Abstract
In order to investigate molecular characteristics of beta-hemolytic streptococcal isolates from western Norway, we analysed the entire emm gene sequences, obtained superantigen gene profiles and determined the prevalence of the gene encoding streptococcal phospholipase A2 (SlaA) of 165 non-invasive and 34 contemporary invasive group A, C and G streptococci (GAS, GCS and GGS). Among the 25 GAS and 26 GCS/GGS emm subtypes identified, only emm3.1 was significantly associated with invasive disease. M protein size variation within GAS and GCS/GGS emm types was frequently identified. Two non-invasive and one invasive GGS possessed emm genes that translated to truncated M proteins as a result of frameshift mutations. Results suggestive of recombinations between emm or emm-like gene segments were found in isolates of emm4 and stG485 types. One non-invasive GGS possessed speC, speG, speH, speI and smeZ, and another non-invasive GGS harboured SlaA. speA and SlaA were over-represented among invasive GAS, probably because they were associated with emm3. speG dys was identified in 83% of invasive and 63% of non-invasive GCS/GGS and correlated with certain emm subtypes. Our results indicate the invasive potential of isolates belonging to emm3, and show substantial emm gene diversity and possible lateral gene transfers in our streptococcal population.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Group A streptococci (Streptococcus pyogenes, GAS) cause human disease ranging from mild skin and throat infections to necrotising fasciitis (NF) and streptococcal toxic shock syndrome (STSS). Group C streptococci (GCS) and group G streptococci (GGS) causing human infections are most often of the species Streptococcus dysgalactiae subsp. equisimilis (SDSE) and are phylogenetically related to S. pyogenes. SDSE has recently emerged as bacteria increasingly associated with invasive human infections resembling those caused by GAS [1, 2, 3]. GAS produces a variety of cell-wall-anchored virulence proteins. Among them is the antiphagocytic M-protein, an alpha-helical coiled-coil dimer anchored in the cell wall and extending from the cell surface. The basic central structure consists of conserved, variable and hypervariable repeat blocks with a seven residue periodicity (labelled C, B and A blocks respectively), and the N-terminal portion terminates in a non-helical hypervariable opsonogenic segment [4]. M proteins of GAS can be divided into class I and class II molecules based on variations in the structure of repeated segments in the conserved C-terminal region [5]. M proteins have also been identified in GCS and GGS associated with human disease and have been shown to have antiphagocytic activity and be structurally similar in their conserved domain to class I molecules of GAS [6, 7]. The emm gene encodes the M protein, and emm typing based on the nucleotide sequence encoding the 50N-terminal amino acids (aa) of the mature protein is a major epidemiological tool in surveys on GAS, GCS and GGS. The M protein is multifunctional, and the variable and conserved domains also seem to play a significant role in the pathogenesis of streptococcal disease [8, 9, 10]. Diversity across the full length of emm genes, including variations in the number of repeats in the hypervariable, variable or conserved regions of emm genes have previously been shown among GAS within types emm6, emm18 and emm28 [11, 12, 13, 14]. Furthermore, in a recent phylogenetic analysis based on the whole surface exposed M protein a high degree of M protein diversity was observed within B- and C-repeats of Belgian GAS isolates [15]. Full-length emm gene analysis is also interesting from a vaccine perspective, as both N-terminal opsonogenic fragments and epitopes from C-repeats of GAS M proteins have formed the basis of different GAS vaccine candidates [16, 17].
Phage genomes are mobile genetic elements, and phages integrated into the bacterial chromosome have accounted for up to 10% of the total genome in GAS [18]. Genes encoding the majority of the virulence-associated exoproteins called streptococcal superantigens (SAgs) are carried on phages, and the gene encoding streptococcal extracellular phospholipase A2 (SlaA) was localised on the same phage as the SAg speK in the M3 strain MGAS315 [19]. Phages are probably the primary means of lateral gene transfer among GAS, GCS and GGS, and genetic recombinations between related streptococcal species are likely to change the pathogenic potential of the recipient strains. The phage-mediated SAgs speA, speC, ssa and speM, the chromosomally encoded smeZ and the speG orthologue speG dys have previously been identified in GCS/GGS isolates [20, 21, 22], but SlaA has not previously been documented in SDSE. Studies on GAS, GCS and GGS epidemiology often include isolates associated with invasive disease only, and over-representation of certain emm types/M serotypes or streptococcal clones could merely reflect the distribution of these in the geographical area under investigation. However, results from studies comparing strains causing mild and serious infections have not been unequivocal in this respect. Although certain streptococcal emm types or clones correlate significantly with invasive disease [23, 24], other studies do not identify strains or emm types significantly associated with severe disease manifestations [25].
Invasive group A streptococcal (iGAS) disease is endemic in our community, and outbreaks of invasive streptococcal disease with different emm/M types have occurred during the last two decades [26, 27]. In order to compare molecular characteristics of isolates associated with mild and severe disease and search for evidence of horizontal gene transfers between related streptococcal strains, we have analysed the full-length emm genes, SAg gene profiles and the prevalence of SlaA in a sample of non-invasive and contemporary invasive GAS, GCS and GGS isolates from the same geographical distribution in western Norway during 2005–2006.
