Abstract
Background
Gastric cancer (GC) is an aggressive malignancy whose mechanisms of development and progression are poorly understood. The identification of prognosis-related genomic loci and genes may suffer from the relatively small case numbers and a lack of systematic validation in previous studies.
Methods
Array-based comparative genomic hybridization (aCGH) coupled with patient clinical information was applied to identify prognosis-related loci and genes with high-frequency recurrent gains in 129 GC patients. The candidate loci and genes were then validated using an independent cohort of 384 patients through branched DNA signal amplification analysis (QuantiGene assays).
Results
In the 129 patients, a copy number gain of three chromosome regions—namely, 8q22 (including ESRP1 and CCNE2), 8q24 (including MYC and TNFRSF11B), and 20q11-q13 (including SRC, MMP9, and CSE1L)—conferred poor survival for patients. In addition, the correlation between the branched DNA signal amplification analysis results and the aCGH results was analyzed in 73 of these 129 patients, and MYC, TNFRSF11B, ESRP1, CSE1L, and MMP9 were found to be well correlated. Further validation using an independent cohort (n = 384) verified that only MYC and TNFRSF11B within 8q24 are related to survival. Patients with gains in both MYC and TNFRSF11B had poorer survival than those with no gains, particularly those with noncardia GC. Gains in both of these genes were also a significant independent prognostic indicator.
Conclusions
Our results revealed that copy number gains in MYC and TNFRSF11B located at 8q24 are associated with survival in GC, particularly noncardia GC.
Similar content being viewed by others
Introduction
Gastric cancer (GC) is one of the commonest causes of cancer-related death worldwide, and develops through a multistage process that involves the accumulation of genetic and epigenetic alterations [1, 2]. Many factors can affect the prognosis of GC patients. DNA copy number amplifications are essential drivers of tumorigenesis and malignant progression of solid tumors, including GC [3–5].
High-throughput, genome-wide profiling of DNA copy number variations (CNVs) has led to the discovery of regions that are recurrently amplified in GC. In addition, these regions house key oncogenes involved in GC progression, including MYC in 8q, SRC and MMP9 in 20q, ERBB2 in 17q, EGFR in 7p, FGFR1 in 8p, and FGFR2 in 10q [6–11]. The focal amplification of these regions and an increased frequency of their amplification throughout the genome are also associated with invasive clinicopathological characteristics and poor disease prognosis. For example, a gain of 1q32.3 has been significantly correlated with lymph node status [12]. The degree of genetic change at the DNA copy number level was found to be well correlated with the tumor stage in diffuse-type GC [13]. Moreover, some genomic imbalance may also be related to the different locations of GC [9, 12, 14, 15], such as cytogenetic abnormalities of 5p+ and 18q−, which were identified to result in significant differences between esophagogastric junction and distal gastric tumors [16]. However, the detection of the amplification of key regions was found to be inconsistent in a previous study, in which the number of cases was relatively small and no systematic validation was performed [10, 17].
To validate the copy number alterations of multiple genes within these key regions, a high-throughput quantitative method, such as branched DNA signal amplification technology (QuantiGene assay), has been used owing to its specificity and detection sensitivity, which demonstrates excellent correlation with traditional quantitative PCR and was adopted in the FDA-launched MicroArray Quality Control project [18]. This assay can be performed on 96-well plates to quantify DNA targets through the multiplexing of three to 80 DNA targets. Using this technology, researchers have found that the CNV of a specific region associated with pancreatic cancer risk is likely due to an effect on the long-term regulation of CDKN2B [19]. Moreover, the QuantiGene assay may be more significant in cancer research owing to its high accuracy and convenience for use with many types of samples, including blood, tissues, and, most importantly, formalin-fixed paraffin-embedded (FFPE) slides, which are easily accessed in a clinical setting [20].
In this study, we used a high-resolution array-based comparative genomic hybridization (aCGH) coupled with patient clinical information to identify prognosis-related chromosome loci and genes that may functionally contribute to tumor progression in a training cohort of 129 GC patients. Then, using an independent validation cohort of 384 patients, the relationship between the copy number gains of selected cancer-related genes within candidate genomic loci detected by the QuantiGene assay from FFPE samples and patient survival was further verified.
