Abstract
Background
Immune checkpoint inhibitors (ICI) are now standard-of-care treatment for patients with metastatic gastric cancer (GC). To guide patient selection for ICI therapy, programmed death ligand-1 (PD-L1) biomarker expression is routinely assessed via immunohistochemistry (IHC). However, with an increasing number of approved ICIs, each paired with a different PD-L1 antibody IHC assay used in their respective landmark trials, there is an unmet clinical and logistical need for harmonization. We investigated the interchangeability between the Dako 22C3, Dako 28–8 and Ventana SP-142 assays in GC PD-L1 IHC.
Methods
In this cross-sectional study, we scored 362 GC samples for PD-L1 combined positive score (CPS), tumor proportion score (TPS) and immune cells (IC) using a multiplex immunohistochemistry/immunofluorescence technique. Samples were obtained via biopsy or resection of gastric cancer.
Results
The percentage of PD-L1-positive samples at clinically relevant CPS ≥ 1, ≥ 5 and ≥ 10 cut-offs for the 28–8 assay were approximately two-fold higher than that of the 22C3 (CPS ≥ 1: 70.3 vs 49.4%, p < 0.001; CPS ≥ 5: 29.1 vs 13.4%, p < 0.001; CPS ≥ 10: 13.7 vs 7.0%, p = 0.004). The mean CPS score on 28–8 assay was nearly double that of the 22C3 (6.39 ± 14.5 vs 3.46 ± 8.98, p < 0.001). At the clinically important CPS ≥ 5 cut-off, there was only moderate concordance between the 22C3 and 28–8 assays.
Conclusion
Our findings suggest that scoring PD-L1 CPS with the 28–8 assay may result in higher PD-L1 scores and higher proportion of PD-L1 positivity compared to 22C3 and other assays. Until stronger evidence of inter-assay concordance is found, we urge caution in treating the assays as equivalent.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Immune checkpoint inhibitors targeting the programmed death-1 (PD-1)/programmed death ligand-1 (PD-L1) pathway are now standard of care for patients with various advanced and metastatic cancers, including gastric cancer (GC). Several studies have demonstrated that GC with higher levels of PD-L1 expression tends to derive higher benefit from treatment with anti-PD-1 blockade [1, 2]. Several randomized phase III trials with pembrolizumab (anti-PD-1 antibody) have been designed using the Dako 22C3 assay to select PD-L1 positive populations [3, 4]. This has led to the Food and Drug Administration (FDA) approving pembrolizumab in these specific indications[5] along with the Dako 22C3 assay as a companion diagnostic [6]. Thus, evaluation of PD-L1 expression via immunohistochemistry (IHC) has become an integral part of the treatment algorithm for patients with metastatic gastric cancer.
Presently, various standardized IHC PD-L1 antibody assays (e.g. Dako 22C3, Dako 28–8 and Ventana SP-142) have been approved as companion diagnostics to predict treatment response to different ICIs in various other tumor types (pembrolizumab, nivolumab and atezolizumab) [7]. Each assay requires a different staining protocol, equipment and cut-offs, leading to a potential source of confusion among clinicians and pathologists, and a logistical hassle for laboratories and hospitals. Importantly, CheckMate-649 was a randomized phase III trial that demonstrated the benefit of the addition of nivolumab to chemotherapy in the first-line treatment of metastatic gastric or esophageal adenocarcinoma [8]. The study analyzed various PD-L1 combined positive score (CPS) subgroups as primary and secondary endpoints. While the study demonstrated benefit in the all-randomized CPS ≥ 1 and CPS ≥ 5 populations, there was significant controversy on the benefit in the PD-L1 low expressing population (CPS < 5) [1, 2, 9]. This led to different regulatory approvals in various parts of the world, with the US Food and Drug Administration (FDA) approving nivolumab regardless of CPS score, and the European Medicines Agency (EMA) approving nivolumab only for patients with a PD-L1 IHC score of CPS ≥ 5. To add further complexity, the antibody used to score CPS in CheckMate-649 was the Dako 28–8 antibody [8]. Much uncertainty exists on the concordance of CPS scores between different PD-L1 assays. With an increasing number of approved ICIs and corresponding companion IHC assays [1, 8], there is an unmet clinical need to demonstrate the concordance between these assays to allow interchangeable utilization in the clinic.