Materials and methods
Study population and bacterial isolates
We included GAS, GCS and GGS isolates associated with non-invasive and invasive infections in western Norway during a 13-month period from February 2005 to March 2006. The non-invasive isolates (n = 165) were the same as in a previous study [28]. The isolates associated with invasive streptococcal disease (n = 34) included all the available contemporary invasive isolates (one per patient) identified in the laboratory of bacteriology, Haukeland University Hospital. Invasive disease was defined by isolation of GAS, GCS or GGS from a normally sterile site, or from a non-sterile site in combination with streptococcal toxic shock syndrome (STSS) or necrotising fasciitis (NF). STSS was defined using criteria originally meant for GAS [29], and NF was defined as described previously [30]. The study was approved by the Privacy Appeals Board and the Regional Committee of Medical Research Ethics.
Out of the 22 isolates associated with iGAS disease, 14 were from blood, 5 were from other sterile sites (synovial fluid, peritoneal fluid or bone) and 3 were obtained from skin or soft tissue in association with NF. We identified 1 GCS and 11 GGS isolates associated with invasive disease; 10 were from blood, 1 GGS isolate was from a soft tissue biopsy in association with NF and the GCS strain was obtained from synovial fluid. All 199 isolates were beta-haemolytic and formed large colonies on blood agar. Group carbohydrate was ascertained using the Streptococcal Grouping kit (Oxoid, Cambridge, UK).
emm typing and sequence analysis
emm typing of the invasive isolates was done as described for the isolates associated with non-invasive infection [28], with previously reported primers [31]. In order to analyse the entire emm genes of the 199 isolates, the primers used for emm amplification were also used for sequencing in both directions. The emm genes predicted to encode truncated M proteins were sequenced twice. The nucleotide and predicted protein sequences downstream of the signal peptide cleavage site were analysed. Alignments of all sequences belonging to the same emm type were obtained using the ClustalW2 software program (http://www.ebi.ac.uk/Tools/clustalw2/index.html) or EMBOSS Pairwise Alignment Algorithms (http://www.ebi.ac.uk/Tools/emboss/align/) when appropriate. The sequences of different alleles of the same emm type were also aligned and analysed manually. Sequence homology to the full length of the sequences was sought using BLASTN (GenBank).
Detection of SAg genes, SlaA and the 16S ribosomal RNA gene
A multiplex PCR with primer pairs for the 11 GAS exotoxin genes speA, speC, speG, speH, speI, speJ, speK, speL, speM, ssa and smeZ was used as described [32]. In order to cover the allelic variations of smeZ, we also used a simplex PCR with an alternative primer pair [23]. Simplex PCR amplifications of speG dys and SlaA were performed with primers previously described [33]. The speG dys primers amplified gene segments of equal size in both speG dys and speG, while the speG primers included in the multiplex PCR only amplified alleles of speG. Thus, all 199 isolates were screened for the presence of the 11 GAS exotoxin genes and SlaA, while only GCS/GGS isolates were subjected to PCR with the speG dys primers. The single non-invasive GGS of type stG6.7 possessed genes encoding speC, speG, speH, speI and smeZ, and 1 of the 3 non-invasive GGS of type stG10.0 possessed SlaA. The SAg genes and SlaA amplified from GGS were sequenced twice in both directions using the same primers as for initial amplification. In order to confirm that these two GGS isolates were of the species SDSE we sequenced their 16S ribosomal RNA genes using a previously reported primer pair [34].
Nucleotide sequence accession numbers
Sequence data were assigned to GenBank accession numbers: FJ531815-FJ5319 (emm4.5, emm4.0–4, emm4.0–1, emm4.0–2, emm4.0–3), FJ531820 (emm12.0-2), FJ531821 (emm12.17), FJ531822 (emm22.3), FJ531823 (emm22.0), FJ531824 (emm28.4), FJ531825 (emm28.0-2), FJ531826 (emm49.3), FJ531827 (emm73.0), FJ531828 (emm75.0), FJ531829 (emm78.3), FJ531830 (emm80.0), FJ531831 (emm80.1), FJ531832 (emm82.0), FJ531833 (emm87.0–1), FJ531834 (emm87.0–2), FJ531835 (emm89.0–1), FJ531836 (emm89.0–3), FJ531837 (stC1400.5), FJ531838 (stC1400.0), FJ531839 (stC74a.0–2), FJ531840 (stC6979.0), FJ531841 (stCK401.3), FJ531842 (stG166b.0–1), FJ531843 (stG166b.0–2), FJ531844 (stG245.0), FJ531845 (stG245.1), FJ531846 (stG480.0), FJ531847 (stG4222.0), FJ531848 (stG485.0–1), FJ531849 (stG485.0–2), FJ531850 (stG4831.0), FJ531851–FJ531857 (stG6.0-1, stG6.0–3, stG6.0–4, stG6.0–2, stG6.3, stG6.4, stG6.5), FJ531858–FJ531862 (stG643.0–1, stG643.0–2, stG643.1–1, stG643.1–3, stG643.1–2), FJ531863–FJ531868 (stG652.0–1, stG652.0–4, stG652.0–2, stG652.0–3, stG652.1, stG652.3), FJ493181 (emm1.0–2), GQ845001 (stC74a.0–1), GQ923927 (stG6.7), GU015026 (stG6792.0), GU015027 (emm11.7), GQ923928–GQ923932 (SDSE smeZ, speC, speG, speH, speI), GQ923933 (stG10.0), GQ923934 (SDSE SlaA). The alleles emm1.0–1, emm2.0, emm3.1, emm9.0, emm12.0–1, emm28.0–1, emm77.0–1, emm77.0–2, emm82.1, emm89.0–2, emm92.0 and stG62647.0 exactly matched emm gene sequences with the following GenBank numbers respectively: CP000017, CP000260, AE014074, EF460485, CP000259, CP000056, DQ010927, AY139399, DQ010928, EU089975, EF460478 and DQ522163.
