Abstract
Background
The prognosis of advanced gastric cancer (GC) invading the gastric serosa remains poor, mainly owing to high incidence of peritoneal recurrence. Patients with peritoneal metastases are often treated with neoadjuvant intraperitoneal and systemic chemotherapies (NIPS). Good responders to NIPS often undergo conversion gastrectomy. This study aims to explore biomarkers predicting the occurrence of peritoneal metastasis (PM) and evaluating the efficacy of NIPS in GC patients.
Methods
We collected six peritoneal lavage (PL) samples from two patients with PM, two without PM, and two with diminished PM after NIPS via intraperitoneal access ports. We equally isolated microRNAs from exosomes derived from PL samples for deep sequencing. Two microRNAs (hsa-let-7g-3p and hsa-miR-10395-3p) were identified, and their expression levels were examined in PL samples of 99 GC patients using qRT-PCR. Moreover, we performed in vivo and in vitro functional assays to investigate effects of these microRNAs on metastasis and chemoresistance of GC cells.
Results
Exosomal microRNA expression profiling of six PL samples indicated that the microRNA signature in exosomes of PLs from patients with diminished PM was similar to that from patients without PM. Expression levels of hsa-let-7g-3p and hsa-miR-10395-3p were associated with PM. In vivo and in vitro functional assays confirmed that hsa-let-7g-3p and hsa-miR-10395-3p are involved in GC metastasis and chemoresistance.
Conclusion
PL-derived exosomes in GC contain large amounts of microRNAs related to PM. Moreover, hsa-let-7g-3p and hsa-miR-10395-3p could be used as biomarkers predicting PM and NIPS efficacy and are involved in GC metastasis and chemoresistance.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Background
Gastric cancer (GC) is the fifth most common cancer and the third leading cause of cancer-related mortality worldwide [1]. Infection with Helicobacter pylori is the main risk factor for non-cardia GC [2] and is associated with bacterial virulence, genetic polymorphism of hosts, and environmental factors [3]. Although the incidence and mortality rates are on a decreasing trend in recent times [4, 5], the prognosis of advanced GC invading the gastric serosa is still poor, with a median overall survival of 10–12 months [6], mainly due to the high incidence of peritoneal recurrence. The peritoneum is the most common site of metastasis and recurrence of GC, and the quality of life of such patients deteriorates rapidly as malignant ascites due to peritoneal dissemination of GC cells develops.
Neoadjuvant chemotherapy (chemotherapy before surgery) increases the opportunity for curable resection, eliminates early microscopic spread, and allows for evaluation of biological response to treatment [3]. Scholars from Japan have designed a therapeutic regimen called neoadjuvant intraperitoneal and systemic chemotherapy (NIPS), which involves weekly intravenous and intraperitoneal paclitaxel (PTX) administration with oral S-1 (tegafur, gimeracil, and oteracil; TGO) twice daily for advanced GC with peritoneal metastases. Good responders with diminished peritoneal metastases undergo conversion gastrectomy [7,8,9]. However, biomarkers that evaluate the efficacy of NIPS in patients with GC with high specificity and sensitivity have not been revealed so far.
Biomarkers derived from liquid biopsies, including cell-free circulating tumor DNA, circulating tumor cells, and exosomes comprehensively reflect the conditions of heterogeneous tumors compared to those in tissue biopsies, as well as can be safely, effectively and conveniently incorporated in the management of patients with metastatic tumors [3, 10,11,12]. Many biomarkers, especially predictive biomarkers, have been postulated in GC, but few have been applied in routine clinical use [13]. Furthermore, there are currently no reliable markers with high specificity and sensitivity for predicting peritoneal metastasis (PM) in GC patients. Hence, exploring biomarkers for predicting PM and the efficacy of NIPS is of utmost importance.
MicroRNAs (miRs) are short noncoding RNAs consisting of 19 to 25 nucleotides that regulate post-transcriptional silencing of target gene expression and participate in various physiological and pathological biological processes, including cell proliferation and apoptosis [14]. Studies have shown that miRs are largely involved in tumor initiation and progression, and serve as oncogenes and suppressors [15,16,17]. MiRs are wrapped in membrane vesicles, especially exosomes, thereby preventing them from degradation, supporting them to accomplish cell-to-cell communication, and participating in miR-based signaling mechanisms [18, 19]. Exosomes are secretory vesicles with diameters ranging from 40 to 160 nm and are derived from nearly all types of cells, and exist in a wide variety of body fluids, including blood, urine, and ascites [20]. Exosomes can deliver tumor-derived antigens that induce cellular immune responses, and exosomal miRs are appropriate therapeutic targets or biomarkers for cancer diagnosis and prognosis [21,22,23]. Nevertheless, to date, few studies have explored the use of exosomal miRs as biomarkers in patients with GC and PM. Thus, exosomal miRs derived from ascites could serve as potential specific biomarkers for GC dissemination in the peritoneum.
