Abstract
Detecting strong species interactions in food webs is often challenging due to difficulties related to adequate experimentation and the prevalence of generalist diets throughout nature. A promising new mathematical technique, empirical dynamic modeling (EDM), has demonstrated the potential to identify trophic interactions between populations by assessing time lags between associated time series. We attempted to analyze trophic linkages both within a subtropical estuary, as well as a simulated, theoretical ecosystem, to determine how energy moves through these systems. Additionally, we intended to evaluate the technique’s ability to detect biological relationships in ecosystems of different complexity. In both datasets, we were able to clearly identify strong consumer—resource interactions, which were generally related to bottom-up drivers. Overall, trophic connections at lower trophic levels were more easily detected than linkages higher in the food web. The ability of EDM to detect food web interactions appeared to be strongly influenced by the degree of observation error exhibited in the data. In the empirical dataset, several examples of bottom-up processes were clearly evident including effects of discharge, nutrients, and/or chlorophyll-a concentrations on anchovies (Anchoa spp.), Gulf flounder (Paralichthys albiguttata), and red drum (Sciaenops ocellatus). We also observed instances where lengths of time lags decreased as trophic level distances between consumers and resources decreased (for example, Anchovies, Gulf flounder, young-of-the-year seatrout). This analysis demonstrates the promising application of EDM to detect energetic pathways in systems of varying complexity.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Highlights
-
Bottom-up dynamics were clearly identified in simulated and empirical data
-
EDM models detected the strongest food web interactions at low trophic levels
-
A negative relationship was evident between observation error and strength of trophic interactions
Introduction
Describing energetic pathways within ecosystems is a foundational objective of ecological research. Although of significant scientific importance, the ability of ecologists to definitively characterize these pathways, and their associated drivers, is rather limited. Two principal reasons for this dilemma include the difficulties in conducting controlled experiments as well as the highly complex nature of real-world ecosystems (Hilborn and Mangel 1997). This complexity results from the abundance of species within nature, the various ways in which they interact (for example, predation, competition, grazing, decomposition), and the different environmental drivers that affect populations. In these dynamical ecosystems, an abiotic factor may directly affect the population of one species, but numerous indirect effects may also be reflected throughout the food web. Understanding these indirect effects and their causal drivers is also challenging due to state-dependent relationships among multifactorial dynamics. State-dependent relationships are nonlinear interactions between variables of a given system, whereby the associated coefficients between variables change in relation to time and the state of the dynamical system (Priestly 1980; Rahmani 2017).
One way to study complex food webs is by assessing time delays in energetic transfer. Biomass production is not instantaneous, so the effects of environmental change on biological communities must inherently be observed at some given time lag. This has been a recognized research topic in both fisheries science and ecology broadly, exhibited by the lagged effects that abiotic factors (for example, temperature and discharge) have on fish recruitment and catch (Pascual and Ellner 2000; Moraes and others 2012; Isaac and others 2016). Changes in environmental conditions can cause bottom-up effects that initially impact lower trophic level taxa, such as planktonic species, then subsequently influence larval fish abundance (Frederiksen and others 2006). These effects can often be represented in a lagged, nonlinear manner (Peebles 2002; Isaac and others 2016). In addition to bottom-up effects, top-down control can be evident in aquatic ecosystems by means of trophic cascades (Carpenter and others 1987). Variability in the populations of high trophic level predators may have ecosystem wide impacts that can be observed throughout the food web (Frank and others 2005). Like bottom-up effects, top-down drivers can create nonlinear changes within ecosystems (Pace and others 1999).
Time lags and nonlinearity can be problematic for more traditional statistical techniques, such as linear regression, or when the relationships are state dependent. A contemporary approach toward addressing these issues is with empirical dynamic modeling (EDM). The technique is a nonlinear modeling tool that relies on time series data to find causation between variables (Sugihara and others 2012; Chang and others 2017). In EDM, predictions are made based on time lags between related time series using Takens’ theorem (Takens 1981). This theorem describes the reconstruction of an attractor manifold from a time series in a dynamical system. This analysis has been utilized in a variety of disciplines for its ability to discover causal time series, as well as its forecasting potential (Deyle and others 2016a, b; Tsonis and others 2015).
In biology, EDM has generally been implemented to analyze simple, direct relationships. Examples include predator—prey relationships between unicellular Didinium and Paramecium as well as chlorophyll-a densities with respect to temperature (Veilleux 1979; Sugihara and others 2012; Ye and others 2015a). However, one of the more complex ecological interactions where EDM has been utilized was between northern anchovy (Engraulis mordax), Pacific sardine (Sardinos sagax), and sea surface temperature (Sugihara and others 2012) along the California coastline. It was previously believed that competition between Pacific sardine and northern anchovy for prey resources led to the observed decadal oscillating patterns of abundance (Murphy and Isaacs 1964), but Sugihara and others (2012) demonstrated using EDM that temperature is potentially the more influential factor driving their populations. Other examples where EDM has been applied to food web interactions include the assessment of bottom-up processes in oligotrophic lakes and the effects of climate change on biodiversity in lentic systems (Frossard and others 2018; Chang and others 2020).
Among studies utilizing EDM, these models are generally applied to lower trophic level taxa, as model performance tends to be associated with lifespan of study species (Ye and others 2015a; Munch and others 2018; Anneville and others 2019; Rogers and others 2020; Cai and others 2020; Luken 2020;). A review by Munch and others (2018) found that predictive ability of EDM was highest for species with shorter generation times.
Some of the key uncertainties in aquatic ecology include the relative importance of bottom-up versus top-down drivers, planktonic versus detrital pathways, and the role of forage fish in ecosystems, with conflicting evidence for different positions. One of the major obstacles that scientists experience when attempting to investigate these food web related hypotheses is the difficulty of conducting experiments that can mechanistically explain changes in predator—prey populations at population scale resolution. This study demonstrates how EDM can be applied to ecological research to identify species interactions and trophic pathways in food webs without the constraints of traditional experimentation.
The objectives of this study were to (1) test whether EDM can detect time lags associated with trophic linkages in a simple, theoretical system forced with bottom-up and top-down drivers, (2) analyze the influence of observation error on EDM performance, and (3) use EDM to determine time lags and species interactions in food webs using real-world long-term monitoring data of fish abundance from a diverse subtropical estuary. We assessed the efficacy of EDM using simulated time series from a dynamic food web model with different combinations of top-down and bottom-up influences and different amounts of assumed observation error added to the simulated data. We utilized EDM and the simulation model to evaluate how well the known time lags and food web interactions could be detected under top-down versus bottom-up control and with varying degrees of observation error in the simulated data. Model performance was also evaluated relative to the trophic level of study species. Following the successful application on the simulation data, we then applied EDM to determine time lags and causality from a long-term fisheries independent monitoring program and assess whether this methodology would be adequate for real world applications in field ecology and fisheries science. Based on these findings, we discuss the utility and limitations of EDM for ecological research in systems with multifactorial, nonlinear dynamics.
Methods
Simulated Datasets for EDM
We constructed the operating model using Ecopath with Ecosim (EwE, Christensen and Walters 2004), because it provides a framework to simulate trophic dynamics, can represent both bottom-up and top-down food web processes, and it has been demonstrated to replicate historical patterns in real-world, complex ecosystems (Chagaris and others 2015a, b; Kao and others 2016; Sinnickson and others 2021). EwE models typically include all trophic levels within an ecosystem, from detritus and primary producers to top predators. In Ecosim, a system of differential equations is utilized to model biological production within each functional group based on a series of input parameters and changes in predator and prey abundances. Consumption by predators in Ecosim is modeled based on foraging arena theory, whereby prey biomass is partitioned into vulnerable and invulnerable states (Christensen and others 2005; Ahrens and others 2012), and the exchange rates between those states are described by the vulnerability parameters, which govern predation rates, energy flows, and biomass dynamics. There is one vulnerability parameter (vij) for each prey i and predator j interaction, and high values (> 10) allow for strong ‘top-down’ dynamics because prey rapidly move into the vulnerable state where they can be consumed by the predator. Low vij values (vij ~ 1.0) restrict flow into the vulnerable pool, resulting in very small changes to baseline predator biomass and predation mortality rates, making the prey item ‘bottom-up’ controlled (Christensen and others 2005). Time lags in EwE are an emergent property of the model that result from the varying input parameters describing biomass production and consumption across trophic levels, such that biomass of short-lived groups (plankton) will respond more quickly to changes in prey than long-lived species. With respect to simulating datasets, the driving factors in EwE models can be controlled. For example, primary production and predator abundances can be altered, and the biomass of each functional group in the model can be simulated under those prescribed treatments. The output provided by EwE includes monthly time series of biomass for all species or functional groups (aggregated groups of species), resulting from the combination of all direct and indirect trophic processes, which provides a good test dataset for EDM.