Statistical analysis
Nominal data were analysed using Stata Statistical Software; version 10 (Stata). Fisher’s exact test was used in order to assess the association between disease type (invasive or non-invasive) and emm type, SAg genes and SlaA. Because multiple comparisons were performed, both unadjusted and Bonferroni corrected p values were calculated. A two-sided p value <= 0.05 was considered significant.
Results
emm types and clinical manifestations
Table 1 shows the distribution of emm types and clinical manifestations associated with GAS, GCS and GGS disease. Seven GAS and seven GCS/GGS emm types were shared by both non-invasive and invasive isolates. Among these 14 types, emm3 was the only one significantly associated with invasive disease (unadjusted p < 0.001, Bonferroni corrected p < 0.016). All emm3 isolates belonged to subtype emm3.1 and accounted for 32% of the invasive and 4% of the non-invasive isolates. The emm sequence of the entire sequenced region was identical in these isolates. emm3, 12 and 28 accounted for 68% of the total iGAS isolates. Among the GCS/GGS isolates, the predominant types were stG485, stG6 and stG643, accounting for 59% of the invasive and 58.5% of the non-invasive isolates. Skin or soft tissue infections were the most frequent primary site of both the total non-invasive (74.5%) and invasive (41%) infections. NF was associated with both GAS (n = 3, emm types emm1.0, emm3.1 or emm28.0) and GGS (n = 1, emm type stC74a.0). STSS developed in 2 patients with NF (emm1.0 or emm3.1) and in 2 patients with primary bacteraemia or skin/soft tissue infection (stG480.0 or stG485.0).
Variations in the entire emm sequences and predicted M proteins
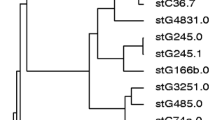
M protein size variations were inferred within 8 out of 13 GAS emm types and 7 out of 11 GCS/GGS emm types identified in two or more strains (Fig. 1). Deletions/insertions of repeated segments occurred mainly in the conserved regions of emm genes in GAS, while such variations were seen in the hypervariable, variable or conserved regions of emm genes in GCS/GGS. Nucleotide polymorphisms resulting in aa variations occurred within 5 GAS emm types and 7 GCS/GGS emm types. Novel subtypes, stG6.5 and stG6.7, were found in 2 GGS isolates, both predicted to encode truncated M proteins of only 56 and 42 residues respectively. The former was associated with severe soft tissue infection together with bacteraemia and the latter with mild skin infection. Both had unique single nucleotide deletions in the repeated segments of the hypervariable region (HVR) compared with other alleles of stG6, causing frameshift and stop codons downstream. A single nucleotide insertion in the emm gene of another non-invasive isolate of type stG652.0 caused a frameshift, and this emm gene was predicted to generate a truncated M protein of 110 aa. Eight alleles (emm1.0–2, emm78.3, emm82.1, emm89.0–1, stC1400.0, stG6.5, stG652.0–3 and stG6792.0) were exclusively found among isolates associated with invasive disease. Four out of 9 non-invasive isolates possessed an emm4 allele (emm4.0–4), which was highly divergent from the other alleles of this subtype in the conserved region, probably as a result of intergenic recombination between emm4 and the emm-like gene enn4 (Fig. 2a). Among the 13 isolates possessing stG485.0, we identified 2 alleles (stG485.0–1 and –2), which were highly divergent downstream of nucleotide 333, indicating lateral genetic transfer between different emm genes (Fig. 2b). All 3 invasive and 6 out of 10 non-invasive isolates belonging to stG485.0 possessed allele stG485.0–2.
Segments of a group A streptococci (GAS) and b group C/group G streptococci (GCS/GGS) M protein sequences differing in size mainly because of variation in the number of repeats in the hypervariable, variable and/or conserved regions. emm alleles encoding the M proteins are italicised in the left margin and alleles shown in bold type are exclusively associated with invasive isolates. Numerals representing the first amino acids of each line are placed to the left of the sequences. Dashes indicate deletions and dots indicate stop codons. To highlight our findings, repeated segments of varying sizes in the hypervariable/variable regions (pink or brown) and C-repeat sub-blocks of 28 (blue) or 7 (red, green) amino acids are shown
Analysis of the full length emm gene sequences of a allele emm4.0–4 and b alleles stG485.0–1 and stg485.0–2. Genes or gene segments deposited in GenBank with close homology to the analysed genes are shown above the lines. The percentage of homology between these gene segments and segments of the genes analysed (indicated by base numbers) is shown below the lines. The GenBank accession numbers of the sequences referred to in the figure are DQ010939 (emm4.0), Z11602 (enn4), AF239717 (stG485.0), DQ522169 (stC74a.0), FJ531842 (stG166b.0), FJ531844 (stG245.0), DQ522164 (stC839) and DQ010924 (stG4222.1). The relatively low homology between the C-terminal segment of stG485.0–1 and other emm types was mainly due to a 21 bp deletion in stG485.0–1 compared with the corresponding segments of emm genes deposited in GenBank
C-repeat analysis among GAS, GCS and GGS
Thirty-four GAS isolates (28%) had 4 C-repeats, 38 isolates (31%) had 3 C-repeats and the remaining 51 GAS isolates (41%) had 2 C-repeats. We identified 4 C-repeats in 34 GCS/GGS isolates (47%), 30 isolates had 3 C-repeats (41%), eight isolates had 5 C-repeats (11%) and only one isolate contained 2 C-repeats. We also checked the C-repeat regions for the presence of J14, a short peptide sequence that forms the basis for GAS vaccine candidates based on conserved M protein epitopes [35]. J14 was identified among all isolates of GAS class 1 M proteins (emm1, emm3, emm12 and emm80). The remaining GAS isolates possessed class II M proteins. All these isolates harboured the J14 homologue J14.1, except for four isolates possessing the recombinant emm4.0–4 allele and a single isolate harbouring emm78.3. The predicted M protein of the latter contained a J14 homologue sharing 13 out of 14 aa with J14.1. Seventy-two out of 73 predicted GCS/GGS M proteins available for C-repeat analysis (i.e. all except the three truncated M proteins) contained J14; the isolate possessing stC6979.0 contained a J14 homologue sharing 13 out of 14 aa with J14. J14.1 was not detected in any of the GCS/GGS M proteins.