Materials and methods
Patients and tumors
This is a cross-sectional study using archival formalin-fixed, paraffin-embedded (FFPE) tissue samples from patients obtained via biopsy or resection of gastric cancer at the National University Hospital (NUH), Singapore, between 1997 and 2019. A majority of the samples were developed into a tissue microarray (TMA) while some samples were used as whole-slides for orthogonal validation. All samples selected for the TMA were derived from surgical resection specimens. The samples selected for whole-slide analysis were obtained from gastric cancer patients treated with immune checkpoint inhibitor therapy, and consisted of both surgical resection specimens and biopsies. Cases recorded to be suitable for research and with sufficient tissue for analysis were identified by the Department of Pathology, NUH. Clinicopathological data (including age at diagnosis, gender, ethnicity, disease stage, degree of differentiation and Lauren classification) was annotated for eligible cases, and de-identified. Sections from the tissue samples were prepared and submitted for conventional IHC and multiplex immunohistochemistry/immunofluorescence (mIHC/IF) analysis.
Patient and public involvement
The National Healthcare Group Domain-Specific Review Board provided ethical approval for the use of patient materials in this study (reference number 2015/00209 and 2020/00189). As this was not an interventional trial, we did not directly involve patients in the design or determination of outcome measures of this study.
mIHC/IF protocol
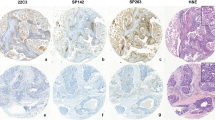
mIHC/IF was performed using an Opal Multiplex fIHC kit (Akoya Biosciences, California), as previously described [10,11,12]. In brief, FFPE tissue sections were cut onto Bond Plus slides (Leica Biosystems, Richmond) and heated at 60 °C for 20 min. The tissue slides were subjected to deparaffinization, rehydration, and heat-induced epitope retrieval using a Leica Bond Max autostainer (Leica Biosystems, Melbourne) before endogenous peroxidase blocking (Leica Biosystems, Newcastle). Next, the slides were incubated with primary antibodies followed by incubation with polymeric HRP-conjugated secondary antibodies (Leica Biosystems, Newcastle) (Fig. 1 and Supplementary Table S2). The samples were incubated with Opal fluorophore-conjugated tyramide signal amplification (TSA) (Akoya Biosciences, California) at 1:100 dilution. The slides were rinsed with wash buffer (BOND Wash Solution 10X Concentrate) after each step. Following TSA deposition, the slides were again subjected to heat-induced epitope retrieval to strip the tissue-bound primary/secondary antibody complexes before further labelling. These steps were repeated until the samples were labelled with all six markers and spectral DAPI (Akoya Biosciences, California) at a 1:10 dilution. Finally, the slides were mounted in ProLong Diamond Anti-fade Mountant (Molecular Probes, Life Technologies, USA) and developed in the dark at room temperature for 24 h. Images were captured for each case under a Vectra 3 pathology imaging system microscope (Akoya Biosciences, California) and then analyzed and scored by a pathologist using inForm software (version 2.4.2; Akoya Biosciences), cellXpress software [13] and HALO™ (Indica Lab). Particularly for PD-L1 scoring formula and algorithm, we followed a detailed protocol that our group previous reported to set up the mIHC/IF-based PD-L1 quantification for multiple PD-L1 simultaneously across cancer types [14].
CPS, TPS and IC scoring
Consecutive tissue sections from tumors were labelled to allow for the separate quantification of CPS, tumor proportion score (TPS) and immune cells (IC) using the EpCAM-labelled sample as a reference to allow tumor cell counts quantification. CPS was calculated as the number of PD-L1 staining tumor and immune cells divided by the total viable tumor cells multiplied by 100 with a maximum score of 100. TPS was calculated as the percentage of tumor cells showing staining relative to all tumor cells present in the sample. IC was calculated as the proportion of tumor area occupied by PD-L1-positive tumor infiltrating immune cells.
Analysis for sample-degradation bias
To study the effect of sample-degradation bias, a subgroup analysis was performed, based on the age of the tissue sample. Samples were divided into two cohorts: those obtained in the last 15 years (2007–2021, new cohort) and those obtained more than 15 years ago (2006 and prior, old cohort).