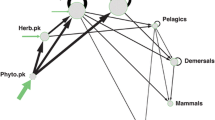
The Florida Estuary (FLE) Model
The FLE model was a simple, hypothetical model consisting of eight functional groups and two fishing fleets (Figure 1). The model included state variables for detritus, phytoplankton, zooplankton, benthic invertebrates, a forage fish, an invertivore fish, a generalist fish, and a piscivorous fish. The basic data inputs for biomass, mortality, production, consumption, and diet were based on both previous modeling experience with EwE (Sinnickson and others 2021) as well as an intuitive understanding of estuaries and energetic requirements for achieving mass-balance of the system. The hypothetical food web in the FLE model was specified to represent different trophic levels and diet breadth. Four of the consumer groups fed on just one prey item, while the piscivore and generalist fish fed on multiple prey times. In the initial stable state, the fishing mortality rates for piscivores, generalists, and forage fish were 0.2, 0.2, and 0.5, respectively. Regarding the vulnerability parameters, these values were scaled to trophic level and ranged from 1.2 to 10. Specifically, vulnerability rates for all prey items of piscivores, generalists, invertivores, forage fish, benthic invertebrates, and zooplankton were 10, 7.7, 5.5, 5.5, 1.2, and 1.2, respectively.
Due to the variable diets, vulnerabilities, and basic input parameters for our study species, we hypothesized that EDM performance may exhibit variability for the different trophic interactions throughout the model food web. To account for the differences in data inputs, we assessed EDM performance with respect to each functional group’s net efficiency. This is a comprehensive term that describes the rate at which a species assimilates consumed food into new biomass (Christensen and others 2005). The equation for net efficiency is the following:
whereby net efficiencies are positively related to production rates and negatively related to respiration rates. Additionally, net efficiencies are also related to trophic levels and turnover rates of functional groups because biomass will inherently decrease higher in the food web to ensure mass balance and fundamental thermodynamic principles. Low trophic level species will have higher production rates to account for high rates of mortality and fast generation times. Consequently, this leads to higher net efficiencies for these groups. The opposite is generally true for higher trophic level taxa, which are longer-lived species with slow turnover rates. For each food web interaction, we assessed EDM performance relative to the net efficiency rate of the respective consumer.
Simulation Scenarios
Environmental and fishing scenarios were implemented in the FLE to simulate data under assumptions of top-down and bottom-up processes on trophic linkages. Experimental treatments were conducted by utilizing forcing functions in Ecosim to drive abundances of top predators and rates of primary production within the FLE. The bottom-up (BU) scenarios were created by driving rates of primary production with an autocorrelated random time series of primary production anomalies that were applied as multipliers on the baseline production rate. In the FLE, the primary production multipliers were applied to the sole primary producer group, phytoplankton. Top-down (TD) scenarios were implemented using fishing mortality time series to drive changes in predator abundances. Variable fishing mortality was applied to the piscivore and generalist functional groups using an autocorrelated random time series of fishing effort. For both BU and TD time series, 600 random numbers were selected from a normal distribution with a mean of one and a standard deviation of 0.3. The means from these distributions were weighted (BU-2, TD-7), to generate the autocorrelation. These 600 values represented 50 years of data at a monthly time step. Simulations were also conducted using a combination of bottom-up and top-down drivers (BU + TD). Each simulation scenario produced 50 years of monthly biomass outputs which were then used as input data for the EDM analysis (Figure 2). These time series were either used directly as input to EDM or were modified to include observation error. This error was incorporated to represent realistic ecological data from an empirically collected monitoring program. The error scenarios were created by adding a normal random deviate to the Ecosim biomass output assuming coefficients of variation of 0.3 and 0.6. We conducted three environmentally driven simulations (BU, TD, BU + TD) and three runs of varying observation error (0.0, 0.3, 0.6) for a total of nine different combined scenarios (Table 1). As the FLE consisted of eight functional groups, the nine model scenarios produced a total of seventy-two simulated biomass time series.
Empirical Datasets for EDM
Long-Term Monitoring Data
In addition to using simulated model output from EwE, we also applied EDM to empirically collected ecological data on fish abundances from a subtropical estuary. Long-term monitoring data were collected by the Fisheries Independent Monitoring Program (FIM) of the Florida Fish and Wildlife Conservation Commission. Sampling was conducted from 1997 to 2018 in the Suwannee estuary using three different gear types. Briefly, two seines, 21.3 m and 183 m in length, were deployed in shallow habitats, generally along shorelines, while offshore seagrass meadows were sampled using a 6.1 m otter trawl. All sampling was conducted monthly in a stratified random design. Due to the use of multiple gear types and stratified random sampling throughout the estuary, this allowed for a robust representation of the entire fish community. Along with measuring relative abundances of estuarine fishes, water temperature data were also collected at haul sites as well.
The fish taxa evaluated in this observed time series analysis included species or aggregated taxonomic groups that were caught in at least five percent of hauls of one gear type or one percent of hauls of all gear types (Table 2). Taxa of particular economic or recreational importance were broken into age specific stanzas based on observed lengths and known age-length relationships. This included three age stanzas (young-of-year, intermediate, adult) for red drum (Sciaenops ocellatus) and spotted seatrout (Cynoscion nebulosus) and two stanzas for mullet (juvenile and adult). Species that were not caught in at least 50 months throughout the study were excluded from the analysis.
In addition to the fisheries independent monitoring data, we also constructed time series of relative fishing effort to analyze the impacts of top-down, anthropogenic drivers. These data were collected monthly by the National Oceanic and Atmospheric Administration’s (NOAA) Marine Recreational Information Program (MRIP) (NOAA 2018) using the raw data and custom domain analysis programs provided by the MRIP website. We partitioned the MRIP data into four fishing fleet time series. These data included the number of trips taken by private and charter fleets, as well as the total trips taken specifically targeting spotted seatrout or red drum. Limitations in the MRIP data prevented us from developing an estuary specific fishing effort time series. Therefore, data from counties surrounding the Suwannee River estuary (for example, Levy, Dixie, and Citrus Counties) were utilized to create relative time series of fishing effort, and we assumed that trends in recreational fishing effort from the adjacent counties would be representative of the effort occurring in the Suwannee River estuary itself.
Environmental data from the Suwannee estuary were also assembled as potential bottom-up drivers, including measurements of discharge (cf/s), total nutrient concentrations (nitrogen and phosphorus, µg/L), and chlorophyll-a (µg/L). Discharge (cf/s) data were collected monthly from the United States Geological Survey (USGS) at its Wilcox station, which is approximately 40 km upstream of the mouth of the Suwannee River (USGS 2020). Nutrient and chlorophyll-a data were both collected by the Project COAST program, which samples monthly at ten fixed locations in the Suwannee estuary (unpublished data Frazer 2018). These environmental metrics were utilized to represent bottom-up drivers on the food web. After assembling all fish and environmental variables and interpolating over months with missing data, the empirical time series ranged from 1997 to 2015 sequenced at monthly time steps.
Fish Abundance Time Series Standardization
Because of variable environmental conditions that affected the catchability of study species, we standardized the fish abundance densities using delta lognormal generalized linear models (GLM) (Lo and others 1992; Maunder and Punt 2004; Grüss and others 2019). Variables for which we controlled included depth, month, location, gear type, year, temperature, dissolved oxygen, salinity, bottom type (for example, mud, oyster, sand, structure, unknown), and shoreline type (for example, emergent vegetation, mangrove, oyster, structure, terrestrial, none). A zero-inflated approach was used by fitting a binomial GLM to estimate the probability of a nonzero catch and fitting a lognormal GLM to the nonzero data to estimate the mean catch-per-unit effort (CPUE). Akaike information criterion (AIC) was used to select the most parsimonious binomial and lognormal GLM’s (Akaike 1974). The “delta” model estimates were then made following Grüss and others (2019) by multiplying the least squared means from both model components to obtain an overall standardized CPUE. Although relatively infrequent, missing values were interpolated using a seasonally adjusted linear interpolation from the “timetk” package for RStudio (Dancho and Vaughan 2021). This methodology was applied to 4% of all biomass time series values.