SAg gene profiles and prevalence of SlaA, related to GAS emm types
Table 2 shows the distribution of SAg genes and SlaA within emm types. Each GAS emm type had a fairly similar SAg gene profile regardless of the source of the isolates. However, it is noteworthy that speC was present in all invasive isolates of five different emm types, but was detected in only some of the non-invasive isolates within 4 of these 5 types. ssa seemed to be more frequently detected among invasive than non-invasive GAS isolates possessing emm12, but there were only 3 invasive isolates. speA, detected in all isolates possessing emm3 or emm1 and in 1 non-invasive isolate harbouring emm28, and was significantly over-represented among invasive strains (unadjusted p = 0.002, Bonferroni corrected p = 0.032). speH and speI, previously reported to be on the same phage [36], were found in all isolates of types emm12, emm82, emm49 and emm73. speG was found in all GAS isolates except in all 8 isolates of emm4.0 and the 2 isolates that belonged to emm77.0, while only isolates of emm2.0 (n = 2) and emm49.3 (n = 1) failed to amplify smeZ. Thus, these isolates either lacked the same speG or smeZ or had allelic variations not detected by our primers. SlaA and speK, identified previously on the same phage [19], were detected in all 11 isolates harbouring emm3, in 1 out of 5 invasive and in 13 out of 22 non-invasive emm28 isolates, and in 2 isolates of either emm80.0 or emm77.0. However, the single isolate of emm80.1 seemed to harbour only speK, as SlaA was negative.
Identification of SAg genes and SlaA in GCS/GGS
speGdys was identified in 10 out of 12 (83%) invasive and 40 out of 64 (63%) non-invasive isolates (unadjusted p = 0.202). As shown in Table 3, the presence of this SAg gene correlated with certain emm types, and within types stC1400, stG6, stG643 and stG652 also with specific subtypes. The GGS isolate of emm type stG6.7 possessed speC, speG, speH, speI and smeZ. One out of three non-invasive GGS isolates of emm type stG10.0 harboured SlaA. The sequenced regions of the speC-H-I and SlaA genes were identical to corresponding genes previously identified in GAS and deposited in GenBank. The speH, speI and SlaA alleles found in GGS were also highly homologous to alleles of these genes identified in Streptococcus equi subsp. equi isolated from horse (Streptococcus equi subsp. equi 4047, complete genome, GenBank accession number FM204883). The sequenced region of the GGS smeZ and speG alleles identified, differs from their closest match in GAS only by a single nucleotide substitution (smeZ-3, GenBank accession number AB046865 and speG, GenBank accession numbers AM295007, CP000261, CP000259 and CP000056). Sequencing of the 16S rRNA gene confirmed the identity of these two GGS isolates as SDSE.
Discussion
To our knowledge, this is the first study comparing the full-length emm genes, the distribution of all known streptococcal SAg genes and the prevalence of SlaA among invasive and contemporary non-invasive GAS, GCS and GGS isolates. Although the relatively small sample size, short study period and lack of geographical diversity do not allow firm conclusions, emm3 seemed to be particularly associated with highly virulent GAS isolates. The prevalence of isolates belonging to other major GAS emm types, like emm12 and emm28, indicated a widespread occurrence of these in the community and not a distinct ability to cause severe disease. emm1, over-represented among isolates associated with iGAS disease in Norway during 1988–2003 and in other western countries [13, 37, 38, 39], was infrequently identified among our GAS isolates. These four types accounted for 46% of the invasive GAS isolates in a recent Strep-EURO report [40], and isolates possessing emm1 or emm3 have previously been associated with severe disease manifestations like STSS and NF [37, 41, 42]. In western Norway, emm1, emm3 and emm6 accounted for 86% of the isolates associated with iGAS disease during 1992–1994 [13], while emm89, emm1 and stG10 were the dominating types during an outbreak of severe streptococcal disease in 2002–2003 [27]. The three most prevalent GCS/GGS emm types among both non-invasive and invasive isolates in our material (stG485, stG6 and stG643) were recently reported to be frequently associated with severe GCS/GGS disease in the United States [43], and stG485 and stG6 were prevalent types among invasive GGS isolates collected in Israel, Taiwan and Japan during the last two decades [1, 2, 33]. stG10, a type that significantly correlated with invasive GCS/GGS disease in Portugal during 1998–2004 [24], was absent from our invasive sample. The relatively low prevalence of emm1 and the absence of emm6 and stG10 among our invasive isolates illustrate that predominant emm types associated with severe streptococcal disease vary with time and geographical location.