Materials and methods
Patients and tissue specimens
All of the patients with GC included in this study received their diagnosis and were surgically treated at Peking University Cancer Hospital between 1999 and 2007 and were followed up to January 2013. The training cohort comprised 129 patients, including 94 males and 35 females (mean age 60 years, range 34–81 years). A total of 129 frozen cancer tissues and 22 matched noncancerous mucosa samples were subjected to aCGH. The validation cohort comprised 384 patients not included in the training cohort, including 264 males and 120 females (mean age 58.4 years, range 22–81 years). This investigation was performed after approval by the Ethics Committee of Peking University. General informed consent was obtained from each patient.
After gastrectomy, resected specimens were processed routinely for macroscopic pathological assessment, and tissues were sampled and snap-frozen in liquid nitrogen. Fresh human tissues were stored at −80 °C and fixed with 10 % formalin in phosphate-buffered saline. To ensure the quality of the tissues, routine histological evaluation was performed for each sample. In brief, the deep-frozen GC tissues and noncancerous mucosa tissue samples were cryosectioned and stained with hematoxylin–eosin, and the tumor tissues containing more than 80 % tumor cells qualified for aCGH. To prepare the FFPE slides, the adjacent noncancerous mucosa tissues were removed, and only tumor tissues were used for the QuantiGene assay.
The depth of tumor invasion, histological grade, lymph node metastasis, liver metastasis, and vascular invasion were obtained from the histopathology reports. The GC stage was classified according to the tumor–node–metastasis (TNM) classification (seventh edition) recommended by the American Joint Committee on Cancer [21].
DNA-based microarray-CGH and data analysis
The DNA from the frozen tissues was isolated according to the standard phenol/chloroform protocol. The extracted DNA was assessed for genomic DNA concentration and purity using a NanoDrop 2000 spectrophotometer (Thermo Scientific, Wilmington, DE, USA). Agarose gel electrophoresis was used to assess the integrity of the genomic DNA, and aCGH was performed using the human genome comparative genomic hybridization microarray kit 244A (Agilent Technologies, Santa Clara, CA, USA). The experiments were performed as recommended by the manufacturer with some modifications [22]. Briefly, DNA (1.5 µg) samples of the cells and control (a pool of normal human blood DNA samples) were differentially labeled with Cy5 and Cy3-dUTP, respectively, purified, and then hybridized to an Agilent human genome comparative genomic hybridization 244K microarray. The gene copy number alterations were measured through log ratios using the BioDiscovery Nexus Copy Number software application (http://www.biodiscovery.com/software/nexus-copy-number/) and a program written in R (http://www.r-project.org/), which detected the same chromosomal aberrations. The gain and amplified segments were analyzed as follows: a segment presenting a recurrent gain was defined as a continuous genome region that satisfied the criteria log ratio of 0.2 or greater and frequency of 30 % or greater; a log ratio greater than 0.5 was regarded to define amplification. The segments that exhibited loss and deletion were analyzed as follows: a segment that presented recurrent loss was defined by a continuous genome region that satisfied the criteria log ratio of −0.2 or lower and frequency of 30 % or greater; a log ratio less than −0.7 was regarded to define deletion.