Empirical Dynamic Modeling
Empirical dynamic modeling is an approach to analyzing nonlinear relationships between time series of a given dynamical system (Sugihara and others 2012; Chang and others 2017; Deyle and others 2018). If causality is evident between two variables, their time series will be reflective of a manifold within the system, thereby allowing cross-mapping between their historic values (Sugihara and others 2012; Chang and others 2017). Empirical dynamic modeling operates by first analyzing the time series and subsequently inferring the associated manifold. In this paper, we focus on simplex projections to describe the complexity of the system’s manifold as represented by the number of embedding dimensions, and convergent cross-mapping (CCM) to assess time-lagged causation and cross-prediction between time series. All EDM analyses were conducted using the “rEDM” package (Ye and others 2016).
Time series Standardization
Separate EDM analyses were run on the simulated data from the FLE model and the empirical data from the Suwannee River estuary, using the same methods to standardize and analyze the data. The time series used in the EDM simulation test included monthly biomass output from the FLE model of each environmental scenario (BU, TD, BU + TD). Regarding the FLE output, three time series were created from each environmental scenario by applying differing degrees of observation error. In total, we utilized simulated time series of biomass for eight functional groups from each of the nine scenarios. In the real-world application of the Suwannee River estuary, the data included thirty-five empirically collected time series of monthly fish abundance, fishing effort, and water quality data assembled from the FIM, USGS, MRIP, and Project COAST sampling programs. Prior to running EDM, each time series of the simulated and empirical datasets was standardized to remove seasonality and non-stationarity (Chang and others 2017) using multiplicative time series decomposition. To determine the stationarity of the biomass metrics, we used an augmented Dickey—Fuller test and differenced the data accordingly. Lastly, each time series was normalized with a z-score transformation.
Simplex Estimation
Simplex is a form of nearest neighbor forecasting, whereby the shortest Euclidean distance is found between separate points in a given state space (Deyle and others 2018). This technique allows for the assessment of a system’s dimensionality (Sugihara and May 1990; Chang and others 2017), known as the embedding dimension (E), which describes the amount of predictor variables (lagged coordinate variables) necessary to adequately describe a given dependent variable (Sugihara and May 1990; Hsieh and others 2005). Simplex calculates Pearson correlation coefficients (ρ) for each number of embedding dimensions, and the highest ρ value generally demonstrates the appropriate value for E. After assembling the time series of fish biomass, 2/3 of the data were subset into the “library” dataset and 1/3 into the “prediction” dataset. The “library” data were the training data for the model, while the “prediction set” were the data on which the model was tested. This was followed by finding the appropriate number of embedding dimensions, the E value with the highest ρ, by using the simplex function. Simplex was applied to each time series described previously.
Convergent Cross-Mapping
Lastly, we used convergent cross-mapping to identify interactions between paired variables, while accounting for time lags that may be unknown. This technique relies on how time delayed, historic values of a certain variable can effectively predict the values of another variable (Sugihara and others 2012; Ye and others 2015a). The CCM approach can identify causal interactions because if an explanatory variable has an influence on a response variable, then the explanatory variable will leave its signature within the historic time series of the response variable. Causality can be inferred if the response variable can accurately predict values of the independent variable within a time series (Sugihara and others 2012). If time lags are present, the independent variable will be mapped at some specified time interval within the time series of the dependent variable (Ye and others 2015a). The appropriate time lag was found by comparing the association between the present state of the response variable and different lags on historic values of the predictor variable using Pearson’s correlation coefficient.
A critical element of CCM which allows for the assessment of interacting time series is that of convergence. If an increase in the length of the analyzed time series results in greater predictive performance of the CCM outputs, then the model displays convergence (Sugihara and others 2012). Convergent cross-mapping can be described by the equation.\(y\left(t+{t}_{p}\right)= F (x (t))\approx \sum_{i=1}^{E+1}y ({t}_{i} +t\text{p})\)(Sugihara and others 2012; Deyle and others 2018):where the response variable is represented by ‘y’ at time ‘t’ and time ‘tp’ is the prediction time. In this analysis, time lags were assessed at a monthly time step. Again, ‘E’ represents the number of embedding dimensions, or the amount of coordinate variables. For the CCM analysis, the optimum E from the simplex assessment was used as the relevant number of embedding dimensions. The response and independent variables used in the CCM analysis consisted of all combinations of functional groups within the simulated and real ecosystems. Each functional group was paired with every other functional group as both a response variable and as a predictor variable, resulting in n*(n − 1) combinations in both the simulation test and empirical application, where n is the number of species or functional groups. In total, there were fifty-six interactions for each of the nine EwE scenarios (n = 8) and 1190 interactions in the empirical dataset (n = 35). In the CCM models of the simulated and empirical datasets, time lags were assessed up to 40 months before the current time step.
We identified potential causal trophic linkages from the CCM analysis by detecting interactions that both had high ρ values (for example, the effect of one species on another taxon was high at a certain lagged interval), and the relationship between ρ and time lag length was distinctly dome shaped. In these interactions, we documented the maximum value of ρ and the corresponding time lag. When comparing results between the environmental simulations, we used the 0.0 observation error scenario as the baseline condition. For the tests on observation error, we utilized the BU + TD simulation as the baseline condition when formulating results and conclusions.
In both the simulation test and empirical analysis, interactions between all time series were assessed. While each interaction from the simulation test is presented, in the empirical analysis, only interactions from select sub-food webs are described. The sub-food webs were obtained from an ecosystem model previously built for the Suwannee system (Sinnickson and others 2021) and are intended to simplify the results and interpretation, while highlighting interactions assumed to be ecologically important based on known food web interactions. These subwebs included energetic pathways from physical factors and primary producers to three predatory fish species [Gulf flounder (Paralichthys albiguttata), red drum, and spotted seatrout] and one pelagic forage fish [anchovies (Anchoa spp.)].
Results
Simulation Test
Detecting Top-Down Versus Bottom-Up Effects
When EDM was applied to the simulated data with no observation error (model runs 1, 4, and 7), we consistently found that interpretable and meaningful trophic linkages were only detected by EDM when bottom-up drivers were implemented on the system (Figure 3) and when these interactions occurred at lower trophic levels. Measurable lags were only identified in model runs 1 (BU) and 7 (BU + TD) with bottom-up processes, as opposed to model run 4 (TD), where these lags were not discernable (Figure 3).
Convergent cross-mapping analysis of the trophic interactions in the FLE. Time lags and ρ values are demonstrated on the x and y-axes, respectively. The bottom-up (BU, black) driven scenario represents model run 1, the top-down (TD, blue) simulation depicts model run 4, and the combined scenarios (BU + TD, orange) represents model run 7. No observation error was utilized in these runs.
In the bottom-up (BU) and combined (BU + TD) scenarios, the strongest interactions with high maximum ρ values (ρ > 0.9) were consistently found at low trophic levels (Figure 3). These included the connections between benthic invertebrates, zooplankton, phytoplankton, and detritus, where lags ranged from 0 to 5 months. Domed shaped relationships between ρ values and time lags were very clearly defined at the base of the food web (Figure 3). For example, benthic invertebrates and zooplankton interacted with their primary dietary items, detritus and phytoplankton, respectively, at lags of 3 and 2 months (ρ > 0.97, ρ > 0.97, Figure 3). Trophic interactions were also evident for the mid-trophic level taxa, forage fish and invertivores, but to a lesser extent. These groups were influenced by detritus, planktonic taxa, and invertebrates at lags of 0–7 months with ρ values ranging between 0.73 and 0.86 (Figure 3). For the predatory groups (piscivores and generalists), interaction strengths tended to be weak. Generalists appeared to be mildly impacted by detritus, planktonic groups, and invertebrates in model run 1 (BU) (ρ > 0.79, lag = 1–9) (Figure 3). Although ρ values were relatively high, the domed relationship between ρ and time lag length was poorly defined (Figure 3).