Therefore, in our study, we aimed to analyze miR expression profiling and identify miR signatures in exosomes of peritoneal lavage (PL) samples from patients with advanced GC having PM receiving NIPS. We also identified two candidate biomarkers (hsa-let-7g-3p and hsa-miR-10395-3p) for the prediction of PM and the evaluation of the efficacy of NIPS. Furthermore, we characterized the functional roles of these miRs in the progression of GC through in vivo and in vitro experiments and explored the mechanisms underlying metastasis and chemoresistance of GC cells.
Materials and methods
Patient samples
We collected PL samples via intraperitoneal access ports from 99 patients at the Ruijin Hospital affiliated with Shanghai Jiaotong University School of Medicine, Shanghai, China. All enrolled patients met the following criteria: (1) pathologically diagnosed with gastric cancer; (2) definitively diagnosed with PM through peritoneoscopy; (3) had a complete clinicopathological and follow-up information; and (4) had no other signs of distant metastasis except PM. All patients involved in this study provided their written informed consent, and the study protocol was approved by the ethics committee of the Ruijin Hospital.
Exosomes and RNAs extraction
To remove adipose tissue and cell debris, PL samples were centrifuged at 1500 rpm for 15 min at 4 °C and filtered through a 0.22-μm filter (Millipore, USA). Total RNA in exosomes was extracted from PL samples using an exoRNeasy Midi Kit (Qiagen, Hilden, Germany) according to the manufacturer’s instructions. The particle size and morphological structure of exosomes were assessed by Umibio Science and Technology (Shanghai, China). The quality and concentration of total exosomal RNA were assessed using NanoPhotoment N120 (Implen, Germany).
Quantitative real-time PCR (qRT-PCR)
Reverse transcription was performed using the Evo M-MLV RT Kit with gDNA clean for qPCR (Accurate Biotechnology, Changsha, China). The qRT-PCR was performed using SYBR® Green Premix Pro Taq HS qPCR Kit (Accurate Biotechnology, Changsha, China) in a real-time fluorescent quantitative PCR machine (Analytik Jena, Germany). The expression level of U6 was used as the reference control miR. The primer and RT primer sequences are shown in Table S4.
Cell culture
GES-1, AGS, NCI-N87 (N87) and other GC cell lines were purchased from the American Type Culture Collection (ATCC, USA). All cell lines were tested for mycoplasma contamination. STR DNA profiling was performed for cell line authentication. Cells were cultivated in an RPMI 1640 medium (Meilunbio, Dalian, China) supplemented with 10% FBS (Gibco, USA) and 1% penicillin–streptomycin in a standard 37℃ incubator with 5% CO2.
Cell transfection and treatment
Mimics and inhibitors of hsa-let-7 g-3p and hsa-miR-10395-3p and their negative controls were synthesized by Bioegene Co., Ltd. (Shanghai, China). Transfections were performed using the Hilymax (DOJINDO, Kumamoto, Japan). The mimics and inhibitors were transfected at a final concentration of 50 nM into a 6-well plate. Cells were harvested 48 h after transfection for further experiments. The mimics and inhibitors sequences are listed in Table S4. GES-1, AGS, and N87 cells were treated with different concentrations of paclitaxel and TGO to determine their IC50 values.
In vivo peritoneal metastasis model
AGS cells transfected with negative control, hsa-let-7 g-3p mimics, and hsa-miR-10395-3p mimics were injected intraperitoneally into 5-week-old male nude BALB/c mice (107 cells/mouse, 4 mice/group). All animal experiments were approved by the Shanghai Jiaotong University School of Medicine Animal Care and Use Committee.
Statistical analysis
GraphPad Prism v8.0.1 and Statistical Package for Social Science (SPSS, v20.0) software were used for statistical analysis. Variables were expressed as mean ± SD. The results of two groups were compared using Student’s t-tests, while multiple comparisons were performed by one-way ANOVA. The survival curve of patients was calculated using the Kaplan–Meier method. The clinicopathological features of patients with different expression levels of hsa-miR-10395-3p or hsa-let-7 g-3p were compared using Pearson’s chi-square test for categorical variables. A P-value less than 0.05 was considered statistically significant. The results of RNA-seq were performed on the Majorbio analysis platform (Shanghai, China).