QuantiGene assay for quantification of DNA in FFPE tissues
Branched DNA signal amplification technology (QuantiGene assay) is a sandwich nucleic acid hybridization assay that provides a unique approach for DNA detection and quantification by amplifying the reporter signal rather than the target sequence. First, tissue homogenates from FFPE slides were prepared according to the procedure described by the manufacturer of the QuantiGene sample processing kit for FFPE tissues (Panomics, Fremont, CA, USA). Briefly, deparaffinized 5-µm-thick sections were incubated with 200 µl of homogenizing solution supplemented with 2 µl of proteinase K (50 µg/µl) overnight at 65 °C. The tissue homogenate was then separated from the debris by brief centrifugation and transferred to a new tube. The target gene DNA in the tissue homogenates was hybridized to the probe set (Table S1). Standard probe design software was used to design specific oligonucleotide probe sets for the target genes in the QuantiGene Plex 2.0 reagent system (Panomics), which provides a 400-fold signal amplification. The QuantiGene Plex 2.0 reagent system was used according to the manufacturer’s recommended protocols (Panomics). Briefly, the oligonucleotide probe set was mixed with the sample, and the mixture was added to an assay well in a 96-well plate. The target and control DNA (pool of normal human blood DNA samples) was captured during an overnight incubation at 54 °C (QuantiGene Plex 2.0). Any unbound material was removed by three washes with 200 µl of wash buffer. This step was followed by the sequential hybridization of DNA amplifier molecules, and preamplifier hybridization, amplifier hybridization, and label probe hybridization were then performed. Finally, streptavidin–phycoerythrin working reagent was added to the wells, and the plate was prepared for analysis. Chr12_p, Chr13_q, Chr16_q, SULT1A1, and RPLP0 were chosen as internal controls for the QuantiGene assay. The gene copy numbers for the cancer samples were calculated by dividing the sample values by the control values: no gain was regarded as a copy number of 2 or less, gain was regarded as a copy number greater than 2 but not greater than 4, and high copy number gain was regarded as a copy number greater than 4.
Statistical analysis
All of the analyses were performed using the program SPSS (version 16.0; SPSS, Chicago, IL, USA). Overall survival was plotted and calculated using the Kaplan–Meier method, and differences between groups were compared by the log-rank test. The Cox proportional hazards model (backward, stepwise) was used to estimate the influence of each variable on survival. The chi-squared test was performed to analyze the correlations between gene co-gain and clinicopathological variables. The Pearson test was used to analyze correlations between the aCGH and QuantiGene results. P values less than 0.05 (two-sided) were considered statistically significant.
Results
Genomic alterations in GCs detected by aCGH and identification of candidate survival-related loci and genes
We applied a genome-wide search for prognosis-related genomic loci and genes based on the results from two cohorts—namely, a training cohort comprising 129 patients and a validation cohort comprising 384 patients. The overall study design is shown in Fig. 1a.
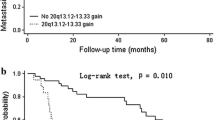
The study design and the results of the copy number variation analysis. a Overview of the study design. b Ideogram of genomic variations in 129 gastric carcinomas. Summary of chromosomal variations in 129 cases of gastric carcinoma. Red bars indicate deletion and green bars) indicate amplification. c Summary of specific regions of 8q and 20q gain in 129 cases. Each dot represents a single gene, and red dots represent gene gain (log2 ratio greater than 0.2). CGH comparative genomic hybridization, GC gastric cancer
First, 129 GC and 22 noncancerous gastric mucosa samples were profiled by aCGH. The clinical and pathological characteristics of the patients included in this training cohort are shown in Table 1. Chromosomal CNVs were observed in all 129 cancer patients, and the ideogram of the GC chromosomal abnormalities is shown in Fig. 1b. The regions that presented gains of at least 30 % in all of the cases were mainly localized on 8p, 8q, 20p, 20q, 7q, 13q, 1q, 19q, 3p, and 3q in order of decreasing frequency, whereas the 22 adjacent noncancerous mucosa samples were almost free of genomic alterations. Representative amplified regions and genes are presented in Table S2. Because DNA copy number gain and amplification are essential drivers of tumorigenesis, particularly in epithelial cancers, such as GC, our study mainly focused on the association between gene gain/amplification and survival. Representative deleted regions and genes are presented in Table S3.