In the assessment of CCM skill with respect to net efficiency, a similar phenomenon was evident. Net efficiencies and CCM ρ were directly related, but both variables generally decreased moving up the food web as higher trophic level species tend to have slower turnover rates and would respond less to the higher frequency dynamics of their prey. Efficiencies ranged between 0.075 (piscivores—highest trophic level consumer) to 0.353 (zooplankton—lowest trophic level consumer) (Figure 4). Overall, we observed a strong, linear relationship between net efficiencies and CCM ρ (Figure 4; BU + TD; ρ = 0.71 + 0.78*Net Efficiency; R2 = 0.73).
Convergent cross-mapping skill (ρ) with respect to net efficiency rates in Ecopath. The net efficiency describes the rate at which food is assimilated into new biomass of the consumer. Net efficiencies are provided for each consumer taxon in each of the trophic interactions. Legend is ordered with respect to trophic level of response group. A strong positive relationship is evident between CCM skill and net efficiency. Confidence intervals (± 95%) are represented in grey. Results are from the BU + TD scenario with no observation error.
When comparing model run 1 (BU) and 7 (BU + TD), the effects of detritus, planktonic groups, and invertebrates on generalists appeared to be dampened when top-down drivers were implemented (Figure 3). For piscivores, the application of top-down processes in model run 7 (BU + TD) appeared to change the direction of the interaction compared to model run 1 (BU). From model run 1 to model run 7, maximum ρ of piscivores predicted by invertebrate, planktonic, and detrital groups shifted from positive to negative lags, demonstrating that these groups only affected piscivores when variable fishing effort was applied (Figure 3).
For the top-down scenario, not only were strong connections not exhibited (model run 4, TD), but even when top-down drivers were combined with bottom-up drivers (model run 7, BU + TD), the results did not differ substantially from the scenario with solely bottom-up processes (Figure 3). Also, when the system was driven by bottom-up processes, detectable trophic interactions were generally observed in the bottom-up direction. Prey populations were found to influence predator populations, but predators did not exhibit significant impacts on prey items, with the exception of the influence of zooplankton grazing upon phytoplankton (ρ > 0.96, lag = 0–1 months) (Figure 3).
Effects of Observation Error on Detecting Lagged Interactions
As expected, we found that EDM performed best when observation error was low, resulting in higher ρ values and more defined time lags (Figures 5, 6). For example, in the BU + TD scenario, ρ values decreased for linkages between detrital, planktonic, and invertebrate groups when observation error increased, but defined interactions were still clearly evident (Figures 5, 6). For most interactions, decreases in ρ were relatively proportional to the degree of error implemented. For the interactions between detrital, planktonic, and benthic invertebrate groups, ρ values decreased by approximately 0.14–0.24 when comparing interactions from the no error and high error scenarios. Regarding the influence of phytoplankton on zooplankton, maximum ρ decreased from 0.97 to 0.73 between the no error and high error simulations (lag = 0–3 months) (Figures 5, 6). Smaller changes were observed in the zooplankton (predictor) and benthic invertebrate (response) relationship, as ρ decreased from 0.98 to 0.84 (lag = 3–6 months) (Figure 5). On the contrary, at high trophic levels, the strength of relationships appeared to weaken in the high error scenario. In the bottom-up direction, generalists and invertivores demonstrated inconsistent and weak relationships with invertebrates, planktonic species, and detritus when high amounts of error were applied (Figure 5). These changes were notably greater higher up the food web, as forage fish, invertivores, and generalists demonstrated large changes in CCM skill with prey species when error was implemented. For example, maximum ρ values depicting the effect of invertebrates on invertivores decreased from 0.78 to 0.44 when comparing the no error (model run 7) and high error scenarios (model run 9) (Figure 5). For the effect of zooplankton on forage fish, maximum ρ decreased from 0.85 to 0.42 in model run 7 to model run 9 (lag = 0–2 months). In some circumstances, such as the relationship between detritus and invertivores and detritus and generalists, the high degree of observation error almost completely eliminated the ability of EDM to detect a relationship (Figure 5). In general, error did not appear to impact the estimated time lags.
Observation error simulations in the BU + TD scenario of the FLE. Plots depict relationships between ρ values and time lags from the assessment of trophic linkages using convergent cross-mapping. The no error (orange), 0.3 error (red), and 0.6 error (black) simulations represent model runs 7, 8, and 9, respectively. Relationships demonstrate how different degrees of observation error have detectable impacts on trophic interactions strengths.
Convergent cross-mapping skill with respect to level of observation error and functional group. Plot represents the effect of phytoplankton on upper trophic level taxa through bottom-up effects in the FLE. Functional groups are colored and ordered in relation to their representative trophic level. Results are from the BU + TD scenario.
Empirical Time Series Analysis
In the empirical dataset, we observed significant lagged interactions in the sub-food webs that describe trophic pathways connecting environmental factors to four specific taxa: anchovies, Gulf flounder, spotted seatrout, and red drum. For anchovies, we observed a strong connection from a bottom-up signal, as abundance interacted with bottom-up processes relating to river discharge (ρ > 0.60; Figure 7), nutrient concentrations (ρ > 0.61; Figure 7), and chlorophyll-a (ρ > 0.60; Figure 7). The effects from the abiotic factors appeared to be longer, as discharge and nutrient concentrations influenced anchovies at lags of approximately 7–8 and 2–4 months, respectively, while impacts from biotic chlorophyll-a levels appeared to be faster, at lags at 0–1 month (Figure 7).
Food web diagram demonstrating energetic transfer to anchovies along with associated CCM results. In the food web diagram, trophic pathways were informed by Sinnickson and others (2021). Sizes of nodes represent amount of biomass in functional groups, while linkages represent energetic transfers through consumption. The CCM plot depicts factors influencing anchovy abundances in the Suwannee River estuary. Time lags (months) are represented on the x-axis, while the strength of the relationship at time lags is depicted by Pearson’s correlation coefficient along the y-axis.
Similar bottom-up signals were also observed when assessing the lagged relationships of Gulf flounder (Figure 8). Interactions between Gulf flounder and its direct prey items constituted the shortest time lags, as blue crabs (ρ > 0.55; Figure 8) and pinfish (ρ > 0.60; Figure 8) populations interacted with flounder abundances at lags of 4–5 and 5–8 months, respectively. Indirect effects relating to eutrophication and primary production were also observed but at longer intervals. For example, concentrations of chlorophyll-a (ρ > 0.29, lag = 9–11; Figure 8) exhibited interactions with Gulf flounder abundance at lags of 9–11 months, while relationships with nutrient concentrations were demonstrated at 9–14-month intervals (ρ > 0.51; Figure 8).
Food web diagram depicting energetic pathways to Gulf flounder along with associated CCM results. The food web diagram was informed by Sinnickson and others (2021). Sizes of nodes and width of linkages represent amounts of biomass. The CCM plot demonstrates ecological factors related to Gulf flounder abundances. The x-axis is represented by time lags (months), while the y-axis is depicted by Pearson’s correlation coefficient. Gulf flounder appear to be affected more quickly by factors that are of a higher trophic level.
When assessing the sub-food web for spotted seatrout, we also observed lagged interactions from bottom-up processes (Figure 9). The abiotic factors of discharge and nutrients demonstrated lagged interactions with seatrout that were notably longer than lags with prey items. For the YOY stanza, correlations with discharge (ρ > 0.56; Figure 9) and nutrients (ρ > 0.33; Figure 9) were exhibited at approximately 15 and 11 months, respectively (Figure 9), while prey items of anchovies (ρ > 0.58; Figure 9) and shrimp (ρ > 0.39; Figure 9) demonstrated interactions at shorter intervals. This was similarly reflected for the intermediate age stanza, as lags from discharge (ρ > 0.63; Figure 9) and nutrients (ρ > 0.41; Figure 9) were exhibited at 9 and 14 months, whereas interactions with clupeids (ρ > 0.43; Figure 9), YOY mullet (ρ > 0.76; Figure 9), and decapods (ρ > 0.70; Figure 9) ranged from 0 to 5 months.