The HVRs of GAS M proteins elicit the production of protective antibodies in the host, and mutations in this region of emm genes could promote escape from immune clearance [44]. It is conceivable that GCS and GGS M proteins also contain opsonic epitopes and that antigenic variation in their HVRs could be the means of evasion from host antibody recognition. The emm type diversity in our region was illustrated by the identification of 25 GAS and 26 GCS/GGS emm subtypes. One of the three isolates with a predicted truncated M protein was from severe soft tissue infection together with bacteraemia, indicating the involvement of virulence factors other than the M protein. The frequently observed in-frame emm gene size mutations within emm types of both GAS and GCS/GGS were probably caused by homologous intragenic recombinations [12] or slipped-strand mispairing, and generated variable numbers of A-, B- and C-repeats. Such size mutations involving B- and C-repeats in emm6 and C-repeats in emm18 and emm28 have previously been reported [11, 13, 14]. It is not shown that B-repeats contain opsonogenic epitopes, but studies on the M5 protein have indicated that these segments are crucial for phagocytosis resistance [10]. The binding sites for the complement regulatory protein CD46 are located within the C-repeat of GAS M protein, and the bound CD46 mediate adherence to keratinocytes and invasion of human lung epithelial cells [8, 9]. Thus, changes in the hypervariable, variable and conserved segments of M proteins may influence many aspects of streptococcal virulence.
Interestingly, the vast majority of GAS and GCS/GCS isolates in the present study harboured either J14 or J14.1. J14 evoked opsonising antibodies against GAS isolates of many emm types, including those that harbour J14.1, in a previous study [35]. J8, a peptide fragment contained within J14, has been proposed as a GAS vaccine candidate. J8 conjugated to diphtheria toxin (DT) induced the production of opsonic antibodies against GAS in a mouse model [16], and both J8-DT- and J14-DT-immunised mice were shown to be protected from challenge with GAS strains expressing J14 or J14.1 (Michael Batzloff, Queensland Institute of Medical Research, unpublished data). In a recent study, GAS and GCS/GGS isolates from Fiji were emm and C-repeat typed. As among our streptococcal isolates, nearly all of those isolates contained either J14 or J14.1 [45]. The 26-valent GAS M protein vaccine composed of epitopes from the hypervariable end [17] would theoretically cover 86% of our invasive and 65% of our non-invasive GAS, but only 26% of the GAS isolates from Fiji. The J8 vaccine candidate would protect against a broader range of emm types, and theoretically could also induce cross-protective immunity against GCS/GGS. Therefore, a vaccine based on conserved M protein epitopes may be an alternative to a multivalent M protein vaccine in areas with a high burden of GAS, GCS and GGS disease.
Genetic recombinations between GAS, GCS and GGS involving SAg genes, neutral genes, group carbohydrate and emm genes have previously been documented [20, 21, 46, 47, 48], and such transfers may create mosaic chromosomal backgrounds and potentially alter the virulence potential of the strains involved. Our data indicate that lateral gene transfer is ongoing in our streptococcal population, although such events did not seem to be particularly associated with highly virulent strains. The identification of multiple SAg genes and SlaA in non-invasive GGS was suggestive of lateral gene transfers from GAS to GGS: The detection of speC-H-I and SlaA in GGS indicate phage-mediated genetic transfers, while we might speculate that the chromosomally encoded speG and smeZ have been transferred from GAS to GGS by conjugation. speH, speI and SlaA have to our knowledge not previously been documented in SDSE, but orthologues of these genes have been identified in strains of Streptococcus equi subsp. equi associated with clinical infection or carriage in horses [49].
The virulence gene profiles were highly conserved within most of the emm types in our material, indicating a link between emm type and phage preference. Results from previous studies have also suggested a correlation between emm type and specific SAg gene profiles in GAS [23, 50]. In line with these reports, we found that speA was highly prevalent among isolates bearing emm1 and emm3, ssa was detected in the majority of isolates of emm3 or emm4, all our emm12 isolates harboured speH, and speC was the most prevalent phage-encoded SAg. Although it is noteworthy that ssa and speC were over-represented among invasive isolates within certain emm types in our material, the small number of isolates involved do not allow firm conclusions to be drawn. The over-representation of speA and SlaA among our invasive GAS isolates probably reflects the genetic armament of isolates belonging to emm3. SlaA seems to be a virulence factor, as an isogenic SlaA mutant attenuated colonisation of epithelial cells and decreased tissue destruction compared with the wild type parental strain in a mouse model [51]. Recently, SlaA was found in the vast majority of contemporary isolates belonging to emm3 and was infrequently identified in isolates of other emm types, including emm28 [52].
Among the GGS/GCS isolates in the present study, speG dys correlated with certain emm subtypes, but was not significantly linked to invasiveness. In two previous studies from Japan, speG dys was identified in 19 out of 28 GCS/GGS isolates associated with STSS, and none of these harboured SAgs previously identified in GAS [33, 53]. Furthermore, culture supernatants from GGS associated with STSS showed no mitogenic activity towards peripheral blood mononuclear cells (PBNC), and recombinant proteins encoded by speG dys from the same bacterial isolates stimulated PBCN only weakly in a recent study [54]. These facts imply that speG dys and other SAgs might not play a major role in the pathogenesis of severe human disease caused by SDSE.