The analysis of the regions that presented gains revealed that the copy number gains of three regions—namely, 8q22, 8q24, and 20q11-q13—were greater than 50 % in all of the cases. Sixty cancer-related genes located at 8q22 and 8q24 and 70 cancer-related genes located at 20q11-q13 were selected for the Kaplan–Meier survival analysis. Our results suggested that almost all of gene gains in 20q11-q13, 8q22, and 8q24 are correlated with high risk in GC patients (P < 0.05 for all except two genes, for which P = 0.06). Some well-recognized oncogenes, such as MYC (chromosome 8, gain of 62 % in all of the cases) and SRC (chromosome 20, gain of 55.8 % in all of the cases), which are also well known as amplification targets at chromosomes 8 and 20 (Fig. 1c), were affected by genomic CNVs. Both MYC and SRC gains were correlated with poor prognosis in GC patients (P = 0.012 and P = 0.035, respectively) (Fig. 2a, e). Moreover, the amplification of some other important genes (Table S1), such as TNFRSF11B, ESRP1, CCNE2, MMP9, CSE1L, BCL2L1, PLAGL2, E2F1, CD40, CEBPB, BCAS4, BMP7, PTK6, SAMD10, SNORA72, STK3, COX6C, YWHAZ, SAMD12, MAPK15, EPPK1, and PARP10, located at the three regions was also associated with poor survival (the survival plots of the first five genes are shown in Fig. 2).
Analysis of seven candidate genes located at 20q11-q13, 8q22, and 8q24 by the QuantiGene assay
Within 20q11-q13, 8q22, and 8q24, we selected seven candidate genes that are generally related to tumorigenesis, as determined in previous studies, for further testing: ESRP1 and CCNE2 at 8q22, TNFRSF11B and MYC at 8q24, and SRC, MMP9, and CSE1L at 20q11-q13. Branched DNA signal amplification, which is also known as the QuantiGene assay, which exhibits high accuracy and precision for the detection of CNVs in FFPE slides, was applied to analyze these genes in 73 of the 129 GC patients included in the training cohort (the characteristics of this test cohort are shown in Table 1). The correlations between the QuantiGene assay results and the aCGH results were analyzed. Our results suggested that there was a significant correlation between the aCGH results and the QuantiGene assay results for the MYC, CSE1L, TNFRSF11B, ESRP1, and MMP9 genes (P = 0.012, P < 0.001, P =0.001, P =0.004, and P =0.005, respectively; Table 2). The other two genes—namely, CCNE2 and SRC—showed poor correlations (P > 0.05; Table 2) and were removed from further validation.
Independent cohort validation of MYC and TNFRSF11B at 8q24
An independent validation cohort was used to validate the association between gains of MYC, TNFRSF11B, ESRP1, MMP9, and CSE1L and patient prognosis. The results including stage IV GC are shown in Fig. S1 and Table S4. To remove any excess statistical confounding factors, patients with TNM stage IV GC were excluded from the validation cohort (the characteristics of this group are shown in Table 1). The general conditions of the patients in the training and validation cohorts are comparable, with the exception of the TNM stage (Table S5). Patients with a gain of either MYC or TNFRSF11B had a shorter median overall survival than those without gain (MYC, median survival 24.92 months vs 48.78 months, P = 0.016; TNFRSF11B, median survival 26.66 months vs 45.07 months, P = 0.030; Fig. 3a, b), whereas there was no significant difference in survival between the patients with a gain of MMP9, CSE1L, or ESRP1 and those without gain (P = 0.243, P = 0.327, and P = 0.668, respectively; data not shown).
Survival analysis in the validation cohort of 384 gastric cancer patients. Two genes, MYC and TNFRSF11B, were validated to be associated with survival. a, b The association of MYC and TNFRSF11B, respectively, with survival. c, d The association of copy number (CN) variation status (high CN gain, CN greater than 4; gain, CN greater than 2 but not greater than 4; no gain, CN of 2 or less) of MYC and TNFRSF11B, respectively, with survival. e The association of concurrent gain and concurrent no gain of MYC and TNFRSF11B with survival. f, g Survival analysis in the cardia cancer subgroup and noncardia cancer subgroup, respectively, of the validation cohort. Minus sign no gain, one plus sign gene gain, two plus signs high CN gain
Moreover, the GC patients with a high copy number gain (copy number greater than 4) of MYC or TNFRSF11B exhibited even poorer overall survival than those with a copy number gain corresponding to a copy number greater than 2 but not greater than 4 and those with no gain (MYC, median survival 12.79 months vs 27.02 months vs 48.78 months, P < 0.001; TNFRSF11B, median survival 17.98 months vs 27.75 months vs 45.07 months, P = 0.012; Fig. 3c, d).