Food web diagram demonstrating energetic pathways to spotted seatrout along with associated CCM results. The food web was informed by Sinnickson and others (2021). Sizes of nodes represent amount of biomass within functional groups, while width of lines depict biomass leaving taxa. The CCM plots depict factors influencing YOY spotted seatrout abundances (left) and intermediate aged seatrout (1–2.5 years) (right). Time lags (months) are represented on the x-axis, while Pearson’s correlation coefficient is represented on the y-axis, demonstrating the strength of the relationship at given time lags. For both age stanzas, direct prey items appear to influence seatrout populations more quickly relative to the physical, abiotic factors.
For the red drum age classes, we observed several bottom-up signals, as well as a potential example of recruitment effects (Figure 10). Both nutrients (ρ > 0.42; Figure 10) and discharge (ρ > 0.42; Figure 10) interacted with YOY red drum at short time lags, ranging from 0 to 2 months. The intermediate aged stanza (1–4 years) also exhibited interactions with a series of variables but at longer time lags relative to YOY red drum. We observed associations between nutrient concentrations and intermediate aged red drum (ρ > 0.52, lag = 30–32 months; Figure 10), as well as from prey items such as pinfish (ρ > 0.58, lag = 24–29 months; Figure 10) and decapods (ρ > 0.51, lag = 21–22 months; Figure 10). In addition to the bottom-up impacts, we also observed a likely effect from recruitment. Intermediate aged red drum appeared to demonstrate a mild causal relationship with YOY red drum at a lag of 30–32 months (ρ > 0.24; Figure 10).
Food web diagram representing energetic transfers to red drum within the Suwannee River estuary and associated CCM results. The food web diagram was informed by Sinnickson and others (2021). Nodes range from detritus and primary producers at the first trophic level, to red drum between the third and fourth trophic levels. Widths of linkages and sizes of nodes represent amounts of biomass. The CCM plots demonstrate environmental factors influencing YOY red drum and intermediate aged red drum (1–4 years). Time lags (months) and Pearson’s correlation coefficient (ρ) are represented on the x and y-axes, respectively. Young-of-year red drum appear to be affected by variables relatively quickly, while the intermediate aged fish are affected at lags ranging from 20 to 33 months.
Discussion
In simulation testing scenarios, several common tendencies were observed. We found that (1) EDM performed best when bottom-up processes were included in the simulations, whereby lower trophic level taxa impacted higher level groups through the process of nutrient inputs, primary, and secondary production; (2) EDM was less capable of detecting lagged connections higher in the food web; and (3) observation error had a significant impact on EDM model performance. In general, we observed that the effects of observation error increased with trophic level. While these trends were evident, these results should be interpreted within the context of a highly simplified system. Even in this confined system where all trophic dynamics were clearly established, EDM was still unable to distinguish certain ecological relationships. This likely reflects the significant complexity that is represented even in simple systems.
An important concept to consider when assessing results from EDM is how to interpret results of modeled parameters and potential causation. For example, while CCM can detect important system interactions, it cannot describe the direction of these relationships. Although this is a significant limitation of these models, recent developments have been made addressing this issue (Deyle and others 2016a, b; Chang and others 2021). Additionally, another potential concern is that EDM may describe causal effects between time series even if mechanisms for such causality do not exist. These models may incorrectly detect causation between synchronous time series that do not influence one another but instead covary over time. Potential examples of this phenomenon may exist in the relationships between discharge with populations of anchovies and YOY seatrout and red drum. This is possible due to the seasonal dynamics of both discharge rates and life-history patterns of these fishes. This concern can easily be circumvented by refining research questions and focusing on known or hypothesized connections. Within ecology, this can be done by highlighting known trophic interactions through simplified sub-food webs, a methodology employed by this study.
Although we found stronger interactions associated with bottom-up processes than top-down forces in the FLE, the discussion over the more influential driver continues throughout the contemporary literature. Historically, some have suggested that ecosystems, particularly aquatic systems, are more influenced by top-down processes (Carpenter and others 1987; Estes and Duggins 1995; Estes 1996; Kay 1998; Frank and others 2005; Heck and others 2007). Although many systems are significantly regulated by top-down control, examples of bottom-up regulation in aquatic environments abound. In particular, high rates of eutrophication have been observed throughout many ecosystems globally, and these changes at the base of the food web have significantly influenced trophic pathways within these respective systems (Rabalais and others 2002; Kemp and others 2005; Chislock and others 2013; Liu and others 2018). This has been particularly evident in subtropical ecosystems, where irradiance levels and nutrient concentrations have been observed as significant drivers of ecosystem dynamics (Lapointe 1997; Bledsoe and Phlips 2000; Lehrter and others 2009). While we more readily detected bottom-up rather than top-down drivers in the FLE model, most real-world ecosystems, especially those supporting fisheries, are likely influenced by a combination of both processes (Mackinson and others 2009).
A notably similar analysis to our study was conducted by Luken (2020), whereby the author utilized CCM to assess both top-down and bottom-up impacts within a reservoir ecosystem. She evaluated relationships between chlorophyll-a, zooplankton, and larval shad (Dorosoma cepedianum), and, like our study, found that bottom-up drivers tended to have stronger impacts on populations than top-down processes. Although chlorophyll-a was found to impact zooplankton biomass and zooplankton biomass affected larval shad densities, the study did not directly assess the impact of chlorophyll-a on shad.
Like the simulated data assessment, in our empirical dataset, we also observed several significant examples of bottom-up drivers influencing food web interactions. This was exemplified by biotic and abiotic factors having strong, lagged interactions with anchovies, Gulf flounder, spotted seatrout, and red drum (Figures 7, 8, 9, 10). In the former three relationships, we found that there was generally an inverse relationship between the trophic level of the predictor groups and the length of the time lag (Figure 11). River discharge and nutrients demonstrated the longest lagged effects, while the biotic variables and prey species affected anchovies, Gulf flounder, and YOY seatrout more quickly (Figure 11). For example, moving up the food web from discharge rates to nutrients levels and subsequently chlorophyll-a concentrations, these factors influenced anchovy densities at lagged intervals of approximately 8, 3, and 0 months, respectively (Figure 5). This phenomenon was also demonstrated with Gulf flounder, as nutrient and chlorophyll-a concentrations affected flounder densities at lags of 13 and 10 months, while prey items such as pinfish and blue crabs exhibited lagged effects at 6 and 4 months (Figure 8).
Scatterplot describing the relationship between the trophic level of predictor taxa and the reflected time lag of the response species. Data were only included from CCM analyses of anchovies, Gulf flounder, and YOY seatrout. In these relationships, we observed decreases in time lags as the trophic level of the predictor increased. Although trophic levels are not generally ascribed to abiotic factors such as discharge and nutrient concentrations, for this assessment, relative approximations were made. Discharge and nutrient levels were given trophic level estimates of 0 and 0.5, respectively, which would demonstrate how nutrients move through food webs sequentially.
One difficulty in detecting top-down interactions was that some of the higher trophic level fish which inhabit the estuary were not able to be included in the analysis. This was due to their seasonal availability patterns marked by missing data that resulted in the removal of these time series from the assessment. Some of these taxa included adult seatrout (2.5+ years), jack crevalle (Caranx hippos), Spanish mackerel (Scomberomorus maculatus), and bonnethead (Sphyrna tiburo).
In all simulated scenarios, we consistently observed that as trophic level increased, the ability to detect lagged interactions decreased. It appeared that the third trophic level, secondary consumers, was the threshold for which strong trophic interactions could be consistently observed. Relationships between tertiary consumers and lower trophic level groups were generally mild or difficult to distinguish. One possible explanation for this phenomenon is system inefficiency. Biological production is not a completely efficient process, as species lose energy through respiration and defecation. Baird and Ulanowicz (1989) demonstrated how energetic efficiency decreases with an increase in trophic level. They found that first and second order trophic level species transferred approximately 85 and 36% of their energy to the next trophic level, respectively. Energetic efficiency then drops to approximately 10% for the following three trophic levels. We may hypothesize that trophic efficiencies need to exceed 30% for EDM to detect energetic pathways. If energy is being lost at each successive trophic level, we should only expect to observe minimal changes in predatory populations when there are significant fluctuations in primary production rates. These small changes in predator densities may not be substantial enough for detection with EDM.