Statistical analysis
Statistical analyses were performed using R, version 4.0.1 (R Foundation, Vienna, Austria) and SPSS, version 25.0 (IBM Corp., Armonk, United States). Contingency tables and χ2 or Fisher’s exact tests (for categorical variables) and t test for comparison of means or Mann–Whitney U test for comparison of medians (for continuous variables) were used to investigate associations of PD-L1 IHC and mIHC/IF results with clinicopathological characteristics and outcomes. Correlations between the three antibody assays (22C3, SP-142 and 28–8) were analyzed using Spearman’s rank correlations. Spearman’s coefficient varies from 0 (no correlation) to 1 (perfect correlation) [15], with ranges interpreted as follows: 0.01–0.30 (negligible correlation), 0.30–0.50 (weak), 0.51–0.70 (moderate), 0.71–0.90 (strong) and 0.90–1.00 (very strong correlation) [16]. Concordance between the three antibody assays (22C3, SP-142 and 28–8) for each of the scoring systems (CPS, TPS and IC) was assessed using Gwet’s kappa (to account for the paradox of unbalanced main diagonal), with the value of kappa ranging from 0 to 1, with 0 representing total disagreement and 1 total agreement [17]. Values of kappa are interpreted as follows: ≤ 0 (no agreement), 0.01–0.20 (none to slight), 0.21–0.40 (fair), 0.41–0.60 (moderate), 0.61–0.80 (substantial), 0.81–1.00 (almost perfect agreement) [18]. At the same CPS cut-offs, we calculated the classification accuracy, defined as the number of correct predictions made (true positive + true negative) divided by the total number of predictions made. Survival analysis was conducted using the Kaplan–Meier method and compared between different PD-L1 expression groups using a Cox proportional hazards model. For analyses involving pairwise comparison between the antibody assays, the 22C3 assay was used as the reference for comparison with 28–8 and SP-142 assays, respectively. A p value < 0.05 was considered to indicate statistical significance unless otherwise stated.
Results
Main cohort
Patient characteristics
We conducted primary analysis on the main cohort of 344 patients which were analyzed on TMA. Most of the patients were males (236/344, 68.6%) and of Chinese ethnicity (291/344, 85.5%). The median age at diagnosis of the cohort was 68 years (IQR 16.25). Most of the patients had gastric cancer that were poorly differentiated (207/344, 60.2%), and of intestinal subtype based on Lauren classification (173/344, 50.3%). Based on a CPS ≥ 1 on the 22C3 assay, 49.4% of the patients (170/344) were PD-L1 positive. When stratified by PD-L1 positivity status based on CPS ≥ 1 using the 22C3 assay, there was no statistically significant difference in median age or distribution based on gender, ethnicity, stage of disease at diagnosis, degree of differentiation or Lauren classification subtype between both groups (Table 1).
Concordance of PD-L1 status between different assays based on CPS cut-offs
The concordance between the three PD-L1 antibody assays were analyzed at the clinically relevant CPS cut-offs of 1, 5 and 10 [1, 2, 4, 8]. Between the 28–8 and 22C3 assays, scoring with 28–8 assay consistently resulted in a higher proportion of PD-L1 positive samples (70.3 vs 49.4%, p < 0.001 at CPS ≥ 1; 29.1 vs 13.4%, p < 0.001 at CPS ≥ 5; 13.7 vs 7.0%, p = 0.004 at CPS ≥ 10) (Table 2).
At CPS ≥ 1, the classification accuracy between the 22C3 and 28–8 assays was 62.2% with only fair concordance (Gwet’s Kappa = 0.276). Notably, the classification accuracy (73.3 and 85.2%) and concordance (Gwet’s Kappa = 0.598 and 0.818) improved with increasing CPS cut-off of CPS ≥ 5 and CPS ≥ 10, respectively.
The classification accuracy and concordance between the 22C3 and SP142 assays was higher than that between the 22C3 and 28–8 assays at all CPS cut-offs (classification accuracy: 65.2 vs 62.2% at CPS ≥ 1, 80.8 vs 73.3% at CPS ≥ 5, 89.8 vs 85.2% at CPS ≥ 10; Gwet’s Kappa: 0.302 vs 0.276 at CPS ≥ 1, 0.735 vs 0.598 at CPS ≥ 5, 0.880 vs 0.818 at CPS ≥ 10). Results are summarized in Table 3.
Mean difference in continuous CPS scores obtained from different PD-L1 assays
The mean CPS score obtained from the 28–8 assay was significantly higher than both the 22C3 assay (6.39 ± 14.5 vs 3.46 ± 8.98, t(343) = − 4.083, p < 0.001) and the SP-142 assay (6.39 ± 14.5 vs 4.08 ± 10.3, t(343) = − 3.370, p = 0.001).