Association of 8q24 gain with survival and clinicopathological features
The survival-related genes MYC and TNFRSF11B are found within a wide range of amplified regions in 8q24, which spans approximately 20 million base pairs. There was a significant correlation between the copy number gains of MYC and TNFRSF11B, with a correlation coefficient of 0.328 (P < 0.001). In addition, 40.36 % of the patients had gains of both MYC and TNFRSF11B. We found that the patient group with gains of both MYC and TNFRSF11B was markedly segregated from the nongain group. The co-gain group had a significantly lower 5-year survival rate than the nongain group (median survival 24.92 months vs 51.94 months, P = 0.008; Fig. 3e).
The statistical analysis showed that the co-gain of MYC and TNFRSF11B was strongly associated with the depth of wall invasion, lymph node metastasis, and TNM stage in both the training cohort and the validation cohort (Table 3). A multivariate Cox proportional hazards model (Table 4) using variables that were found to be associated with survival in our study revealed that a co-gain in MYC and TNFRSF11B was a significant independent prognostic indicator in both the training cohort and the validation cohort (P = 0.004 and P = 0.032, respectively).
Association of 8q24 gain with survival in GC at different locations
Moreover, the analysis of patients with noncardia GC revealed that the patients with a co-gain of MYC and TNFRSF11B had a shorter overall survival than those with a nongain gene signature (median survival 23.08 months vs 54.57 months, P = 0.014; Fig. 3f), whereas the analysis of the patients with cardia GC revealed that the overall survival did not differ significantly between those patients with a co-gain signature and those with a nongain signature (median survival 27.02 months vs 37.11 months, P = 0.391; Fig. 3g).
Discussion
In this study, the genomic CNVs of gastric carcinoma were well characterized by aCGH using a very large training cohort of 129 patients. Our results suggested that the regions that presented gain in GC were mainly focused on chromosome 8 (p11, q11-q24), chromosome 20 (p11-p13, q12-q13.33), chromosome 7 (q22), chromosome 13 (q32.1-q34), chromosome 1 (q21.1-q22), chromosome 19 (q13), and chromosome 3 (p22, q26.31), and particularly in the regions 20q13, 20q11, 8q24, and 20p12. Our results are partially consistent with the results from two previous studies that used aCGH. In those studies, which included 30 patients [10] and 43 patients [17], respectively, the results revealed that the commonest gains observed in GC were in 7q (35 %), 8q (35 %), 7p (28 %), 1q (26 %), 13q (26 %), and 20q (21 %). However, our results are more precise owing to the inclusion of a higher number of patient samples.
Although the association between copy number gain of genomic loci and genes and survival has been widely studied by aCGH in other cancer types, a similar study in GC has not been performed. For example, 8q gains were associated with tumor progression and poor prognosis in prostate cancer [23], [24], and a gain of 8q has been associated with metastases and poor survival in patients with clear cell renal cell carcinoma [25]. Moreover, some genes located in 8p have been associated with progression and patient survival in hepatocellular carcinoma [26] and breast cancer [27]. Our study provides the first investigation of the association of gene gain and survival of GC patients by aCGH using a larger sample size (n = 129). In addition, our aCGH results suggested that 20q11-13, 8q22, and 8q24 confer poor prognosis on GC patients. An association between gains of 8q and poor survival has also been reported in prostate cancer [23, 24], renal cell carcinoma [25], and breast cancer [27].
Furthermore, the correlations between CNVs and prognosis were validated by the QuantiGene assay using a large independent cohort (n = 384). Because the treatment of TNM stage IV GC is mostly palliative and prognosis-related factors are farraginous and confounding owing to a complicated clinical intervention rather than the genetic background, patients with this stage of GC were not included in the validation cohort. With the exception of the TNM stage, the other clinicopathological characteristics of the patients in the training and validation cohorts were comparable, similarly to the designs used in previous studies [28]. Our validation results verified that gains of TNFRSF11B and MYC located at 8q24 were associated with poor survival, whereas gains of CSE1L, MMP9, and SRC located at 20q13 were not. The co-gain of TNFRSF11B and MYC was strongly associated with TNM stage and was found to be a significant independent prognostic indicator. Among these regions, MYC, a well-known oncogene that is altered in some primary tumors [29, 30], was previously reported to be associated with late-onset, intestinal-type, advanced-stage tumor, and the presence of distant metastasis in GC [31, 32]. TNFRSF11B, another gene located at 8q24 and that encodes osteoprotegerin (OPG), is a member of the tumor necrosis factor (TNF) receptor superfamily. OPG has been identified to be a prognostic marker in various cancer types, including myeloma, breast cancer, prostate cancer [33–35], and GC [36]. A number of studies have suggested that OPG protects tumor cells from the effects of TNF-related apoptosis-inducing ligand (TRAIL) [37, 38] and may thereby provide a survival advantage to OPG-producing tumor cells and contribute to carcinogenesis and cancer cell survival by driving the expression of OPG through the Wnt/β-catenin pathway [39].
