema M, Jonker MJ, Bel K, Brul S, Ter Kuile BH (2018) Genome rearrangements in Escherichia coli during de novo acquisition of resistance to a single antibiotic or two antibiotics successively. BMC Genomics 19(1):973. https://doi.org/10.1186/s12864-018-5353-y
Ibrahim DR, Dodd CE, Stekel DJ, Ramsden SJ, Hobman JL (2016) Multidrug resistant, extended spectrum β-lactamase (ESBL)-producing Escherichia coli isolated from a dairy farm. FEMS Microbiol Ecol 92(4):fiw013. https://doi.org/10.1093/femsec/fiw013
Izdebski R, Baraniak A, Fiett J, Adler A, Kazma M, Salomon J, Lawrence C, Rossini A, Salvia A, Vidal Samso J, Fierro J, Paul M, Lerman Y, Malhotra-Kumar S, Lammens C, Goossens H, Hryniewicz W, Brun-Buisson C, Carmeli Y, Gniadkowski M, WP5 Study Groups (2013) Clonal structure, extended-spectrum β-lactamases, and acquired AmpC-type cephalosporinases of Escherichia coli populations colonizing patients in rehabilitation centers in four countries. Antimicrob Agents Chemother 57(1):309–316. https://doi.org/10.1128/AAC.01656-12
Kurpiel PM, Hanson ND (2011) Association of IS5 with divergent tandem blaCMY-2 genes in clinical isolates of Escherichia coli. J Antimicrob Chemother 66(8):1734–1738. https://doi.org/10.1093/jac/dkr212
Larsen MV, Cosentino S, Rasmussen S, Friis C, Hasman H, Marvig RL, Jelsbak L, Sicheritz-Pontén T, Ussery DW, Aarestrup FM, Lund O (2012) Multilocus sequence typing of total-genome-sequenced bacteria. J Clin Microbiol 50(4):1355–1361. https://doi.org/10.1128/JCM.06094-11
Lavakhamseh H, Mohajeri P, Rouhi S, Shakib P, Ramazanzadeh R, Rasani A, Mansouri M (2016) Multidrug-resistant Escherichia coli strains isolated from patients are associated with class 1 and 2 integrons. Chemotherapy 61(2):72–76. https://doi.org/10.1159/000438666
Liu X, Thungrat K, Boothe DM (2015) Multilocus sequence typing and virulence profiles in uropathogenic Escherichia coli isolated from cats in the United States. PLoS ONE 10(11):e0143335. https://doi.org/10.1371/journal.pone.0143335
Li W, Raoult D, Fournier PE (2009) Bacterial strain typing in the genomic era. FEMS Microbiol Rev 33(5):892–916. https://doi.org/10.1111/j.1574-6976.2009.00182.x
Mukherjee M, Basu S, Majumdar M (2011) Detection of bla TEM and bla CTX-M genes by multiplex polymerase chain reaction amongst uropathogenic Escherichia coli strains isolated from hospitalized patients in Kolkata. India Int J Biosci 1:64–69
Mukherjee SK, Mandal RS, Das S, Mukherjee M (2018) Effect of non-β-lactams on stable variants of inhibitor-resistant TEM β-lactamase in uropathogenic Escherichia coli: implication for alternative therapy. J Appl Microbiol 124(3):667–681. https://doi.org/10.1111/jam.13671
Najafi A, Hasanpour M, Askary A, Aziemzadeh M, Hashemi N (2018) Distribution of pathogenicity island markers and virulence factors in new phylogenetic groups of uropathogenic Escherichia coli isolates. Folia Microbiol (praha) 63(3):335–343. https://doi.org/10.1007/s12223-017-0570-3
Naziri Z, Derakhshandeh A, SoltaniBorchaloee A, Poormaleknia M, Azimzadeh N (2020) Treatment failure in urinary tract infections: a warning witness for virulent multi-drug resistant ESBL-producing Escherichia coli. Infect Drug Resist 13:1839–1850. https://doi.org/10.2147/IDR.S256131
Ojer-Usoz E, González D, Vitas AI (2017) Clonal diversity of ESBL-producing Escherichia coli isolated from environmental, human and food samples. Int J Environ Res Public Health 14(7):676. https://doi.org/10.3390/ijerph14070676
Parra GI, Squires RB, Karangwa CK, Johnson JA, Lepore CJ, Sosnovtsev SV, Green KY (2017) Static and Evolving. Norovirus genotypes: implications for epidemiology and immunity. PLoS Pathog 13(1):e1006136. https://doi.org/10.1371/journal.ppat.1006136
Pérez-Etayo L, Berzosa M, González D, Vitas AI (2018) Prevalence of integrons and insertion sequences in ESBL-producing E. coli isolated from different sources in Navarra, Spain. Int J Environ Res Public Health 15(10):2308. https://doi.org/10.3390/ijerph15102308