There was, however, no significant difference between mean CPS scores obtained on 22C3 and SP-142 (3.46 ± 8.98 vs 4.08 ± 10.3, t(343) = − 1.130, p = 0.259) (Fig. 2A).
PD-L1 scoring obtained from 22C3, 28 to 8 and SP-142 assays. A Log-transformed violin plot of CPS scores on PD-L1 obtained from 22C3, 28 to 8 and SP-142 assays among the main cohort of gastric cancer patients (n = 344). B Heat map representation of correlation between PD-L1 scoring of CPS, TPS and IC obtained from 22C3, 28 to 8 and SP-142 assays among the main cohort of gastric cancer patients (n = 344). C Heat map representation of correlation between PD-L1 scoring of CPS, TPS and IC obtained from 22C3, 28 to 8 and SP-142 assays among the additional cohort of ICI-treated gastric cancer patients (n = 18)
Correlation between continuous PD-L1 scores obtained from different PD-L1 assays
Between the 28–8 and 22C3 assays, the Spearman’s correlation values were 0.392, 0.381 and 0.230 for CPS, TPS and IC scores, respectively, suggesting consistently weak correlation (Fig. 2B and Supplementary Table S1A).
The correlations between the 28–8 and SP-142 assay were also consistently weak (Spearman’s values of 0.213, 0.180 and 0.133 for CPS, TPS, IC, respectively).
Similar to the trend in concordance analyses, correlations between 22C3 and SP142 were noted to be marginally higher (Spearman’s values of 0.409, 0.417 and 0.347 for CPS, TPS, and IC, respectively).
A subgroup analysis was performed to study the effect of sample-degradation bias found that the inter-assay variability between the various assays persisted across subgroups (Supplementary Tables S3–5), with similar differences in mean CPS scores obtained on the 28–8 and 22C3 assays in both the old (7.46 vs 3.72) and new (5.50 vs 3.23) cohorts, respectively.
Additional cohort
Patient characteristics
In this additional cohort of 18 ICI-treated GC patients (Supplementary Table S6), CPS, TPS and IC scoring with the three assays were performed on whole-slide tissue samples, to compare with the TMA samples. 83.3% of patients (15/18) were treated with nivolumab, and the remaining 16.7% (3/18) with pembrolizumab.
Orthogonal validation of PD-L1 scores obtained from different PD-L1 assays
Spearman’s value suggested weak correlation between the 22C3 and 28–8 assays, similar to the trend observed in the main cohort. (Fig. 2C and Supplementary Table S1B).
Additionally, we assessed the correlation between PD-L1 scores from the three antibody assays in a combined analysis comprising both the main and additional cohort (Supplementary Table S1C). The correlations between CPS scores obtained on the 22C3 and 28–8 assays were similar across all three analyses (TMA only = 0.392, whole-slide only = 0.360, TMA and whole-slide combined = 0.414) (Supplementary Tables S1A–S1C). In this cohort, 11 (61.1%) samples were derived from endoscopic biopsy while the rest were derived from surgical resection. The proportion of PD-L1 positivity did not differ significantly between biopsy and resected specimens (Supplementary Table S7).
Correlation between CPS scores and survival with immunotherapy treatment
In the main TMA cohort, CPS positivity status did not guide the treatment modality as this patient database predated the advent of mainstream immunotherapy in gastric adenocarcinoma treatment. Therefore, to assess any relation between PD-L1 status and survival, we also performed an exploratory survival analysis in this additional cohort of 18 ICI-treated patients.
In these patients treated with immunotherapy, though statistical significance was not observed in this small cohort, univariate Cox regression demonstrated a trend toward improved overall survival in PD-L1-positive patients at the CPS cut-off of ≥ 5 on the 22C3 and 28–8 assays. (Supplementary Figure S1 and Supplementary Table S8).
Discussion
To address the interchangeability of the various commercially available assays, we performed one of the largest comparisons of PD-L1 assays in gastric adenocarcinoma with more than 350 samples. To our knowledge, our study is the first large-scale study to harness mIHC to comprehensively score PD-L1 expression on a single slide and assess the interchangeability of the different PD-L1 assays in a gastric cancer dataset. Given the spatial heterogeneity of gastric cancer [19], there are advantages of performing mIHC on a single slide compared to conventional IHC on consecutive slides to study the various PD-L1 assays.