Furthermore, the analysis of patients with noncardia GC revealed that those patients with a co-gain of MYC and TNFRSF11B at 8q24 had shorter overall survival than those without these alterations. A previous study also suggested that a genomic imbalance of 5p+ and 18q- may be related to the location of the GC, such as esophagogastric junction and distal gastric tumors [16]. These abnormalities may distinguish distinct tumor subtypes with identical histological features.
In conclusion, through a global correlation analysis of aCGH data coupled with GC clinicopathological information from a large cohort and further validation using a large independent cohort, our study revealed that copy number gains of MYC and TNFRSF11B located at 8q24 are associated with survival in GC, particularly noncardia GC.
References
Chen W, Gao N, Shen Y, Cen JN. Hypermethylation downregulates Runx3 gene expression and its restoration suppresses gastric epithelial cell growth by inducing p27 and caspase3 in human gastric cancer. J Gastroenterol Hepatol. 2010;25:823–31.
Tada M, Kanai F, Tanaka Y, Sanada M, Nannya Y, Tateishi K, et al. Prognostic significance of genetic alterations detected by high-density single nucleotide polymorphism array in gastric cancer. Cancer Sci. 2010;101:1261–9.
Yoshida S, Matsumoto K, Arao T, Taniguchi H, Goto I, Hanafusa T, et al. Gene amplification of ribosomal protein S6 kinase-1 and -2 in gastric cancer. Anticancer Res. 2013;33:469–75.
Marenne G, Real FX, Rothman N, Rodriguez-Santiago B, Perez-Jurado L, Kogevinas M, et al. Genome-wide CNV analysis replicates the association between GSTM1 deletion and bladder cancer: a support for using continuous measurement from SNP-array data. BMC Genomics. 2012;13:326.
Uchida M, Tsukamoto Y, Uchida T, Ishikawa Y, Nagai T, Hijiya N, et al. Genomic profiling of gastric carcinoma in situ and adenomas by array-based comparative genomic hybridization. J Pathol. 2010;221:96–105.
Peng DF, Sugihara H, Mukaisho K, Tsubosa Y, Hattori T. Alterations of chromosomal copy number during progression of diffuse-type gastric carcinomas: metaphase- and array-based comparative genomic hybridization analyses of multiple samples from individual tumours. J Pathol. 2003;201:439–50.
Morohara K, Nakao K, Tajima Y, Nishino N, Yamazaki K, Kaetsu T, et al. Analysis by comparative genomic hybridization of gastric cancer with peritoneal dissemination and/or positive peritoneal cytology. Cancer Genet Cytogenet. 2005;161:57–62.
Leung SY, Ho C, Tu IP, Li R, So S, Chu KM, et al. Comprehensive analysis of 19q12 amplicon in human gastric cancers. Mod Pathol. 2006;19:854–63.
Kang JU, Kang JJ, Kwon KC, Park JW, Jeong TE, Noh SM, et al. Genetic alterations in primary gastric carcinomas correlated with clinicopathological variables by array comparative genomic hybridization. J Korean Med Sci. 2006;21:656–65.
Tsukamoto Y, Uchida T, Karnan S, Noguchi T, Nguyen LT, Tanigawa M, et al. Genome-wide analysis of DNA copy number alterations and gene expression in gastric cancer. J Pathol. 2008;216:471–82.