Ranjbar R, Tabatabaee A, Behzadi P, Kheiri R (2017) Enterobacterial repetitive intergenic consensus polymerase chain reaction (ERIC-PCR) genotyping of Escherichia coli strains isolated from different animal stool specimens. Iran J Pathol 12(1):25–34
Safwat Mohamed D, Farouk Ahmed E, Mohamed Mahmoud A, Abd El-Baky RM, John J (2018) Isolation and evaluation of cocktail phages for the control of multidrug-resistant Escherichia coli serotype O104: H4 and E. coli O157: H7 isolates causing diarrhea. FEMS Microbiol Lett 365(2). https://doi.org/10.1093/femsle/fnx275
Salem Mounir M, Muharram M, Alhosiny Ibrahim M (2010) Distribution of classes 1 and 2 integrons among multidrug resistant E. coli isolated from hospitalized patients with urinary tract infection in Cairo. Egypt. BTAIJ 4(3):145
Salvador E, Wagenlehner F, Köhler CD, Mellmann A, Hacker J, Svanborg C, Dobrindt U (2012) Comparison of asymptomatic bacteriuria Escherichia coli isolates from healthy individuals versus those from hospital patients shows that long-term bladder colonization selects for attenuated virulence phenotypes. Infect Immun 80(2):668–678. https://doi.org/10.1128/IAI.06191-11
Srivastava S, Agarwal J, Mishra B, Srivastava R (2016) Virulence versus fitness determinants in Escherichia coli isolated from asymptomatic bacteriuria in healthy nonpregnant women. Indian J Med. Microbiol 34(1):46–51. https://doi.org/10.4103/0255-0857.174103
van der Mee-Marquet N, Diene SM, Chopin F, Goudeau A, François P (2016) Enigmatic occurrence of NDM-7 enzyme in the community. Int J Antimicrob Agents 47(6):505–507. https://doi.org/10.1016/j.ijantimicag.2016.04.002
van Elsas JD, Semenov AV, Costa R, Trevors JT (2011) ISME J 5(2):173–83. https://doi.org/10.1038/ismej.2010.80
Venkatesan KD, Chander S, Loganathan K, Victor K (2017) Study on asymptomatic bacteriuria in diabetic patients. Int J Contemp Med Res 4(2):480–483
Wang Y, Zhou J, Li X, Ma L, Cao X, Hu W, Zhao L, Jing W, Lan X, Li Y, Gong X, Chen Q, Stipkvits L, Szathmary S, Tarasiuk K, Pejsak Z, Liu Y (2020) Genetic diversity, antimicrobial resistance and extended-spectrum β-lactamase type of Escherichia coli isolates from chicken, dog, pig and yak in Gansu and Qinghai Provinces, China. J Glob Antimicrob Resist 22:726–732. https://doi.org/10.1016/j.jgar.2020.06.028
Wilson LA, Sharp PM (2006) Enterobacterial repetitive intergenic consensus (ERIC) sequences in Escherichia coli: evolution and implications for ERIC-PCR. Mol Biol Evol 23(6):1156–1168. https://doi.org/10.1093/molbev/msj125
Wirth T, Falush D, Lan R, Colles F, Mensa P, Wieler LH, Karch H, Reeves PR, Maiden MC, Ochman H, Achtman M (2006) Sex and virulence in Escherichia coli: an evolutionary perspective. Mol Microbiol 60(5):1136–1151. https://doi.org/10.1111/j.1365-2958.2006.05172.x
Wright MH, Adelskov J, Greene AC (2017) Bacterial DNA extraction using individual enzymes and phenol/chloroform separation. J Microbiol Biol Educ 18(2):18.2.48. https://doi.org/10.1128/jmbe.v18i2.1348
Xiao L, Wang X, Kong N, Zhang L, Cao M, Sun M, Wei Q, Liu W (2019) Characterization of beta-lactamases in bloodstream-infection Escherichia coli: dissemination of blaADC-162 and blaCMY-2 among bacteria via an IncF plasmid. Front Microbiol 10:2175. https://doi.org/10.3389/fmicb.2019.02175
Yadav S (2018) Correlation analysis in biological studies. J Pract Cardiovasc Sci 4:116–121
Zhang J, Poh CL (2018) Regulating exopolysaccharide gene wcaF allows control of Escherichia coli biofilm formation. Sci Rep 8(1):13127. https://doi.org/10.1038/s41598-018-31161-7
Zheng B, Feng C, Xu H, Yu X, Guo L, Jiang X, Song X (2019) Detection and characterization of ESBL-producing Escherichia coli expressing mcr-1 from dairy cows in China. J Antimicrob Chemother 74(2):321–325. https://doi.org/10.1093/jac/dky446
Acknowledgements
The authors would like to express their sincere gratitude to Director and Head of Department of Tropical Medicine, School of Tropical Medicine, Kolkata West Bengal, India, for their kind support. The authors would also like to thank Mr. Snehashis Koley, for his assistance in acquiring access to the BioNumerics version 7.6 software, Applied Maths, Sint-Martens-Latem, Belgium.