One of the first FDA approvals for ICI in GC was pembrolizumab as third-line treatment for GC patients with CPS ≥ 1. This was based on the results of the KEYNOTE-059 study, with the 22C3 assay approved as the companion diagnostic [2]. More recently, the EMA approved nivolumab in combination with chemotherapy for the first-line treatment of metastatic GC in patients with CPS ≥ 5. This approval was based on the results of the CheckMate 649 study [8], which utilized the Dako 28–8 assay. As pathology labs across the globe were already performing GC PD-L1 CPS score using the 22C3 assay, we paid particular attention to the inter-assay concordance at CPS ≥ 5 between 22C3 and 28–8 [20]. Considering the significantly higher PD-L1 positivity rate using 28–8 compared to 22C3 especially at CPS ≥ 5, our findings suggest that using the 22C3 assay in lieu of the 28–8 assay will likely result in fewer patients eligible for first-line nivolumab if prescribed as per EMA indication. However, it must be noted that there remains significant controversy on the use of ICI in PD-L1 low expressing tumors (CPS < 10), and the clinical impact of this difference between assays needs to be established [9]. This is probably best done by performing both the 22C3 and 28–8 assays in the pivotal trials that have been conducted such as CheckMate-649 or KEYNOTE-590, a trial which led to the indication of first-line pembrolizumab in esophageal cancer in CPS ≥ 10, independent of histology and, therefore, includes adenocarcinomas of the gastro-esophageal junction [21].
This variability in PD-L1 scores observed between the different assays may thus present a dilemma for clinicians in determining patient eligibility for ICI therapy. This is confounded by the difference in the IHC antibody assays used in landmark clinical trials. Notably, trials utilizing different PD-L1 assays have reported PD-L1 positivity proportions that are remarkably consistent with our study. The CheckMate 649 study, which used the 28–8 assay, reported double the prevalence (60%) of patients with CPS ≥ 5, compared to the KEYNOTE-061 study (31%), which utilized the 22C3 assay [2, 8].
Furthermore, in a recent smaller study by Ahn et al. (n = 55), though the 22C3 and 28–8 assays were found to be comparable at various CPS cut-offs, they similarly reported that the CPS scores on 28–8 assay was more frequently higher than that of the 22C3 assay, than vice versa (29% vs 4%) and the 28–8 assay more often detected PD-L1 expression in immune cells [7]. While other factors such as heterogeneity in patient population and tumor characteristics may also play a role [22, 23], this phenomenon is likely explained by the analytical discordance between the various assays, especially since similar observations were made in other tumor types with the same antibody assays [24, 25].
Overall, these findings hence do not support the interchangeability of the assays in determining the PD-L1 status of gastric adenocarcinoma. Despite the logistical concerns associated with conducting different assays on the same patient, it may still be necessary to use the distinct assays as companion diagnostics to predict treatment response to their respective ICIs.
Some limitations of our study must be acknowledged. First, our main cohort mIHC analyses were conducted using TMA instead of whole slides. There may be concern that the small tumor area technique may not capture the true heterogeneity in biomarker expression as accurately as larger volume samples [26]. To mitigate this limitation, we included a second cohort of whole-slide mIHC analysis as an orthogonal validation, which reaffirmed the comparability of TMA and whole-slide analyses in our study. Second, marker intensity measured via mIHC may be less robust compared to conventional IHC [27]. These drawbacks can be mitigated by a number of steps during mIHC use, including (1) the confirmation of tumor classification activity, (2) the exclusion of abundant macrophage presence, (3) avoidance of slides with large areas of spurious staining, and (4) confirmation of lower threshold sensitivity levels [28]. This is consistent with our group’s previous demonstration that PD-L1 scoring results obtained from mIHC are concordant to those obtained through conventional IHC methods [14]. Therefore, it is unlikely for the use of mIHC to have substantially biased our results. The age of tissue sample may play a role in sample-degradation bias. A study has previously reported that PD-L1 expression in freshly obtained biopsies (≤ 42 days) had higher positivity rates compared to older samples, in particular those > 900 days [29]. As the samples from our cohort were of varying ages, we performed a subgroup analysis, based on the age of the samples and confirmed that the inter-assay variability between the 22C3 and 28–8 assays persisted across subgroups. Further, the frequency of PD-L1 positivity in our cohort is not dissimilar to those reported in other studies and trials (CPS ≥ 1: our cohort 49 to 70%, CheckMate-649 [8] 60%, Ahn et al. [7] 46 to 49%). However, as CheckMate-649 was a first-line study, it is likely the samples analyzed were freshly obtained which might explain the slightly higher PD-L1 positivity rates. Finally, inter-assay concordance may not necessarily translate to equivalent survival outcomes with immunotherapy treatment—this should be investigated in future cohorts.