Results
The microRNA signature in exosomes of PL samples from patients with diminished PM was similar to that in patients without PM
We collected PL samples via intraperitoneal access ports from two patients with PM (P, positive), two without PM (N, negative) and two with diminished PM after NIPS (PN, negative after NIPS) (Table S1). The particle size and morphological structure were measured to characterize the exosomes derived from the PL samples (Fig. 1a, b). The molecular markers of exosomes, such as CD9, CD81, and CD63, were detected through western blotting (Fig. 1c). MiRs in exosomes derived from PL samples were isolated for deep sequencing. MiR profiling showed that 482 miRs were constitutively expressed in all enrolled patients with GC (Fig. 1d). Interestingly, correlation analysis showed that the miR signature of exosomes derived from the PL of patients with diminished PM was similar to that of patients without PM (P < 0.05, Spearman’s correlation, Fig. 1e, f). To identify exosomal miR markers for predicting PM and the efficacy of NIPS, a group of 11 miRs with two-fold higher expression levels in exosomes derived from patients with PM (P) than those from patients without PM (N) or with diminished PM (PN) were screened (Table S2, Fig. 1g). Among them, the top two miRs, hsa-let-7g-3p and hsa-miR-10395-3p, were selected for further investigation.
MiR signature in exosomes derived from PL samples in Negative, Positive, and Negative after NIPS groups. a Particle size distribution graphs of exosomes extracted from PL samples show a mean diameter of 100 nm. b Representative transmission electron microscopy images of exosomes (scale bar, 100 nm). c Western blotting confirmed the presence of exosome-associated markers, CD81, CD9 and CD63, and the absence of endoplasmic reticulum marker Calnexin. The cell lysate group was considered as the negative control. d Venn diagram shows 482 miRs constitutively expressed in all three groups. There are 76, 83, and 43 specific miRs, respectively, expressed in the Negative, Positive, and Negative after NIPS groups. e Heatmap shows differential expression of these miRs. f Correlation heatmap of three groups. Item correlations are color graded. The closer the correlation coefficient is to 1, the greater the similarity of these samples. g Heatmap based on the expression of 11 candidate miRs in six samples from three groups
Expression levels of hsa-let-7g-3p and hsa-miR-10395-3p are associated with PM
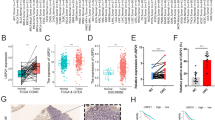
Based on the PL samples of 99 patients with GC (66 with PM and 33 without PM) from our hospital (Table S3), qRT-PCR was performed to detect the expression levels of hsa-let-7g-3p and hsa-miR-10395-3p. Hsa-let-7g-3p and hsa-miR-10395-3p were expressed at higher levels in GC samples with PM than in those without PM (Fig. 2a). Moreover, higher expression of hsa-let-7g-3p and hsa-miR-10395-3p was associated with PM and positive peritoneal lavage cytology (CY) (Fig. 2b). Consistently, univariate and multivariate analyses indicated that the expression levels of hsa-let-7g-3p and hsa-miR-10395-3p were strongly associated with PM in GC patients (Table 1). Interestingly, higher hsa-let-7g-3p expression significantly correlated with shorter overall survival rate (log-rank test, P = 0.04; Fig. 2c), while hsa-miR-10395-3p showed no statistical significance (log-rank test, P = 0.19; Fig.S1a in the Electronic supplementary material, ESM). More intriguingly, patients with high expression of hsa-let-7g-3p and hsa-miR-10395-3p showed worse overall survival time (log-rank test, P = 0.003; Fig. 2c). Moreover, multivariate Cox regression analysis indicated that the expression of hsa-let-7g-3p could be an independent prognostic factor of overall survival in patients with GC (HR = 2.040, P = 0.043, Table 2). The expression levels of these miRs were also examined in longitudinal PL samples from 12 patients who underwent NIPS. The results indicated that the expression of hsa-let-7g-3p and hsa-miR-10395-3p decreased after NIPS treatment (Fig. 2d). Furthermore, 27 patients who received NIPS were selected and divided into two groups: one group contained 23 patients with diminished PM after NIPS (PN, negative after NIPS), and the other group included 4 patients with no improvement in PM (PP, positive after NIPS). The expression levels of hsa-let-7g-3p and hsa-miR-10395-3p in the PL samples were associated with the efficacy of NIPS. The lower the expression of miRs, the higher the possibility of PM negativity after NIPS. Furthermore, patients with absent hsa-let-7g-3p or hsa-miR-10395-3p in PL were always PM-negative after NIPS, implying that miRs could be strict markers of PM (Fig. 2e). In addition, the ROC curves of hsa-let-7g-3p showed ordinary predictive power (AUC = 0.6401); however, the combination of these two miRs exhibited a higher AUC (AUC = 0.7308) in predicting the efficacy of NIPS (Fig. 2f). Thus, hsa-let-7g-3p and hsa-miR-10395-3p could be potential biomarkers for predicting the efficacy of NIPS, thereby guiding conversion gastrectomy in patients with GC who have received or are receiving NIPS.