Takada H, Imoto I, Tsuda H, Sonoda I, Ichikura T, Mochizuki H, et al. Screening of DNA copy-number aberrations in gastric cancer cell lines by array-based comparative genomic hybridization. Cancer Sci. 2005;96:100–10.
Weiss MM, Kuipers EJ, Postma C, Snijders AM, Pinkel D, Meuwissen SG, et al. Genomic alterations in primary gastric adenocarcinomas correlate with clinicopathological characteristics and survival. Cell Oncol. 2004;26:307–17.
Jun KH, Kim SY, Yoon JH, Song JH, Park WS. Amplification of the UQCRFS1 gene in gastric cancers. J Gastric Cancer. 2012;12:73–80.
Park CH, Rha SY, Jeung HC, Kang SH, Ki DH, Lee WS, et al. Identification of novel gastric cancer-associated CNVs by integrated analysis of microarray. J Surg Oncol. 2010;102:454–61.
Varis A, van Rees B, Weterman M, Ristimaki A, Offerhaus J, Knuutila S. DNA copy number changes in young gastric cancer patients with special reference to chromosome 19. Br J Cancer. 2003;88:1914–9.
Stocks SC, Pratt N, Sales M, Johnston DA, Thompson AM, Carey FA, et al. Chromosomal imbalances in gastric and esophageal adenocarcinoma: specific comparative genomic hybridization-detected abnormalities segregate with junctional adenocarcinomas. Genes Chromosomes Cancer. 2001;32:50–8.
Gumus-Akay G, Unal AE, Elhan AH, Bayar S, Karadayt K, Sunguroglu A, et al. DNA copy number changes in gastric adenocarcinomas: high resolution-comparative genomic hybridization study in Turkey. Arch Med Res. 2009;40:551–60.
Canales RD, Luo Y, Willey JC, Austermiller B, Barbacioru CC, Boysen C, et al. Evaluation of DNA microarray results with quantitative gene expression platforms. Nat Biotechnol. 2006;24:1115–22.
Huang LYD, Wu C, Zhai K, Jiang G, Cao G, Wang C, Liu Y, Sun M, Li Z, Tan W, Lin D. Copy number variation at 6q13 functions as a long-range regulator and is associated with pancreatic cancer risk. Carcinogenesis. 2011;33:94–100.
van Heumen BW, Roelofs HM, te Morsche RH, Nagengast FM, Peters WH. Duodenal mucosal risk markers in patients with familial adenomatous polyposis: effects of celecoxib/ursodeoxycholic acid co-treatment and comparison with patient controls. Orphanet J Rare Dis. 2013;8:181.
Ajani JA, Bentrem DJ, Besh S, D’Amico TA, Das P, Denlinger C, et al. Gastric cancer, version 2.2013: featured updates to the NCCN guidelines. J Natl Compr Cancer Netw. 2013;11:531–46.
Yu S, Bittel DC, Kibiryeva N, Zwick DL, Cooley LD. Validation of the Agilent 244 K oligonucleotide array-based comparative genomic hybridization platform for clinical cytogenetic diagnosis. Am J Clin Pathol. 2009;132:349–60.
El Gammal AT, Bruchmann M, Zustin J, Isbarn H, Hellwinkel OJ, Kollermann J, et al. Chromosome 8p deletions and 8q gains are associated with tumor progression and poor prognosis in prostate cancer. Clin Cancer Res. 2010;16:56–64.
Ribeiro FR, Jeronimo C, Henrique R, Fonseca D, Oliveira J, Lothe RA, et al. 8q gain is an independent predictor of poor survival in diagnostic needle biopsies from prostate cancer suspects. Clin Cancer Res. 2006;12:3961–70.
Klatte T, Kroeger N, Rampersaud EN, Birkhauser FD, Logan JE, Sonn G, et al. Gain of chromosome 8q is associated with metastases and poor survival of patients with clear cell renal cell carcinoma. Cancer. 2012;118:5777–82.
Roessler S, Long EL, Budhu A, Chen Y, Zhao X, Ji J, et al. Integrative genomic identification of genes on 8p associated with hepatocellular carcinoma progression and patient survival. Gastroenterology. 2012;142:957–66.e12.