Conclusion
In a large cohort of gastric adenocarcinoma patients, the percentage of PD-L1-positive samples at various CPS cut-offs for the 28–8 assay were approximately twofold higher than that of the 22C3 assay, with only moderate concordance between the 22C3 and 28–8 assays at CPS ≥ 5. These findings do not support the interchangeability of the assays for determining the PD-L1 status of gastric adenocarcinoma, at the clinically relevant CPS cut-off of ≥ 5. Until stronger evidence of inter-assay concordance is found, we urge caution in treating the various assays as equivalent.
References
Shitara K, Ozguroglu M, Bang YJ, Di Bartolomeo M, Mandala M, Ryu MH, et al. Pembrolizumab versus paclitaxel for previously treated, advanced gastric or gastro-oesophageal junction cancer (KEYNOTE-061): a randomised, open-label, controlled, phase 3 trial. Lancet. 2018;392(10142):123–33. https://doi.org/10.1016/S0140-6736(18)31257-1 (Epub 2018/06/09 PubMed PMID: 29880231).
Fuchs CS, Doi T, Jang RW, Muro K, Satoh T, Machado M, et al. Safety and efficacy of pembrolizumab monotherapy in patients with previously treated advanced gastric and gastroesophageal junction cancer: phase 2 clinical KEYNOTE-059 trial. JAMA Oncol. 2018;4(5):e180013. https://doi.org/10.1001/jamaoncol.2018.0013 (Epub 2018/03/16 PubMed PMID: 29543932; PubMed Central PMCID: PMCPMC5885175).
Shitara K, Van Cutsem E, Bang YJ, Fuchs C, Wyrwicz L, Lee KW, et al. Efficacy and safety of pembrolizumab or pembrolizumab plus chemotherapy vs chemotherapy alone for patients with first-line, advanced gastric cancer: the KEYNOTE-062 phase 3 randomized clinical trial. JAMA Oncol. 2020;6(10):1571–80. https://doi.org/10.1001/jamaoncol.2020.3370 (Epub 2020/09/04 PubMed PMID: 32880601; PubMed Central PMCID: PMCPMC7489405).
Sun JM, Shen L, Shah MA, Enzinger P, Adenis A, Doi T, et al. Pembrolizumab plus chemotherapy versus chemotherapy alone for first-line treatment of advanced oesophageal cancer (KEYNOTE-590): a randomised, placebo-controlled, phase 3 study. Lancet. 2021;398(10302):759–71. https://doi.org/10.1016/S0140-6736(21)01234-4 (Epub 2021/08/30 PubMed PMID: 34454674).
Drugs@FDA: FDA-approved drugs. https://www.accessdata.fda.gov/scripts/cder/daf/index.cfm?event=overview.process&varApplNo=125514. Accessed 4 Nov 2021
List of cleared or approved companion diagnostic devices (In Vitro and Imaging Tools) U.S. Food and Drug Administration. https://www.fda.gov/medical-devices/in-vitro-diagnostics/list-cleared-or-approved-companion-diagnostic-devices-in-vitro-and-imaging-tools. Accessed 15 July 2021
Ahn S, Kim KM. PD-L1 expression in gastric cancer: interchangeability of 22C3 and 28–8 pharmDx assays for responses to immunotherapy. Mod Pathol. 2021;34(9):1719–27. https://doi.org/10.1038/s41379-021-00823-9 (Epub 2021/05/19 PubMed PMID: 34002009).
Janjigian YY, Shitara K, Moehler M, Garrido M, Salman P, Shen L, et al. First-line nivolumab plus chemotherapy versus chemotherapy alone for advanced gastric, gastro-oesophageal junction, and oesophageal adenocarcinoma (CheckMate 649): a randomised, open-label, phase 3 trial. Lancet. 2021;398(10294):27–40. https://doi.org/10.1016/S0140-6736(21)00797-2 (Epub 2021/06/09 PubMed PMID: 34102137; PubMed Central PMCID: PMCPMC8436782).