Expression of hsa-let-7g-3p and hsa-miR-10395-3p in GC patients and clinical significance of the expression. a Expression of hsa-let-7g-3p and hsa-miR-10395-3p in PL samples from GC patients. The expression levels are described as ΔCt (Cttarget-CtU6) values. Smaller ΔCt values indicate higher miR expression. If no expression was detected, the Ct values were equated with total cycle number (50 cycles). Error bars represent the mean values ± standard deviation. b Stacked bar graphs indicate the proportion of GC patients with or without PM in the positive or negative group of hsa-let-7g-3p or hsa-miR-10395-3p, and the relationship between peritoneal lavage cytology (CY) and the expression of hsa-let-7g-3p and hsa-miR-10395-3p. Negative: no expression detected in 50 cycles; Positive: expression detected in 50 cycles; CY0: negative peritoneal lavage cytology; CY1: positive peritoneal lavage cytology. c Prognostic analysis of hsa-let-7g-3p and combined hsa-let-7g-3p and hsa-miR-10395-3p using survival data of 76 patients from Ruijin Hospital. d Before and after plots of the expression (ΔCt) of hsa-let-7g-3p or hsa-miR-10395-3p before and after NIPS treatment (n = 12). Smaller ΔCt values indicate higher miR expression. e Stacked bar graphs indicate the proportion of GC patients with or without PM after NIPS in positive or negative group of hsa-let-7g-3p or hsa-miR-10395-3p. f ROC curve shows predictive accuracy of hsa-let-7g-3p (AUC = 0.6401, 95% CI 0.4688 to 0.8114) and combined hsa-let-7g-3p and hsa-miR-10395-3p (AUC = 0.7308, 95% CI 0.6191–0.8424). 7g, hsa-let-7g-3p; 10,395, hsa-miR-10395-3p
hsa-let-7g-3p and hsa-miR-10395-3p contribute to the proliferation, migration, and chemoresistance of GC cells
To explore the function of hsa-let-7g-3p and hsa-miR-10395-3p in tumor metastasis and chemoresistance, in vitro functional assays, including CCK8, foci formation, wound healing, and transwell assays, were performed. First, the expression levels of hsa-let-7g-3p and hsa-miR-10395-3p in GC cell lines were examined using qRT-PCR (Fig. 3a). Of note, N87 (N87), MKN-28 (28) and HGC-27 (27) cell lines were originally derived from GC metastasis, which have higher expression levels of these two miRs, whereas two GC cell lines, AGS and GES-1, with relatively low expression, were selected for overexpression via the transfection of miR mimics, while N87 was selected to suppress the expression of hsa-let-7g-3p or hsa-miR-10395-3p via miR inhibitors (Fig. 3b). On one hand, overexpression of hsa-let-7g-3p and hsa-miR-10395-3p significantly promoted the proliferation and migration of GC cells (Fig. 3c–i). On the other hand, as PTX and TGO are the main chemotherapeutic drugs of NIPS, we studied the effects of hsa-let-7g-3p and hsa-miR-10395-3p on apoptosis and cell death induced by PTX and TGO. The 50% inhibitory concentrations (IC50) of PTX and TGO in the three GC cell lines were determined (Fig. 4a). Transfected GC and control cells were exposed to PTX and TGO. After 72 h treatment with PTX and TGO, apoptotic and dead cells were evaluated using flow cytometry. As seen in Fig. 4b–e, the apoptotic and dead index of hsa-let-7g-3p or hsa-miR-10395-3p overexpressed cells was significantly lower than that of control cells. Furthermore, increased percentages of apoptotic and dead cells were observed in cells transfected with inhibitors of hsa-let-7g-3p or hsa-miR-10395-3p (Fig. 4f, g). In vivo studies further demonstrated that the overexpression of hsa-let-7g-3p and hsa-miR-10395-3p significantly promoted tumor metastasis. MiR mimics and negative control (NC) cells were intraperitoneally administered to male nude mice. Among the three groups, the mimics groups showed different degrees of ascites and splenomegaly that were more severe than those in the NC group (Fig. 5a, Fig.S1b, c in the ESM). Fluorescence in situ hybridization (FISH) analysis of mimic groups exhibited higher expression of hsa-let-7g-3p/hsa-miR-10395-3p and bigger size of spleen (Fig. S1d, e in the ESM). The tumors formed by mimic-transfected cells in the peritoneal cavity were larger than those in the NC group (Fig. 5b). Moreover, immunohistochemistry (IHC) of tumor tissue resected from nude mice showed that tumors with high expression of hsa-let-7g-3p or hsa-miR-10395-3p exhibited higher Ki-67 expression and fewer apoptotic cells (Fig. 5c–f).