Funding
Consumable support to carry out the research was provided from an extramural grant from the Science and Engineering Research Board (SERB), Department of Science and Technology, Government of India (Grant No. EMR/2016/001670 dated August 22, 2017). Arunita Ghosh was the recipient of the INSPIRE fellowship grant from the Department of Science and Technology, New Delhi, Government of India, No.-DST/INSPIRE Fellowship/2016/ IF160069 dated October 31, 2018, and October 10, 2019.
Author information
Authors and Affiliations
Contributions
This work was performed as collaboration among all the authors. MM and AG conceived the study. BG participated in the study design. AG performed all the experiments, participated in statistical and bioinformatics analysis, and also wrote the first draft of the manuscript. BG participated only in the PCR-based phylogenetic assays. MM supervised the entire study and also reviewed and finalized the manuscript. All of the authors had read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval
The present study was approved by the Clinical Research Ethics Committee, School of Tropical Medicine, Kolkata (CREC-STM), Ref No. CREC-STM/317 dated March 29, 2016.
Consent to participate
Informed consent was obtained from all patients for being included in this study.
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Supplementary Fig. 1 Cluster analysis performed on Heat maps generated using R software package (version 3.2.5)., based on the presence and absence three β-lactamase genes in the plasmid DNA of each of the individual isolate from (a) Asymptomatic UPEC group and (b) Symptomatic UPEC group. Numbers in the text box provided on the righthand side represented sample ID of the E. coli isolates considered in each group. Colour key represented the variation in colours from red to white illustrating the complete absence of a particular gene to its complete presence respectively. Supplementary Fig. 2 Cluster analysis performed on Heat maps generated using R software package (version 3.2.5)., based on the presence and absence three β-lactamase genes in the genomic DNA of each of the individual isolate from (a) Asymptomatic UPEC group and (b) Symptomatic UPEC group. Numbers in the text box provided on the righthand side represented sample ID of the E. coli isolates considered in each group. Colour key represented the variation in colours from red to white illustrating the complete absence of a particular gene to its complete presence respectively. Supplementary Fig. 3 Graphical representation of correlation coefficient values computed with confidence level of 95% (p values ≤ 0.05) using GraphPad Prism version 9 (Prism software package) based on the correlation of three different β-lactamase genes in the genomic DNA of each of individual (a) asymptomatic and (b) symptomatic UPECs. Different β-lactamases were represented by interleaved symbols with varied colours. Dotted lines were used to differentiate correlation of each of the β-lactamase gene with two others. Supplementary Fig. 4 Dendogram generated on ERIC-PCR profiles of (a) asymptomatic (n=20) and (b) symptomatic (n=20) UPECs. Dice similarity coefficient values were used to generate the dendogram by UPGMA method of clustering using SPSS version 21.0 software. Isolates were distinctly unrelated at a coefficient of similarity value ≥ 96% (indicated by a solid line). ERIC-banding pattern of 20 each of ABU and Symptomatic UPECs respectively were represented as grouped individual lanes that contained the amplicons of each isolate. Five and six clonal groups were identified based on the cluster analysis of the individual ERIC profiles of asymptomatic and symptomatic isolates respectively. The extreme left column represented respective ESBL /BLIR phenotype of the isolates. Immediately adjacent to that was the phylogroup of the isolates.
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Ghosh, A., Ghosh, B. & Mukherjee, M. Epidemiologic and molecular characterization of β-lactamase-producing multidrug-resistant uropathogenic Escherichia coli isolated from asymptomatic hospitalized patients. Int Microbiol 25, 27–45 (2022). https://doi.org/10.1007/s10123-021-00187-9
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10123-021-00187-9