Another explanation for weaker signals at high trophic levels could relate to diet compositions. During the construction of the FLE, lower trophic level taxa were specialists, while predatory fish had more generalist diets. This could in part explain the lack of strong interactions between forage fish and invertivores with predators. This variability in diet, combined with the high trophic level of predators, creates a series of different energetic pathways for biomass to accumulate up the food web. In the FLE, biomass was transferred to piscivores and generalists through five and three energetic pathways, respectively, while all lower trophic level groups obtained energy from a single pathway. It is likely that predators are not strongly linked to individual species at lower trophic levels because energy can transfer through many different taxa to reach tertiary consumers. Understanding the implications of energetic inefficiencies, diet compositions, and trophic pathways will be important for applying EDM to real-world ecosystems. Even in relatively simple systems with low biodiversity, there can still be a significant amount of trophic complexity. Future research should consider amending our EwE simulation model in order create alternate scenarios, whereby predatory species have more constrained diets, such as in cold-water systems.
It is likely that our assessment of the relationship between net efficiencies and CCM skill successfully elucidates the impacts of both system inefficiencies and Ecopath parameterization on EDM performance. Net efficiencies will inherently incorporate both factors, and our assessment revealed a strong, linear relationship between net efficiency and CCM ρ (Figure 4; BU + TD; ρ = 0.71 + 0.78*Net Efficiency; R2 = 0.73). Upper trophic level taxa exhibited lower net efficiencies, which may have been a function of higher respiration rates and lower production rates. We believe that EDM application will be most effective for species that demonstrate high gross food conversion efficiencies and therefore have higher turnover rates, allowing them to covary with prey items and make repeated returns to the same state-space.
When interpreting the results from the simulated TD scenario and the higher trophic levels, it is important to consider the underlying methodologies of EDM and how they could potentially impact model results. A foundational principle that may illuminate these results is the concept of recurrences (Kantz and Schreiber 2004; see Munch and others 2022). Empirical dynamic modeling functions by making recurring visits to a given neighborhood within the state-space of a time series. Because species with short generation times have significantly more recurrences relative to longer-lived animals, differences in model results should be expected. The high variability exhibited in the time series of the low trophic level species should subsequently result in frequent visits to the same location in state-space. Conversely, our high trophic level taxa would have made limited recurrences because of the limited variability of their populations, which also occurred on longer timescales. These characteristics are the most plausible explanation for our results. While this may initially appear as a limitation of EDM models, it may actually represent a concern specifically for input time series data. Biologists intending to utilize EDM in future research should consider the generation times and turn-over rates of study species before formal model development.
This phenomenon has also been documented in the current literature, most notably in a comprehensive meta-analysis conducted by Munch and others (2018). Of the 185 study species, the authors found that the predictive ability of EDM was positively related to the frequency of generations exhibited in time series. As a result, the authors recommended the application of EDM for shorter-lived species with faster generation times. Subsequent publications utilizing EDM models document the selection of short-lived species as ideal study taxa (Methot 2000; Giron-Nava and others 2017; Johnson and others 2021; Daugaard and others 2022; Tsai and others 2022).
In our analysis, another notable finding was that observation error had a significant impact on EDM model performance. In the scenarios of 0.3 and 0.6 observation error, variability clearly influenced the ability of EDM to detect the true lagged effect. Throughout fisheries research, significant levels of observation error, sampling inefficiency, and gear bias are common, which can cause bias and obscure the true patterns (Hayward and others 1989; Dunham and others 2001; Rosenberger and Dunham 2005; Breen and Ruetz III 2006; Meyer and others 2014). Monitoring programs aimed at developing indices of abundance in stock assessments frequently target a CV of no more than 0.3 (Hilborn and Liermann 1998). Within the broader discipline of population biology, levels of observation error have been estimated at 32% (Meir and Fagan 2000). For populations that are sampled with high levels of observation error, EDM may not be an effective way to identify food web dynamics. Conversely, in monitoring programs with low observation error, such as the FIM program, EDM would likely be more effective. This supposition is supported by the results of this study, as we observed numerous distinct trophic linkages exhibiting high ρ values using the real-world dataset. A review of the relevant literature on this topic demonstrates conflicting evidence of the effect of error on EDM performance. Similar to our assessment, Mønster and others (2017) investigated this topic and found a negative, linear relationship between noise and CCM correlation coefficients. In contrast, BozorgMagham and others (2015) describe CCM as robust to changes in error when applying Guassian white noise to modeled systems.
Additionally, some of the topics pertaining to the higher trophic levels may also have relevance when interpreting the effects from observation error. Although equal amounts of observation error were added to all taxa, the resulting signal to noise ratios (SNR) of functional groups differed across population sizes. When error is applied, this signal decreases more substantially for taxa with less biomass at higher trophic levels. Subsequently, this would result in fewer visits to the same neighborhood state-space. This discrepancy would be an obstacle for making definitive conclusions on the true effect of observation error across the food web. One way to potentially address this concern would be to use time series of longer lengths, as the predictive ability of EDM S-maps has been found to be directly related to time series lengths (Giron-Nava and others 2017).
While other studies have utilized EDM to analyze direct trophic interactions, this study may provide insight into how to apply EDM to real-world, highly complex ecosystems that include indirect effects, trophic feedbacks, and energetic pathways with low efficiency. In general, EDM tends to be employed in population biology to assess direct predator—prey, grazing, and nutrient uptake interactions (Ye and others 2015a; Anneville and others 2019; Cai and others 2020; Luken 2020). Exceptions would include studies analyzing the relationship between environmental variables and animal populations, but in these interactions, the environment can directly impact populations (Sugihara and others 2012; Ye and others 2015b; Kuriyama and others 2020). Certain studies have addressed bottom-up effects, but these analyses have been focused on the interactions between nutrients and planktonic groups (Frossard and others 2018; Chang and others 2020).
Our study provides new insights as to how bottom-up effects can indirectly impact higher trophic level species at lagged intervals. This would be represented by the relationship between forage fish and phytoplankton biomass. In our models, forage fish did not directly consume phytoplankton, but phytoplankton indirectly affected forage fish population by transferring energy to zooplankton.
One significant similarity between our results and other EDM analyses was that most strong interactions were observed at low trophic levels. Although our study did not detect many strong relationships at high trophic levels, most EDM studies only analyze species that are planktonic or secondary consumers (Ye and others 2015a; Anneville and others 2019; Rogers and others 2020; Cai and others 2020; Luken 2020). The sole study that we found assessing the dynamics of a high trophic level predator was conducted on sockeye salmon (Oncorhynchus nerka) recruitment (Ye and others 2015b). Our study indicated that EDM is not highly effective at identifying interactions between lower and upper trophic level species. Overall, the general lack of studies applying EDM to assess predatory fish populations likely indicates the techniques limited performance in these scenarios.
Our analysis demonstrated that EDM has the potential to identify trophic linkages, particularly at the base of food webs, where there are fewer, and shorter, energetic pathways, and when data are collected with minimal observation error. This has substantial implications in fisheries research and ecology more broadly. Identifying potential causation in ecosystems where experimentation is impractical can help to elucidate frequently debated topics in ecology such as the effects of bottom-up versus top-down processes, the importance of forage fish in aquatic food webs, and whether human harvests or environmental conditions have stronger impacts on populations of commercially valued species (Skud 1975; Hunter and Price 1992; Hilborn and others 2018). By understanding ecological time lags, scientists would be capable of designing more informative research programs and population models, as there would be a better understanding of when responses would manifest. Further developments in this line of research would not only be transformative in ecology but likely within the field of applied natural resource management as well.
References
Ahrens RN, Walters CJ, Christensen V. 2012. Foraging arena theory. Fish Fish 13(1):41–59.
Akaike H. 1974. A new look at the statistical model identification. IEEE Trans Autom Control 19(6):716–723.
Anneville O, Chang CW, Dur G, Souissi S, Rimet F, Hsieh CH. 2019. The paradox of re-oligotrophication: the role of bottom–up versus top–down controls on the phytoplankton community. Oikos 128(11):1666–1677.
Baird D, Ulanowicz RE. 1989. The seasonal dynamics of the Chesapeake Bay ecosystem. Ecol Monogr 59(4):329–364.