hsa-let-7g-3p and hsa-miR-10395-3p promote proliferation and migration of GC cells. a Expression of hsa-let-7g-3p and hsa-miR-10395-3p in nine GC cell lines. b Overexpression or suppression efficiency of mimics/inhibitors in AGS, GES-1, and N87 cell lines. The proliferation ability of AGS, GES-1, and N87 cell lines was tested using the c CCK8 assay, and d, e plate colony assay. The migration capability was measured using f, g wound healing assay and h, i transwell assay (Scale bar: 200 μm, 10x). Error bars represent the mean values ± standard deviation; * P < 0.05, ** P < 0.01, *** P < 0.001, **** P < 0.0001; ns, not significant; 7g, hsa-let-7g-3p; 10395, hsa-miR-10395-3p
hsa-let-7g-3p and hsa-miR-10395-3p suppress cell death after chemotherapies. a Relative cell viability of GES-1, AGS, and N87 cell lines after treatment with different concentrations of PTX or TGO for 48 h. b-e Mimics of hsa-let-7g-3p and hsa-miR-10395-3p suppress cell apoptosis and death of GES-1 and AGS cells treated with PTX or TGO. f, g Inhibitors of hsa-let-7g-3p and hsa-miR-10395-3p promote cell apoptosis and death in N87 cells. The cells were exposed to PTX or TGO for 48 h at the concentration of 200% IC50. Percentage of apoptotic and dead cells = early apoptotic cells (Q1-LR) + late apoptotic cells (Q1-UR) + dead cells (Q1-UL). All data are representative of at least three independent experiments. Error bars represent the mean values ± standard deviation; * P < 0.05, ** P < 0.01, *** P < 0.001, **** P < 0.0001; 7g, hsa-let-7g-3p; 10395, hsa-miR-10395-3p
Overexpression of hsa-let-7g-3p and hsa-miR-10395-3p promotes tumor metastasis in peritoneum in vivo. a Representative images of nude mice with ascites formed by mimics-transfected or NC AGS cells implantation to the peritoneal cavity. b Tumor tissues generated in peritoneum of nude mice from 7g or 10395 mimics and NC group (n = 4/group). Tumors were collected 6 weeks after abdominal injection. c Representative H&E staining images of tumor tissues. d-f IHC analysis of tumor tissues incubated with Ki-67 antibody and TUNEL staining. Scale bar: 200 μm, 10x; 40 μm, 40x. Error bars represent the mean values ± standard deviation; * P < 0.05, ** P < 0.01, *** P < 0.001; 7g, hsa-let-7g-3p; 10395, hsa-miR-10395-3p
hsa-let-7g-3p and hsa-miR-10395-3p promote metastasis and chemoresistance of GC cells by indirectly regulating the TGFβ signaling pathway
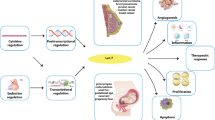
To further explore the mechanisms by which hsa-let-7g-3p and hsa-miR-10395-3p promote GC progression, we performed RNA sequencing of differentially expressed genes between hsa-miR-10395-3p mimic-transfected AGS cells and control cells (Fig. 6a). We then selected four downregulated genes, which were previously shown to be involved in tumorigenicity. Thereafter, their expression levels were examined in GC cells transfected with hsa-let-7g-3p and hsa-miR-10395-3p mimics. The qRT-PCR results demonstrated that RAPGEF3 could be a common downstream target of hsa-let-7g-3p and hsa-miR-10395-3p (Fig. 6b). Gene expression profiling and interactive analyses (GEPIA) showed that the expression of RAPGEF3 was relatively lower in GC tissues than in normal tissues as shown by the STAD cohort of the cancer genome atlas (TCGA) database (Fig. 6c) [24, 25]. Moreover, the binding sites of hsa-let-7g-3p or hsa-miR-10395-3p with the 3’UTR of RAPGEF3 were predicted using RNAhybrid (Methods) and validated by dual-luciferase reporter assays (Fig. 6d, e). As RAPGEF3 inhibits TGFβ signaling at the level of TβRI receptor activation, thereby preventing TGFβ-dependent stimulation of cell migration and inhibition of cell adhesion [26], we hypothesized that hsa-let-7g-3p and hsa-miR-10395-3p could activate the TGFβ signaling pathway by downregulating its expression. As expected, hsa-let-7g-3p and hsa-miR-10395-3p mimics subsequently activated the TGFβ signaling pathway in GC cell lines (Fig. 6f, g, Fig. S1f). Therefore, we believe that hsa-let-7g-3p and hsa-miR-10395-3p promote GC metastasis through the activation of the TGFβ signaling pathway.