Zhao JJ, Yap DWT, Chan YH, Tan BKJ, Teo CB, Syn NL, et al. Low programmed death-ligand 1-expressing subgroup outcomes of first-line immune checkpoint inhibitors in gastric or esophageal adenocarcinoma. J Clin Oncol. 2022;40(4):392–402. https://doi.org/10.1200/JCO.21.01862 (Epub 2021/12/04 PubMed PMID: 34860570).
Lim JCT, Yeong JPS, Lim CJ, Ong CCH, Wong SC, Chew VSP, et al. An automated staining protocol for seven-colour immunofluorescence of human tissue sections for diagnostic and prognostic use. Pathology. 2018;50(3):333–41. https://doi.org/10.1016/j.pathol.2017.11.087 (Epub 2018/02/13 PubMed PMID: 29429740).
Yeong J, Thike AA, Lim JC, Lee B, Li H, Wong SC, et al. Higher densities of Foxp3(+) regulatory T cells are associated with better prognosis in triple-negative breast cancer. Breast Cancer Res Treat. 2017;163(1):21–35. https://doi.org/10.1007/s10549-017-4161-4 (Epub 2017/02/25 PubMed PMID: 28233108).
Cheung CCL, Goh D, Lim X, Tien TZ, Lim JCT, Lee JN, et al. Residual SARS-CoV-2 viral antigens detected in GI and hepatic tissues from five recovered patients with COVID-19. Gut. 2021. https://doi.org/10.1136/gutjnl-2021-324280.
Laksameethanasan D, Tan R, Toh G, Loo L-H. cellXpress: a fast and user-friendly software platform for profiling cellular phenotypes. BMC Bioinform. 2013. https://doi.org/10.1186/1471-2105-14-S16-S4 (Epub 10/22 PubMed PMID: 24564609).
Yeong J, Tan T, Chow ZL, Cheng Q, Lee B, Seet A, et al. Multiplex immunohistochemistry/immunofluorescence (mIHC/IF) for PD-L1 testing in triple-negative breast cancer: a translational assay compared with conventional IHC. J Clin Pathol. 2020;73(9):557–62. https://doi.org/10.1136/jclinpath-2019-206252 (Epub 2020/01/24 PubMed PMID: 31969377).
Mukaka MM. Statistics corner: a guide to appropriate use of correlation coefficient in medical research. Malawi Med J. 2012;24(3):69–71 (PubMed PMID: 23638278; PubMed Central PMCID: PMCPMC3576830).
DE Hinkle WW, Jurs SG. Applied statistics for the behavioral sciences. 5th ed. Boston: Houghton Mifflin; 2003.
Krippendorff K (2011) Computing Krippendorff's Alpha-Reliability. Retrieved from https://repository.upenn.edu/asc_papers/43.
McHugh ML. Interrater reliability: the kappa statistic. Biochem Med (Zagreb). 2012;22(3):276–82 (Epub 2012/10/25. PubMed PMID: 23092060; PubMed Central PMCID: PMCPMC3900052).
Sundar R, Liu DH, Hutchins GG, Slaney HL, Silva AN, Oosting J, et al. Spatial profiling of gastric cancer patient-matched primary and locoregional metastases reveals principles of tumour dissemination. Gut. 2021;70(10):1823–32. https://doi.org/10.1136/gutjnl-2020-320805 (Epub 2020/11/25 PubMed PMID: 33229445; PubMed Central PMCID: PMCPMC8458060).
Bristol myers squibb receives European commission approval for opdivo (nivolumab) + chemotherapy for patients with HER2 negative, advanced or metastatic gastric, gastroesophageal junction or esophageal adenocarcinoma 2021. https://news.bms.com/news/corporate-financial/2021/Bristol-Myers-Squibb-Receives-European-Commission-Approval-for-Opdivo-nivolumab--Chemotherapy-for-Patients-with-HER2-Negative-Advanced-or-Metastatic-Gastric-Gastroesophageal-Junction-or-Esophageal-Adenocarcinoma-/default.aspx. Accessed 22 Nov 2021
Kato K, Shah MA, Enzinger P, Bennouna J, Shen L, Adenis A, et al. KEYNOTE-590: phase III study of first-line chemotherapy with or without pembrolizumab for advanced esophageal cancer. Future Oncol. 2019;15(10):1057–66. https://doi.org/10.2217/fon-2018-0609 (Epub 2019/02/09 PubMed PMID: 30735435).