Bledsoe EL, Phlips EJ. 2000. Relationships between phytoplankton standing crop and physical, chemical, and biological gradients in the Suwannee River and plume region, USA. Estuaries 23(4):458–473.
BozorgMagham AE, Motesharrei S, Penny SG, Kalnay E. 2015. Causality analysis: identifying the leading element in a coupled dynamical system. PLoS One 10(6):e0131226.
Breen MJ, Ruetz CR III. 2006. Gear bias in fyke netting: evaluating soak time, fish density, and predators. N Am J Fish Manag 26(1):32–41.
Cai J, Hodoki Y, Ushio M, Nakano SI. 2020. Influence of potential grazers on picocyanobacterial abundance in Lake Biwa revealed with empirical dynamic modeling. Inland Waters 10(3):386–396.
Carpenter SR, Kitchell JF, Hodgson JR, Cochran PA, Elser JJ, Elser MM, Lodge DM, Kretchmer D, He X, von Ende C. 1987. Regulation of lake primary productivity by food web structure. Ecology 68(6):1863–1876.
Chagaris D, Mahmoudi B, Muller-Karger F, Cooper W, Fischer K. 2015a. Temporal and spatial availability of Atlantic Thread Herring, Opisthonema oglinum, in relation to oceanographic drivers and fishery landings on the Florida Panhandle. Fish Oceanogr 24(3):257–273.
Chagaris DD, Mahmoudi B, Walters CJ, Allen MS. 2015b. Simulating the trophic impacts of fishery policy options on the West Florida Shelf using Ecopath with Ecosim. Mar Coast Fish 7(1):44–58.
Chang CW, Ushio M, Hsieh CH. 2017. Empirical dynamic modeling for beginners. Ecol Res 32(6):785–796.
Chang CW, Ye H, Miki T, Deyle ER, Souissi S, Anneville O, Adrian R, Chiang YR, Ichise S, Kumagai M, Matsuzaki SI, Shiah FK, Wu JT, Hsieh CH, Sugihara G. 2020. Long-term warming destabilizes aquatic ecosystems through weakening biodiversity-mediated causal networks. Glob Change Biol 26(11):6413–6423.
Chang CW, Miki T, Ushio M, Ke PJ, Lu HP, Shiah FK, Hsieh CH. 2021. Reconstructing large interaction networks from empirical time series data. Ecol Lett 24(12):2763–2774.
Chislock MF, Doster E, Zitomer RA, Wilson AE. 2013. Eutrophication: causes, consequences, and controls in aquatic ecosystems. Nat Educ Knowl 4(4):10.
Christensen V, Walters CJ. 2004. Ecopath with Ecosim: methods, capabilities and limitations. Ecol Model 172(2–4):109–139.
Christensen V, Walters CJ, Pauly D. 2005. Ecopath with Ecosim: a user’s guide. Vancouver: Fisheries Centre, University of British Columbia,. p 154.
Dancho M, Vaughan D. 2021. timetk: a tool kit for working with time series in R, r package version 2.6.2. https://CRAN.R-project.org/package=timetk
Daugaard U, Munch SB, Inauen D, Pennekamp F, Petchey OL. 2022. Forecasting in the face of ecological complexity: number and strength of species interactions determine forecast skill in ecological communities. Ecol Lett 25(9):1974–1985.
Deyle ER, Maher MC, Hernandez RD, Basu S, Sugihara G. 2016a. Global environmental drivers of influenza. Proc Natl Acad Sci 113(46):13081–13086.
Deyle ER, May RM, Munch SB, Sugihara G. 2016b. Tracking and forecasting ecosystem interactions in real time. Proc R Soc B Biol Sci 283(1822):20152258.
Deyle E, Schueller AM, Ye H, Pao GM, Sugihara G. 2018. Ecosystem-based forecasts of recruitment in two menhaden species. Fish Fish 19(5):769–781.
Dunham J, Rieman B, Davis K. 2001. Sources and magnitude of sampling error in redd counts for bull trout. N Am J Fish Manag 21(2):343–352.
Estes JA, Duggins DO. 1995. Sea otters and kelp forests in Alaska: generality and variation in a community ecological paradigm. Ecol Monogr 65(1):75–100.
Estes JA. 1996. Predators and ecosystem management. Wildl Soc Bull 24(3):390–396.
Frank KT, Petrie B, Choi JS, Leggett WC. 2005. Trophic cascades in a formerly cod-dominated ecosystem. Science 308(5728):1621–1623.
Frazer, T. 2018. Unpublished data.
Frossard V, Rimet F, Perga ME. 2018. Causal networks reveal the dominance of bottom-up interactions in large, deep lakes. Ecol Model 368:136–146.
Frederiksen M, Edwards M, Richardson AJ, Halliday NC, Wanless S. 2006. From plankton to top predators: bottom-up control of a marine food web across four trophic levels. J Anim Ecol 75(6):1259–1268.
Giron-Nava A, James CC, Johnson AF, Dannecker D, Kolody B, Lee A, Nagarkar M, Pao GM, Ye H, Johns DG, Sugihara G. 2017. Quantitative argument for long-term ecological monitoring. Mar Ecol Prog Ser 572:269–274.
Grüss A, Walter JF III, Babcock EA, Forrestal FC, Thorson JT, Lauretta MV, Schirripa MJ. 2019. Evaluation of the impacts of different treatments of spatio-temporal variation in catch-per-unit-effort standardization models. Fish Res 213:75–93.
Hayward RS, Margraf FJ, Knight CT, Glomski DJ. 1989. Gear bias in field estimation of the amount of food consumed by fish. Can J Fish Aquat Sci 46(5):874–876.
Heck KL, Valentine JF. 2007. The primacy of top-down effects in shallow benthic ecosystems. Estuaries Coasts 30(3):371–381.
Hilborn R, Mangel M. 1997. The ecological detective: confronting models with data. Vol. 28. Princeton: Princeton University Press.
Hilborn R, Liermann M. 1998. Standing on the shoulders of giants: learning from experience in fisheries. Rev Fish Biol Fish 8(3):273–283.
Hilborn R, Amoroso RO, Bogazzi E, Jensen OP, Parma AM, Szuwalski C, Walters CJ. 2018. Response to Pikitch et al.
Hsieh CH, Glaser SM, Lucas AJ, Sugihara G. 2005. Distinguishing random environmental fluctuations from ecological catastrophes for the North Pacific Ocean. Nature 435(7040):336–340.
Hunter MD, Price PW. 1992. Playing chutes and ladders: heterogeneity and the relative roles of bottom-up and top-down forces in natural communities. Ecology 572:724–732.
Isaac VJ, Castello L, Santos PRB, Ruffino ML. 2016. Seasonal and interannual dynamics of river-floodplain multispecies fisheries in relation to flood pulses in the Lower Amazon. Fish Res 183:352–359.
Johnson B, Gomez M, Munch SB. 2021. Leveraging spatial information to forecast nonlinear ecological dynamics. Methods Ecol Evol 12(2):266–279.
Kantz H, Schreiber T. 2004. Nonlinear time series analysis. Vol. 7. Cambridge: Cambridge U.
Kao YC, Adlerstein SA, Rutherford ES. 2016. Assessment of top-down and bottom-up controls on the collapse of alewives (Alosa pseudoharengus) in Lake Huron. Ecosystems 19(5):803–831.
Kay CE. 1998. Are ecosystems structured from the top-down or bottom-up: a new look at an old debate. Wildl Soc Bull 26:484–498.
Kemp WM, Boynton WR, Adolf JE, Boesch DF, Boicourt WC, Brush G, Cornwell JC, Fisher TR, Gilbert PM, Hagy JJ, Harding LW, Houde ED, Kimmel DG, Miller WD, Newell RIE, Roman MR, Smith EM, Stevenson JC. 2005. Eutrophication of Chesapeake Bay: historical trends and ecological interactions. Mar Ecol Prog Ser 303:1–29.
Kuriyama PT, Sugihara G, Thompson AR, Semmens BX. 2020. Identification of shared spatial dynamics in temperature, salinity, and ichthyoplankton community diversity in the California current system with empirical dynamic modeling. Front Mar Sci.