hsa-let-7g-3p and hsa-miR-10395-3p bind to the 3’UTR of RAPGEF3 and indirectly regulating the TGFβ signaling pathway. a Heatmap of differential expression genes between mimics of 10,395 and NC in AGS cells using RNA-Seq data (P < 0.05). b Relative expressions of 4 candidate downstream genes in 7g or 10395 mimics and NC of AGS cells. c. Expression of RAPGEF3 in GC and normal tissues using the data from TCGA. d Predicted binding sites between 7g or 10395 with 3’UTR of RAPGEF3, validated by e dual-luciferase reporter assay. f, g Protein expressions of TGFβ pathway-associated factors in AGS, GES-1 and N87 cells validated by western blotting. Quantitative analyses of main differentially expressed proteins (TGFβ-RI, p-Smad2 and p-Smad3) were shown. Error bars represent the mean values ± standard deviation; * P < 0.05, ** P < 0.01, *** P < 0.001, **** P < 0.0001; ns, not significant; Mut, mutant sequence; WT, wild-type sequence corresponding to the mutated region; 7g, hsa-let-7g-3p; 10395, hsa-miR-10395-3p; mi, mimics; in, inhibitors; TβRI, TGFβ-RI
Discussion
By analyzing miR expression profiling, we revealed that miR signatures in exosomes of PL samples from patients with advanced GC having diminished PM were similar to those from patients without PM. Among the differentially expressed miRs between the positive (with PM) and negative groups (without PM and with diminished PM), the top two candidates, hsa-let-7 g-3p and hsa-miR-10395-3p were selected for this study.
Hsa-let-7g-3p is associated with poor prognosis in human papillomavirus-negative head and neck squamous cell carcinoma [27]. Asakage M et al. also demonstrated that hsa-let-7g-3p is a potential biomarker for uveitis [28]. To date, hsa-miR-10395-3p has not been studied in cancer research. Although a large number of researchers have focused on the application of miRs as biomarkers for the diagnosis and prognosis of cancers, there is still a need for studies focusing on the relationship between miRs in malignant ascites and the PM of GC. In this study, we found that hsa-let-7g-3p and hsa-miR-10395-3p could act as potential biomarkers for assessing the efficacy of NIPS and predicting the survival and conversion outcomes of GC patients with PM.
We enrolled 99 patients into two groups: 66 patients with PM and 33 without PM. The expression levels of hsa-let-7g-3p and hsa-miR-10395-3p were associated with PM. High expression levels of hsa-let-7g-3p and hsa-miR-10395-3p adversely influenced patient survival. Moreover, 27 patients undergoing NIPS were divided into 23 patients with diminished PM after NIPS and 4 patients with the remaining PM. The ROC curve of hsa-let-7g-3p demonstrated an ordinary predictive power for PM of GC due to the small sample. Nevertheless, it still shows higher AUCs than routinely used markers, such as CEA, CA12-5, and CA19-9, in clinical practice [29, 30]. A decrease in hsa-let-7g-3p and hsa-miR-10395-3p levels after NIPS could reflect the response to NIPS, predict the efficacy of NIPS and the feasibility of conversion gastrectomy.
Hsa-let-7g-3p and hsa-miR-10395-3p were demonstrated to contribute to the proliferation, migration, and chemoresistance of GC cells in vitro and in vivo. Furthermore, integrated analysis of RNA sequencing and qPCR results revealed that RAPGEF3 is a common downstream target of hsa-let-7g-3p and hsa-miR-10395-3p. RAPGEF3, also known as EPAC1, has been reported to be an oncogene or tumor suppressor in the process of cancer initiation and progression [31,32,33,34]. However, the effects of RAPGEF3 on PM remain unclear. Conrotto P et al. found that RAPGEF3 inhibits TGFβ signaling at the level of TβRI receptor activation, thereby preventing TGFβ-dependent stimulation of cell migration and inhibition of cell adhesion [26, 35]. Thus, hsa-let-7g-3p and hsa-miR-10395-3p could activate the TGFβ signaling pathway by downregulating RAPGEF3 (Fig. 7).
However, we encountered some limitations during the study process. Firstly, PL samples were not collected at the beginning of every cycle of NIPS, and did not comprehensively reflect the efficacy of NIPS throughout the disease span. Secondly, our study preliminarily explored a single signaling pathway regulated by hsa-let-7g-3p and hsa-miR-10395-3p. Therefore, it is necessary to explore the underlying mechanism of hsa-let-7g-3p and hsa-miR-10395-3p in tumor progression and other pathways. Finally, the sample size undergoing NIPS treatment was insufficient to demonstrate the robustness of the two markers in clinical application; thus, these findings need to be confirmed in larger sample sizes in the future.
In summary, PL-derived exosomes from patients with GC contain a large number of miRs related to PM. Hsa-let-7g-3p and hsa-miR-10395-3p can be used as potential biomarkers to evaluate the efficacy of NIPS, predict the appropriate time to perform conversion gastrectomy, and the survival and conversion outcomes of GC patients with PM. Mechanistically, hsa-let-7g-3p and hsa-miR-10395-3p activated the TGFβ signaling pathway by downregulating RAPGEF3, thereby promoting metastasis and chemoresistance of GC cells. This study provides novel insights into predicting biomarkers for PM of GC and provides important clues to delineate underlying molecular mechanism for further studies and clinical trials.