In conclusion, we found substantial emm gene diversity and possible lateral transfers of phage-and chromosomally encoded virulence genes in the natural population of GAS, GCS and GGS. The over-representation of emm3 among invasive GAS isolates in this small sample collected during a limited time period, calls for continuous epidemiological surveillance of the streptococcal population in our community and further research into the pathogenesis associated with this emm type.
References
Cohen-Poradosu R, Jaffe J, Lavi D, Grisariu-Greenzaid S, Nir-Paz R, Valinsky L, Dan-Goor M, Block C, Beall B, Moses AE (2004) Group G streptococcal bacteremia in Jerusalem. Emerg Infect Dis 10:1455–1460
Liao CH, Liu LC, Huang YT, Teng LJ, Hsueh PR (2008) Bacteremia caused by group G streptococci, Taiwan. Emerg Infect Dis 14:837–840
Rantala S, Vuopio-Varkila J, Vuento R, Huhtala H, Syrjanen J (2009) Clinical presentations and epidemiology of beta-haemolytic streptococcal bacteraemia: a population-based study. Clin Microbiol Infect 15:286–288
Fischetti VA, Parry DA, Trus BL, Hollingshead SK, Scott JR, Manjula BN (1988) Conformational characteristics of the complete sequence of group A streptococcal M6 protein. Proteins 3:60–69
Bessen D, Jones KF, Fischetti VA (1989) Evidence for two distinct classes of streptococcal M protein and their relationship to rheumatic fever. J Exp Med 169:269–283
Bisno AL, Collins CM, Turner JC (1996) M proteins of group C streptococci isolated from patients with acute pharyngitis. J Clin Microbiol 34:2511–2515
Collins CM, Kimura A, Bisno AL (1992) Group G streptococcal M protein exhibits structural features analogous to those of class I M protein of group A streptococci. Infect Immun 60:3689–3696
Okada N, Liszewski MK, Atkinson JP, Caparon M (1995) Membrane cofactor protein (CD46) is a keratinocyte receptor for the M protein of the group A streptococcus. Proc Natl Acad Sci USA 92:2489–2493
Rezcallah MS, Hodges K, Gill DB, Atkinson JP, Wang B, Cleary PP (2005) Engagement of CD46 and alpha5beta1 integrin by group A streptococci is required for efficient invasion of epithelial cells. Cell Microbiol 7:645–653
Sandin C, Carlsson F, Lindahl G (2006) Binding of human plasma proteins to Streptococcus pyogenes M protein determines the location of opsonic and non-opsonic epitopes. Mol Microbiol 59:20–30
Green NM, Beres SB, Graviss EA, Allison JE, McGeer AJ, Vuopio-Varkila J, LeFebvre RB, Musser JM (2005) Genetic diversity among type emm28 group A Streptococcus strains causing invasive infections and pharyngitis. J Clin Microbiol 43:4083–4091
Hollingshead SK, Fischetti VA, Scott JR (1987) Size variation in group A streptococcal M protein is generated by homologous recombination between intragenic repeats. Mol Gen Genet 207:196–203
Mylvaganam H, Bjorvatn B, Hofstad T, Osland A (2000) Emm gene polymorphism among temporally clustered group A streptococcal isolates in western Norway. APMIS 108:303–312
Smoot JC, Korgenski EK, Daly JA, Veasy LG, Musser JM (2002) Molecular analysis of group A Streptococcus type emm18 isolates temporally associated with acute rheumatic fever outbreaks in Salt Lake City, Utah. J Clin Microbiol 40:1805–1810
Smeesters PR, Mardulyn P, Vergison A, Leplae R, Van Melderen L (2008) Genetic diversity of group A Streptococcus M protein: implications for typing and vaccine development. Vaccine 26:5835–5842
Batzloff MR, Hayman WA, Davies MR, Zeng M, Pruksakorn S, Brandt ER, Good MF (2003) Protection against group A Streptococcus by immunization with J8-diphtheria toxoid: contribution of J8- and diphtheria toxoid-specific antibodies to protection. J Infect Dis 187:1598–1608
Mcneil SA, Halperin SA, Langley JM, Smith B, Warren A, Sharratt GP, Baxendale DM, Reddish MA, Hu MC, Stroop SD, Linden J, Fries LF, Vink PE, Dale JB (2005) Safety and immunogenicity of 26-valent group A Streptococcus vaccine in healthy adult volunteers. Clin Infect Dis 41:1114–1122
Ferretti JJ, Ajdic D, McShan WM (2004) Comparative genomics of streptococcal species. Indian J Med Res 119 Suppl]:1–6
Beres SB, Sylva GL, Barbian KD, Lei B, Hoff JS, Mammarella ND, Liu MY, Smoot JC, Porcella SF, Parkins LD, Campbell DS, Smith TM, McCormick JK, Leung DYM, Schlievert PM, Musser JM (2002) Genome sequence of a serotype M3 strain of group A Streptococcus: phage-encoded toxins, the high-virulence phenotype, and clone emergence. Proc Natl Acad Sci USA 99:10078–10083
Igwe EI, Shewmaker PL, Facklam RR, Farley MM, Van Beneden C, Beall B (2003) Identification of superantigen genes speM, ssa, and smeZ in invasive strains of beta-hemolytic group C and G streptococci recovered from humans. FEMS Microbiol Lett 229:259–264
Kalia A, Bessen DE (2003) Presence of streptococcal pyrogenic exotoxin A and C genes in human isolates of group G streptococci. FEMS Microbiol Lett 219:291–295