Lapointe BE. 1997. Nutrient thresholds for bottom-up control of macroalgal blooms on coral reefs in Jamaica and southeast Florida. Limnol Oceanogr 42(5part2):1119–1131.
Lehrter JC, Murrell MC, Kurtz JC. 2009. Interactions between freshwater input, light, and phytoplankton dynamics on the Louisiana continental shelf. Cont Shelf Res 29(15):1861–1872.
Liu Z, Hu J, Zhong P, Zhang X, Ning J, Larsen SE, Chen D, Yiming G, He H, Jeppesen E. 2018. Successful restoration of a tropical shallow eutrophic lake: strong bottom-up but weak top-down effects recorded. Water Res 146:88–97.
Lo NCH, Jacobson LD, Squire JL. 1992. Indices of relative abundance from fish spotter data based on delta-lognormal models. Can J Fish Aquat Sci 49(12):2515–2526.
Luken HG. 2020. Long-term response of zooplankton biomass and phenology to environmental variability in a eutrophic reservoir (Doctoral dissertation, Miami University).
Mackinson S, Daskalov G, Heymans JJ, Neira S, Arancibia H, Zetina-Rejón M, Jiang H, Cheng HQ, Coll M, Arrenguin-Sanchez F, Keeble K, Shannon L. 2009. Which forcing factors fit? Using ecosystem models to investigate the relative influence of fishing and changes in primary productivity on the dynamics of marine ecosystems. Ecol Model 220(21):2972–2987.
Maunder MN, Punt AE. 2004. Standardizing catch and effort data: a review of recent approaches. Fish Res 70(2–3):141–159.
Meir ELI, Fagan WF. 2000. Will observation error and biases ruin the use of simple extinction models? Conserv Biol 14(1):148–154.
Methot RD. 2000. Technical description of the stock synthesis assessment program. US Dept. Commer. NOAA Tech. Memo. NMFS-NWFSC, 43, 46.
Meyer KA, Garton EO, Schill DJ. 2014. Bull trout trends in abundance and probabilities of persistence in Idaho. N Am J Fish Manag 34(1):202–214.
Mønster D, Fusaroli R, Tylén K, Roepstorff A, Sherson JF. 2017. Causal inference from noisy time-series data—testing the convergent cross-mapping algorithm in the presence of noise and external influence. Future Gener Comput Syst 73:52–62.
Moraes LE, Paes E, Garcia A, Möller O Jr, Vieira J. 2012. Delayed response of fish abundance to environmental changes: a novel multivariate time-lag approach. Mar Ecol Prog Ser 456:159–168.
Munch SB, Giron-Nava A, Sugihara G. 2018. Nonlinear dynamics and noise in fisheries recruitment: a global meta-analysis. Fish Fish 19(6):964–973.
Munch SB, Rogers TL, Sugihara G. 2022. Recent developments in empirical dynamic modelling. Methods Ecol Evol 14:732–745.
Murphy GI, Isaacs JD. 1964. Species replacement in marine ecosystems with reference to the California current. San Diego: Scripps Institution of Oceanography.
National Oceanic and Atmospheric Administration. Data from: Recreational Fisheries Statistics Queries. 2018. https://www.st.nmfs.noaa.gov/recreational-fisheries/data-and-documentation/queries/index.
Pace ML, Cole JJ, Carpenter SR, Kitchell JF. 1999. Trophic cascades revealed in diverse ecosystems. Trends Ecol Evol 14(12):483–488.
Pascual M, Ellner SP. 2000. Linking ecological patterns to environmental forcing via nonlinear time series models. Ecology 81(10):2767–2780.
Peebles EB. 2002. Temporal resolution of biological and physical influences on bay anchovy Anchoa mitchilli egg abundance near a river-plume frontal zone. Mar Ecol Prog Ser 237:257–269.
Priestley MB. 1980. State-dependent models: a general approach to non-linear time series analysis. J Time Ser Anal 1(1):47–71.
Rabalais NN, Turner RE, Wiseman WJ Jr. 2002. Gulf of Mexico hypoxia, aka “The dead zone.” Annu Rev Ecol Syst 33(1):235–263.
Rahmani, D. 2017. Bayesian singular spectrum analysis with state dependent models (Doctoral dissertation, Bournemouth University).
Rogers TL, Munch SB, Stewart SD, Palkovacs EP, Giron-Nava A, Matsuzaki SIS, Symons CC. 2020. Trophic control changes with season and nutrient loading in lakes. Ecol Lett 23(8):1287–1297.
Rosenberger AE, Dunham JB. 2005. Validation of abundance estimates from mark–recapture and removal techniques for rainbow trout captured by electrofishing in small streams. N Am J Fish Manag 25(4):1395–1410.
Sinnickson D, ChagarisAllen DMS. 2021. Exploring impacts of river discharge on forage fish and predators using Ecopath with Ecosim. Front Mar Sci 8:702.
Skud BE. 1975. Revised estimates of halibut abundance and the Thompson-Burkenroad debate. Seattle: International Pacific Halibut Commission.
Sugihara G, May RM. 1990. Nonlinear forecasting as a way of distinguishing chaos from measurement error in time series. Nature 344(6268):734–741.
Sugihara G, May R, Ye H, Hsieh CH, Deyle E, Fogarty M, Munch S. 2012. Detecting causality in complex ecosystems. Science 338(6106):496–500.
Takens, F. 1981. Detecting strange attractors in turbulence. In: Dynamical systems and turbulence, Warwick 1980. Berlin: Springer. pp. 366–81.
Tsai CH, Munch SB, Masi MD, Pollack AG. 2022. Predicting nonlinear dynamics of short-lived penaeid shrimp species in the Gulf of Mexico. Can J Fish Aquat Sci 80:57–68.
Tsonis AA, Deyle ER, May RM, Sugihara G, Swanson K, Verbeten JD, Wang G. 2015. Dynamical evidence for causality between galactic cosmic rays and interannual variation in global temperature. Proc Natl Acad Sci 112(11):3253–3256.
United States Geological Survey. USGS 02323500 Suwannee River Near Wilcox, Fla. 2020. https://waterdata.usgs.gov/usa/nwis/uv?site_no=02323500.
Veilleux BG. 1979. An analysis of the predatory interaction between Paramecium and Didinium. J Anim Ecol 48:787–803.
Ye H, Deyle ER, Gilarranz LJ, Sugihara G. 2015a. Distinguishing time-delayed causal interactions using convergent cross mapping. Sci Rep 5:14750.
Ye H, Beamish RJ, Glaser SM, Grant SC, Hsieh CH, Richards LJ, Schnute JT, Sugihara G. 2015b. Equation-free mechanistic ecosystem forecasting using empirical dynamic modeling. Proc Natl Acad Sci 112(13):E1569–E1576.
Ye H, Clark A, Deyle E, Sugihara G. 2016. rEDM: an R package for empirical dynamic modeling and convergent cross-mapping. cran. r-project.org.
Acknowledgements
We would like to acknowledge Dr. Stephan Munch and Dr. Hao Ye for valuable comments and advice. We would also like to thank the Florida Fish and Wildlife Research Institute for providing long-term estuary and fishery independent monitoring datasets used in this study. Funding for this research was provided by the University of Florida School of Forestry, Fisheries, and Geomatic Sciences, the National Oceanic and Atmospheric Administration RESTORE project, and the Florida Forage Fish Coalition consisting of the International Game Fish Association, Pew Charitable Trusts, Fish Florida, Angler Action Foundation, Florida Wildlife Federation, American Sportfishing Association and Wild Oceans. The views expressed herein are those of the authors and do not necessarily reflect the views of the Florida Forage Fish Coalition.
Author information
Authors and Affiliations
Corresponding author
Additional information
Author Contributions: Dr. Dylan Sinnickson performed the overall research, analyzed data, and wrote the manuscript. Dr. Holden Harris contributed new methods and models. Dr. David Chagaris and Dr. Dylan Sinnickson both designed the study and made manuscript revisions throughout the writing process.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Sinnickson, D., Harris, H.E. & Chagaris, D. Assessing Energetic Pathways and Time Lags in Estuarine Food Webs. Ecosystems 26, 1468–1488 (2023). https://doi.org/10.1007/s10021-023-00845-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10021-023-00845-1