Data availability
The data that support the findings of this study are available from the corresponding author, CY, upon reasonable request.
Abbreviations
- GC:
-
Gastric cancer
- NIPS:
-
Neoadjuvant intraperitoneal and systemic chemotherapy
- PM:
-
Peritoneal metastasis
- PL:
-
Peritoneal lavage
- PTX:
-
Paclitaxel
- TGO:
-
Tegafur, gimeracil, and oteracil
- miR:
-
MicroRNA
- N87:
-
NCI-N87
- 7g:
-
Hsa-let-7g-3p
- 10395:
-
Hsa-miR-10395-3p
References
Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424.
Plummer M, Franceschi S, Vignat J, Forman D, de Martel C. Global burden of gastric cancer attributable to Helicobacter pylori. Int J Cancer. 2015;136:487–90.
Smyth EC, Nilsson M, Grabsch HI, van Grieken NC, Lordick F. Gastric cancer. Lancet. 2020;396:635–48.
Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin. 2019;69:7–34.
Siegel RL, Miller KD, Jemal A. Cancer statistics, 2020. CA Cancer J Clin. 2020;70:7–30.
Digklia A, Wagner AD. Advanced gastric cancer: current treatment landscape and future perspectives. World J Gastroenterol. 2016;22:2403–14.
Ishigami H, Yamaguchi H, Yamashita H, Asakage M, Kitayama J. Surgery after intraperitoneal and systemic chemotherapy for gastric cancer with peritoneal metastasis or positive peritoneal cytology findings. Gastric Cancer. 2017;20(Suppl 1):128–34.
Kitayama J, Ishigami H, Yamaguchi H, Yamashita H, Emoto S, Kaisaki S, et al. Salvage gastrectomy after intravenous and intraperitoneal paclitaxel (PTX) administration with oral S-1 for peritoneal dissemination of advanced gastric cancer with malignant ascites. Ann Surg Oncol. 2014;21:539–46.
Kitayama J, Ishigami H, Yamaguchi H, Yamashita H, Emoto S, Kaisaki S. S-1 plus intravenous and intraperitoneal paclitaxel for gastric cancer with peritoneal metastasis. Gastrointest Cancer Res. 2012;5:S10–3.
Chen G, Peng J, Xiao Q, Wu HX, Wu X, Wang F, et al. Postoperative circulating tumor DNA as markers of recurrence risk in stages II to III colorectal cancer. J Hematol Oncol. 2021;14:80.
Sartore-Bianchi A, Pietrantonio F, Lonardi S, Mussolin B, Rua F, Crisafulli G, et al. Circulating tumor DNA to guide rechallenge with panitumumab in metastatic colorectal cancer: the phase 2 CHRONOS trial. Nat Med. 2022;28:1612–8.
Maron SB, Chase LM, Lomnicki S, Kochanny S, Moore KL, Joshi SS, et al. Circulating tumor DNA sequencing analysis of gastroesophageal adenocarcinoma. Clin Cancer Res. 2019;25:7098–112.
Quezada-Marín JI, Lam AK, Ochiai A, Odze RD, Washington KM, Fukayama M, et al. Gastrointestinal tissue-based molecular biomarkers: a practical categorisation based on the 2019 World Health Organization classification of epithelial digestive tumours. Histopathology. 2020;77:340–50.
Lu TX, Rothenberg ME. MicroRNA. J Allergy Clin Immunol. 2018;141:1202–7.
Mishra S, Yadav T, Rani V. Exploring miRNA based approaches in cancer diagnostics and therapeutics. Crit Rev Oncol Hematol. 2016;98:12–23.
Ruan K, Fang X, Ouyang G. MicroRNAs: novel regulators in the hallmarks of human cancer. Cancer Lett. 2009;285:116–26.
Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–8.
Sun Z, Shi K, Yang S, Liu J, Zhou Q, Wang G, et al. Effect of exosomal miRNA on cancer biology and clinical applications. Mol Cancer. 2018;17:147.
Kosaka N, Iguchi H, Yoshioka Y, Takeshita F, Matsuki Y, Ochiya T. Secretory mechanisms and intercellular transfer of microRNAs in living cells. J Biol Chem. 2010;285:17442–52.
He C, Zheng S, Luo Y, Wang B. Exosome theranostics: biology and translational medicine. Theranostics. 2018;8:237–55.
Kalluri R, LeBleu VS. The biology, function, and biomedical applications of exosomes. Science. 2020. https://doi.org/10.1126/science.aau6977.