Sachse S, Seidel P, Gerlach D, Gunther E, Rodel J, Straube E, Schmidt KH (2002) Superantigen-like gene(s) in human pathogenic Streptococcus dysgalactiae, subsp equisimilis: genomic localisation of the gene encoding streptococcal pyrogenic exotoxin G (speG dys). FEMS Immunol Med Microbiol 34:159–167
Darenberg J, Luca-Harari B, Jasir A, Sandgren A, Pettersson H, Schalen C, Norgren M, Romanus V, Norrby-Teglund A, Henriques-Normark B (2007) Molecular and clinical characteristics of invasive group A streptococcal infection in Sweden. Clin Infect Dis 45:450–458
Pinho MD, Melo-Cristino J, Ramirez M (2006) Clonal relationships between invasive and noninvasive Lancefield group C and G streptococci and emm-specific differences in invasiveness. J Clin Microbiol 44:841–846
Johnson DR, Wotton JT, Shet A, Kaplan EL (2002) A comparison of group A streptococci from invasive and uncomplicated infections: are virulent clones responsible for serious streptococcal infections? J Infect Dis 185:1586–1595
Chelsom J, Halstensen A, Haga T, Hoiby EA (1994) Necrotizing fasciitis due to group A streptococci in western Norway: incidence and clinical features. Lancet 344:1111–1115
Mylvaganam H, Bruun T, Vindenes HA, Langeland N, Skrede S (2009) Molecular epidemiological investigation of an outbreak of invasive beta-haemolytic streptococcal infection in western Norway. Clin Microbiol Infect 15:245–252
Kittang BR, Langeland N, Mylvaganam H (2008) Distribution of emm types and subtypes among noninvasive group A, C and G streptococcal isolates in western Norway. APMIS 116:457–464
The Working Group on Severe Streptococcal Infections (1993) Defining the group A streptococcal toxic shock syndrome. Rationale and consensus definition. JAMA 269:390–391
Calandra T, Cohen J (2005) The international sepsis forum consensus conference on definitions of infection in the intensive care unit. Crit Care Med 33:1538–1548
Musser JM, Kapur V, Szeto J, Pan X, Swanson DS, Martin DR (1995) Genetic diversity and relationships among Streptococcus pyogenes strains expressing serotype M1 protein: recent intercontinental spread of a subclone causing episodes of invasive disease. Infect Immun 63:994–1003
Lintges M, Arlt S, Uciechowski P, Plumakers B, Reinert RR, Al-Lahham A, Lütticken R, Rink L (2007) A new closed-tube multiplex real-time PCR to detect eleven superantigens of Streptococcus pyogenes identifies a strain without superantigen activity. Int J Med Microbiol 297:471–478
Ikebe T, Murayama S, Saitoh K, Yamai S, Suzuki R, Isobe J, Tanaka D, Katsukawa C, Tamaru A, Katayama A, Fujinaga Y, Hoashi K, Watanabe H (2004) Surveillance of severe invasive group-G streptococcal infections and molecular typing of the isolates in Japan. Epidemiol Infect 132:145–149
Petti CA, Bosshard PP, Brandt ME, Clarridge III JE, Feldblyum TV, Foxall P, Furtado MR, Pace N, Procop G (2008) Interpretive criteria for identification of bacteria and fungi by DNA target sequencing; approved guideline. Clinical and Laboratory Standard Institute, CLSI Document MM18-A
Vohra H, Dey N, Gupta S, Sharma AK, Kumar R, McMillan D, Good MF (2005) M protein conserved region antibodies opsonise multiple strains of Streptococcus pyogenes with sequence variations in C-repeats. Res Microbiol 156:575–582
Ferretti JJ, McShan WM, Ajdic D, Savic DJ, Savic G, Lyon K, Primeaux C, Sezate S, Suvorov AN, Kenton S, Lai HS, Lin SP, Qian Y, Jia HG, Najar FZ, Ren Q, Zhu H, Song L, White J, Yuan X, Clifton SW, Roe BA, McLaughlin R (2001) Complete genome sequence of an M1 strain of Streptococcus pyogenes. Proc Natl Acad Sci USA 98:4658–4663
Meisal R, Hoiby EA, Aaberge IS, Caugant DA (2008) Sequence type and emm type diversity in Streptococcus pyogenes isolates causing invasive disease in Norway between 1988 and 2003. J Clin Microbiol 46:2102–2105
O’Loughlin RE, Roberson A, Cieslak PR, Lynfield R, Gershman K, Craig A, Albanese BA, Farley MM, Barrett NL, Spina NL, Beall B, Harrison LH, Reingold A, van Beneden C (2007) The epidemiology of invasive group A streptococcal infection and potential vaccine implications: United States, 2000–2004. Clin Infect Dis 45:853–862
Wahl RU, Lutticken R, Stanzel S, van der Linden M, Reinert RR (2007) Epidemiology of invasive Streptococcus pyogenes infections in Germany, 1996–2002: results from a voluntary laboratory surveillance system. Clin Microbiol Infect 13:1173–1178
Luca-Harari B, Darenberg J, Neal S, Siljander T, Strakova L, Tanna A, Creta R, Ekelund K, Koliou M, Tassios PT, van der Linden M, Straut M, Vuopio-Varkila J, Bouvet A, Efstratiou A, Schalén C, Henriques-Normark B, the Strep-EURO Study Group, Jasir A (2009) Clinical and microbiological characteristics of severe Streptococcus pyogenes disease in Europe. J Clin Microbiol 47:1155–1165