Helms MW, Kemming D, Pospisil H, Vogt U, Buerger H, Korsching E, et al. Squalene epoxidase, located on chromosome 8q24.1, is upregulated in 8q+ breast cancer and indicates poor clinical outcome in stage I and II disease. Br J Cancer. 2008;99:774–80.
Kratz JR, He J, Van Den Eeden SK, Zhu ZH, Gao W, Pham PT, et al. A practical molecular assay to predict survival in resected non-squamous, non-small-cell lung cancer: development and international validation studies. Lancet. 2012;379:823–32.
Leal MF, Calcagno DQ, de Fátima Ferreira Borges da Costa J, Silva TC, Khayat AS, Chen ES, et al. MYC, TP53, and chromosome 17 copy-number alterations in multiple gastric cancer cell lines and in their parental primary tumors. J Biomed Biotechnol. 2011;2011:631268.
Calcagno DQ, Freitas VM, Leal MF, de Souza CR, Demachki S, Montenegro R, et al. MYC, FBXW7 and TP53 copy number variation and expression in gastric cancer. BMC Gastroenterol. 2013;13:141.
de Souza CR, Leal MF, Calcagno DQ, Costa Sozinho EK, Borges Bdo N, Montenegro RC, et al. MYC deregulation in gastric cancer and its clinicopathological implications. PLoS One. 2013;8:e64420.
Zhang L, Hou Y, Ashktorab H, Gao L, Xu Y, Wu K, et al. The impact of C-MYC gene expression on gastric cancer cell. Mol Cell Biochem. 2010;344:125–35.
Holen I, Cross SS, Neville-Webbe HL, Cross NA, Balasubramanian SP, Croucher PI, et al. Osteoprotegerin (OPG) expression by breast cancer cells in vitro and breast tumours in vivo—a role in tumour cell survival? Breast Cancer Res Treat. 2005;92:207–15.
Holen I, Shipman CM. Role of osteoprotegerin (OPG) in cancer. Clin Sci (Lond). 2006;110:279–91.
Chen G, Sircar K, Aprikian A, Potti A, Goltzman D, Rabbani SA. Expression of RANKL/RANK/OPG in primary and metastatic human prostate cancer as markers of disease stage and functional regulation. Cancer. 2006;107:289–98.
Ito R, Nakayama H, Yoshida K, Kuraoka K, Motoshita J, Oda N, et al. Expression of osteoprotegerin correlates with aggressiveness and poor prognosis of gastric carcinoma. Virchows Arch. 2003;443:146–51.
Neville-Webbe HL, Cross NA, Eaton CL, Nyambo R, Evans CA, Coleman RE, et al. Osteoprotegerin (OPG) produced by bone marrow stromal cells protects breast cancer cells from TRAIL-induced apoptosis. Breast Cancer Res Treat. 2004;86:269–79.
Lane D, Matte I, Rancourt C, Piche A. Osteoprotegerin (OPG) protects ovarian cancer cells from TRAIL-induced apoptosis but does not contribute to malignant ascites-mediated attenuation of TRAIL-induced apoptosis. J Ovarian Res. 2012;5:34.
De Toni EN, Thieme SE, Herbst A, Behrens A, Stieber P, Jung A, et al. OPG is regulated by β-catenin and mediates resistance to TRAIL-induced apoptosis in colon cancer. Clin Cancer Res. 2008;14:4713–8.
Acknowledgments
This work was partly funded by the National Natural Science Foundation of China (no. 81101879), the National Key Technology R&D Program (2012AA02A203-B01, 2012AA02A504-B01, and 2012AA020101), and the Beijing Municipal Science & Technology Commission (Z121100007512010 and D131100005313010).
Author information
Authors and Affiliations
Corresponding authors
Additional information
X. Wang, Y. Liu, D. Shao, and Z. Qian contributed equally to this work as co-first authors.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Wang, X., Liu, Y., Shao, D. et al. Recurrent amplification of MYC and TNFRSF11B in 8q24 is associated with poor survival in patients with gastric cancer. Gastric Cancer 19, 116–127 (2016). https://doi.org/10.1007/s10120-015-0467-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10120-015-0467-2