Kalluri R. The biology and function of exosomes in cancer. J Clin Invest. 2016;126:1208–15.
Tang XH, Guo T, Gao XY, Wu XL, Xing XF, Ji JF, et al. Exosome-derived noncoding RNAs in gastric cancer: functions and clinical applications. Mol Cancer. 2021;20:99.
Tang Z, Li C, Kang B, Gao G, Li C, Zhang Z. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017;45:98–102.
Tang Z, Kang B, Li C, Chen T, Zhang Z. GEPIA2: an enhanced web server for large-scale expression profiling and interactive analysis. Nucleic Acids Res. 2019;47:556–60.
Conrotto P, Yakymovych I, Yakymovych M, Souchelnytskyi S. Interactome of transforming growth factor-beta type I receptor (TbetaRI): inhibition of TGFbeta signaling by Epac1. J Proteome Res. 2007;6:287–97.
Hess J, Unger K, Maihoefer C, Schüttrumpf L, Wintergerst L, Heider T, et al. A five-microRNA signature predicts survival and disease control of patients with head and neck cancer negative for HPV infection. Clin Cancer Res Off J Am Assoc Cancer Res. 2019;25:1505–16.
Asakage M, Usui Y, Nezu N, Shimizu H, Tsubota K, Yamakawa N, et al. Comprehensive miRNA analysis using serum from patients with noninfectious uveitis. Invest Ophthalmol Vis Sci. 2020;61:4.
Zhang X, Wang X, Li W, Sun T, Diao D, Dang C. Predictive value of neutrophil-to-lymphocyte ratio for distant metastasis in gastric cancer patients. Sci Rep. 2022;12:10269.
Wang K, Jiang X, Ren Y, Ma Z, Cheng X, Li F, et al. The significance of preoperative serum carcinoembryonic antigen levels in the prediction of lymph node metastasis and prognosis in locally advanced gastric cancer: a retrospective analysis. BMC Gastroenterol. 2020;20:100.
Wehbe N, Slika H, Mesmar J, Nasser SA, Pintus G, Baydoun S, et al. The role of epac in cancer progression. Int J Mol Sci. 2020;21.
Kumar N, Gupta S, Dabral S, Singh S, Sehrawat S. Role of exchange protein directly activated by cAMP (EPAC1) in breast cancer cell migration and apoptosis. Mol Cell Biochem. 2017;430:115–25.
Kumar N, Prasad P, Jash E, Jayasundar S, Singh I, Alam N, et al. cAMP regulated EPAC1 supports microvascular density, angiogenic and metastatic properties in a model of triple negative breast cancer. Carcinogenesis. 2018;39:1245–53.
Ichikawa H, Itsumi M, Kajioka S, Maki T, Lee K, Tomita M, et al. Overexpression of exchange protein directly activated by cAMP-1 (EPAC1) attenuates bladder cancer cell migration. Biochem Biophys Res Commun. 2018;495:64–70.
Derynck R, Zhang YE. Smad-dependent and Smad-independent pathways in TGF-beta family signalling. Nature. 2003;425:577–84.
Acknowledgements
This research was supported by grants from National Natural Science Foundation of China [Grant number 81772518, 82002463 and 82173219] and Multicenter Clinical Trial of Shanghai Jiao Tong University School of Medicine [Grant number DLY201602]. The authors thank Bingya Liu, Jianfang Li and Qu Cai from Shanghai Institute of Digestive Surgery for their collection of the samples and guidance in writing the article. We would like to thank Editage (www.editage.cn) for English language editing.
Author information
Authors and Affiliations
Contributions
J.L., L.J. and C.Y. initiated and designed the experiments; J.L, C.H., M.S., Z.Y. and M.S. performed the experiments; L.J., C.H. and M.S. analyzed the data and interpreted the data; C.H., Z.Y., S.L. and C.L. provided the GC clinical samples and the relevant clinical information; J.Z., M.Y. and Z.Z. provided valuable comments; J.L. draft the manuscript; C.Y. and L.J. revised the manuscript, and supervised the study. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no competing conflict of interest.
Ethics approval and consent to participate
All institutional and national guidelines for the care and use of laboratory animals were followed. All procedures followed were in accordance with the ethical standards of the responsible committee on human experimentation (institutional and national) and with the Helsinki Declaration of 1964 and later versions. Informed consent to be included in the study, or the equivalent, was obtained from all patients.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Luo, J., Jiang, L., He, C. et al. Exosomal hsa-let-7g-3p and hsa-miR-10395-3p derived from peritoneal lavage predict peritoneal metastasis and the efficacy of neoadjuvant intraperitoneal and systemic chemotherapy in patients with gastric cancer. Gastric Cancer 26, 364–378 (2023). https://doi.org/10.1007/s10120-023-01368-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10120-023-01368-3