Luca-Harari B, Ekelund K, van der Linden M, Staum-Kaltoft M, Hammerum AM, Jasir A (2008) Clinical and epidemiological aspects of invasive Streptococcus pyogenes infections in Denmark during 2003 and 2004. J Clin Microbiol 46:79–86
Tyrrell GJ, Lovgren M, Kress B, Grimsrud K (2005) Invasive group A streptococcal disease in Alberta, Canada (2000 to 2002). J Clin Microbiol 43:1678–1683
Ahmad Y, Gertz RE Jr, Li Z, Sakota V, Broyles LN, Van Beneden C, Facklam R, Shewmaker PL, Reingold A, Farley MM, Beall BW (2009) Genetic relationships deduced from emm and multilocus sequence typing of invasive Streptococcus dysgalactiae subsp. equisimilis and S. canis recovered from isolates collected in the United States. J Clin Microbiol 47:2046–2054
Demalmanche SA, Martin DR (1994) Protective immunity to the group A Streptococcus may be only strain-specific. Med Microbiol Immunol 183:299–306
Steer AC, Magor G, Jenney AW, Kado J, Good MF, McMillan D, Batzloff M, Carapetis JR (2009) emm and C-repeat region molecular typing of beta-hemolytic streptococci in a tropical country: implications for vaccine development. J Clin Microbiol 47:2502–2509
Kalia A, Enright MC, Spratt BG, Bessen DE (2001) Directional gene movement from human-pathogenic to commensal-like streptococci. Infect Immun 69:4858–4869
Simpson WJ, Musser JM, Cleary PP (1992) Evidence consistent with horizontal transfer of the gene (emm12) encoding serotype M12 protein between group A and group G pathogenic streptococci. Infect Immun 60:1890–1893
Tanaka D, Isobe J, Watahiki M, Nagai Y, Katsukawa C, Kawahara R, Endoh M, Okuno R, Kumagai N, Matsumoto M, Morikawa Y, Ikebe T, Watanabe H (2008) Genetic features of clinical isolates of Streptococcus dysgalactiae subsp. equisimilis possessing Lancefield's group A antigen. J Clin Microbiol 46:1526–1529
Holden MT, Heather Z, Paillot R, Steward KF, Webb K, Ainslie F, Jourdan T, Bason NC, Holroyd NE, Mungall K, Quail MA, Sanders M, Simmonds M, Willey D, Brooks K, Aanensen DM, Spratt BG, Jolley KA, Maiden MCJ, Kehoe M, Chanter N, Bentely SD, Robinson C, Maskell DJ, Parkhill J, Waller AS (2009) Genomic evidence for the evolution of Streptococcus equi: host restriction, increased virulence, and genetic exchange with human pathogens. PLoS Pathog 5:e1000346
Vlaminckx BJ, Mascini EM, Schellekens J, Schouls LM, Paauw A, Fluit AC, Novak R, Verhoef J, Schmitz FJ (2003) Site-specific manifestations of invasive group A streptococcal disease: type distribution and corresponding patterns of virulence determinants. J Clin Microbiol 41:4941–4949
Sitkiewicz I, Nagiec MJ, Sumby P, Butler SD, Cywes-Bentley C, Musser JM (2006) Emergence of a bacterial clone with enhanced virulence by acquisition of a phage encoding a secreted phospholipase A2. Proc Natl Acad Sci USA 103:16009–16014
Nagiec MJ, Lei B, Parker SK, Vasil ML, Matsumoto M, Ireland RM, Beres SB, Hoe NP, Musser JM (2004) Analysis of a novel prophage-encoded group A Streptococcus extracellular phospholipase A2. J Biol Chem 279:45909–45918
Hashikawa S, Iinuma Y, Furushita M, Ohkura T, Nada T, Torii K, Hasegawa T, Ohta M (2004) Characterization of group C and G streptococcal strains that cause streptococcal toxic shock syndrome. J Clin Microbiol 42:186–192
Zhao J, Hayashi T, Saarinen S, Papageorgiou AC, Kato H, Imanishi K, Kirikae T, Abe R, Uchiyama T, Miyoshi-Akiyama T (2007) Cloning, expression, and characterization of the superantigen streptococcal pyrogenic exotoxin G from Streptococcus dysgalactiae. Infect Immun 75:1721–1729
Acknowledgements
This work was supported by the Institute of Medicine, University of Bergen.
We thank Bjørn Blomberg for helpful discussions and Rebecca E. Breistein for technical assistance. We sincerely acknowledge Shiranee Sriskandan and Mark Peter Gerhard van der Linden for providing the GAS isolates that served as positive controls in the multiplex PCR used in this study, and Michael Batzloff for sharing unpublished results with us.
Conflicts of interest
The authors declare that they have no conflicts of interest.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
Open Access This is an open access article distributed under the terms of the Creative Commons Attribution Noncommercial License (https://creativecommons.org/licenses/by-nc/2.0), which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
About this article
Cite this article
Kittang, B.R., Skrede, S., Langeland, N. et al. emm gene diversity, superantigen gene profiles and presence of SlaA among clinical isolates of group A, C and G streptococci from western Norway. Eur J Clin Microbiol Infect Dis 30, 423–433 (2011). https://doi.org/10.1007/s10096-010-1105-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10096-010-1105-x