Fassan M, Brignola S, Pennelli G, Alberti G, Angerilli V, Bressan A, et al. PD-L1 expression in gastroesophageal dysplastic lesions. Virchows Arch. 2020;477(1):151–6. https://doi.org/10.1007/s00428-019-02693-8 (Epub 2019/11/15 PubMed PMID: 31724072).
Lei M, Siemers NO, Pandya D, Chang H, Sanchez T, Harbison C, et al. Analyses of PD-L1 and inflammatory gene expression association with efficacy of Nivolumab +/- Ipilimumab in gastric cancer/gastroesophageal junction cancer. Clin Cancer Res. 2021;27(14):3926–35. https://doi.org/10.1158/1078-0432.CCR-20-2790 (Epub 2021/03/31 Epub 2021/03/31).
Brunnstrom H, Johansson A, Westbom-Fremer S, Backman M, Djureinovic D, Patthey A, et al. PD-L1 immunohistochemistry in clinical diagnostics of lung cancer: inter-pathologist variability is higher than assay variability. Mod Pathol. 2017;30(10):1411–21. https://doi.org/10.1038/modpathol.2017.59 (Epub 2017/07/01 PubMed PMID: 28664936).
Rimm DL, Han G, Taube JM, Yi ES, Bridge JA, Flieder DB, et al. A prospective, multi-institutional, pathologist-based assessment of 4 immunohistochemistry assays for PD-L1 expression in non-small cell lung cancer. JAMA Oncol. 2017;3(8):1051–8. https://doi.org/10.1001/jamaoncol.2017.0013 (Epub 2017/03/10 PubMed PMID: 28278348; PubMed Central PMCID: PMCPMC5650234).
Behling F, Schittenhelm J. Tissue microarrays - translational biomarker research in the fast lane. Expert Rev Mol Diagn. 2018;18(10):833–5. https://doi.org/10.1080/14737159.2018.1522252 (Epub 2018/09/11 PubMed PMID: 30198797).
Taube JM, Akturk G, Angelo M, Engle EL, Gnjatic S, Greenbaum S, et al. The Society for immunotherapy of cancer statement on best practices for multiplex immunohistochemistry (IHC) and immunofluorescence (IF) staining and validation. J Immunother Cancer. 2020. https://doi.org/10.1136/jitc-2019-000155 (Epub 2020/05/18 PubMed Central PMCID: PMCPMC7239569).
Humphries MP, Bingham V, Abdullahi Sidi F, Craig SG, McQuaid S, James J, et al. Improving the diagnostic accuracy of the PD-L1 test with image analysis and multiplex hybridization. Cancers (Basel). 2020. https://doi.org/10.3390/cancers12051114 (Epub 2020/05/06 PubMed PMID: 32365629; PubMed Central PMCID: PMCPMC7281311).
Fashoyin-Aje L, Donoghue M, Chen H, He K, Veeraraghavan J, Goldberg KB, et al. FDA approval summary: pembrolizumab for recurrent locally advanced or metastatic gastric or gastroesophageal junction adenocarcinoma expressing PD-L1. Oncologist. 2019;24(1):103–9. https://doi.org/10.1634/theoncologist.2018-0221.
Funding
RS is supported by the National Medical Research Council (NMRC) (NMRC/Fellowship/0059/2018 and NMRC/MOH/000627). All other authors have no funding to declare. As part of the Singapore Gastric Cancer Consortium, the study is also partially funded by the National Medical Research Council Open Fund-Large Collaborative Grant (OFLCG18May-0023).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
RS has received honoraria from Bristol-Myers Squibb, Lilly, Roche, Taiho, Astra Zeneca, DKSH and MSD; has advisory activity with Bristol-Myers Squibb, Merck, Eisai, Bayer, Taiho, Novartis, MSD and AstraZeneca; received research funding from MSD and Paxman Coolers; and has received travel grants from AstraZeneca, Eisai, Roche and Taiho Pharmaceutical. WPY has advisory activity with Abbvie, Amgen, AstraZeneca, BMS, Ipsen, Novartis, MSD; is a speaker with Bayer, Eisai, Lilly, Sanofi, Taiho. The rest of the authors have no conflicts of interest to declare.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Yeong, J., Lum, H.Y.J., Teo, C.B. et al. Choice of PD-L1 immunohistochemistry assay influences clinical eligibility for gastric cancer immunotherapy. Gastric Cancer 25, 741–750 (2022). https://doi.org/10.1007/s10120-022-01301-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10120-022-01301-0