Abstract
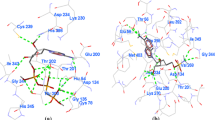
S-adenosyl-L-homocysteine hydrolase (SAHase) is an important regulator in the methylation reactions in many organisms and thus is crucial for numerous cellular functions. In recent years, SAHase has become one of the popular targets for drug design, and SAHase inhibitors have exhibited potent antiviral activity. In this study, we established the complex-based pharmacophore models based on the known crystal complex of SAHase (PDB ID: 1A7A) to screen the drug-likeness compounds of ChEMBL database. Then, three molecular docking programs were used to validate the reliability of compounds, involving Libdock, CDOCKER, and AutoDock Vina programs. The four promising hit compounds (CHEMBL420751, CHEMBL346387, CHEMBL1569958, and CHEMBL4206648) were performed molecular dynamics simulations and MM-PBSA calculations to evaluate their stability and binding-free energy in the binding site of SAHase. After screening and analyzing, the hit compounds CHEMBL420751 and CHEMBL346387 were suggested to further research to obtain novel potential SAHase inhibitors.
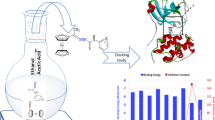
Graphical abstract
A series of computer-aided drug design methods, including pharmacophore, molecular docking, molecular dynamics simulation and MM-PBSA calculations, were employed in this study to identity novel inhibitors of S-adenosyl-L-homocysteine hydrolase (SAHase). Some compounds from virtual screening could form various interactions with key residues of SAHase. Among them, compounds CHEMBL346387 and CHEMBL420751 exhibited potent binding affinity from molecular docking and MM-PBSA, and maintained good stability at the binding site during molecular dynamics simulations as well. All these results indicated that the selected compounds might have the potential to be novel SAHase inhibitors.












Similar content being viewed by others
Data availability
The data sets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Code availability
The software used is commercially available.
References
Gao J, Cahill CM, Huang X (2018) S-Adenosyl methionine and transmethylation pathways in neuropsychiatric diseases throughout life. Neurotherapeutics 15:156–175. https://doi.org/10.1007/s13311-017-0593-0
Dai X, Ren T, Zhang Y, Nan N (2021) Methylation multiplicity and its clinical values in cancer. Expert Rev Mol Med 23:1–10. https://doi.org/10.1017/erm.2021.4
Kachroo P, Morrow JD, Vyhlidal CA, Gaedigk R, Silverman EK, Weiss ST, Tantisira KG, DeMeo DL (2021) DNA methylation perturbations may link altered development and aging in the lung. Aging 13:1742–1764. https://doi.org/10.18632/aging.202544
Adams JM, Cory S (1975) Modified nucleosides and bizarre 5′-termini in mouse myeloma mRNA. Nature 255:28–33. https://doi.org/10.1038/255028a0
Banerjee AK (1980) 5′-terminal cap structure in eucaryotic messenger ribonucleic acids. Microbiol Rev 44:175–205. https://doi.org/10.1128/mr.44.2.175-205.1980
Both GW, Banerjee AK, Shatkin AJ (1975) Methylation-dependent translation of viral messenger RNAs in vitro. Proc Natl Acad Sci 72:1189–1193. https://doi.org/10.1073/pnas.72.3.1189
Roje S (2006) S-Adenosyl-L-methionine: beyond the universal methyl group donor. Phytochemistry 67:1686–1698. https://doi.org/10.1016/j.phytochem.2006.04.019
Youngblood B, Shieh FK, Buller F, Bullock T, Reich NO (2007) S-adenosyl-L-methionine-dependent methyl transfer: observable precatalytic intermediates during DNA cytosine methylation. Biochemistry 46:8766–8775. https://doi.org/10.1021/bi7005948
Lee HO, Wang L, Kuo YM, Andrews AJ, Gupta S, Kruger WD (2018) S-adenosylhomocysteine hydrolase over-expression does not alter S-adenosylmethionine or S-adenosylhomocysteine levels in CBS deficient mice. Mol Genet Metab Rep 15:15–21. https://doi.org/10.1016/j.ymgmr.2018.01.002
Xiao Y, Su X, Huang W, Zhang J, Peng C, Huang H, Wu X, Huang H, Xia M, Ling W (2015) Role of S-adenosylhomocysteine in cardiovascular disease and its potential epigenetic mechanism. Int J Biochem Cell Biol 67:158–166. https://doi.org/10.1016/j.biocel.2015.06.015
Altintas E, Sezgin O (2004) S-adenosylhomocysteine hydrolase, S-adenosylmethionine, S-adenosylhomocysteine: correlations with ribavirin induced anemia. Med Hypotheses 63:834–837. https://doi.org/10.1016/j.mehy.2004.03.031
Brzezinski K (2020) S-adenosyl-l-homocysteine hydrolase: a structural perspective on the enzyme with two Rossmann-fold domains. Biomolecules 10:1682. https://doi.org/10.3390/biom10121682
Converso A, Hartingh T, Fraley ME, Garbaccio RM, Hartman GD, Huang SY, Majercak JM, McCampbell A, Na SJ, Ray WJ, Savage MJ, Wolffe C, Yeh S, Yu Y, White R, Zhang R (2014) Adenosine analogue inhibitors of S-adenosylhomocysteine hydrolase. Bioorg Med Chem Lett 24:2737–2740. https://doi.org/10.1016/j.bmcl.2014.04.034
Turner MA, Yang X, Yin D, Kuczera K, Borchardt RT, Howell PL (2000) Structure and function of S-adenosylhomocysteine hydrolase. Cell Biochem Biophys 33:101–125. https://doi.org/10.1385/CBB:33:2:101
Liu S, Wolfe MS, Borchardt RT (1992) Rational approaches to the design of antiviral agents based on S-adenosyl-L-homocysteine hydrolase as a molecular target. Antiviral Res 19:247–265. https://doi.org/10.1016/0166-3542(92)90083-h
Malanovic N, Streith I, Wolinski H, Rechberger G, Kohlwein SD, Tehlivets O (2008) S-adenosyl-L-homocysteine hydrolase, key enzyme of methylation metabolism, regulates phosphatidylcholine synthesis and triacylglycerol homeostasis in yeast: implications for homocysteine as a risk factor of atherosclerosis. J Biol Chem 283:23989–23999. https://doi.org/10.1074/jbc.M800830200
Qian G, Chen C, Zhou R (2014) A thermostable S-adenosylhomocysteine hydrolase from Thermotoga maritima: properties and its application on S-adenosylhomocysteine production with enzymatic cofactor regeneration. Enzyme Microb Technol 64:33–37. https://doi.org/10.1016/j.enzmictec.2014.06.007
Chiang PK (1998) Biological effects of inhibitors of S-adenosylhomocysteine hydrolase. Pharmacol Ther 77:115–134. https://doi.org/10.1016/S0163-7258(97)00089-2
Tan X, Wang P, Nian S, Wang G (2014) Design and synthesis of amide derivatives as S-adenosyl-L-homocysteine hydrolase inhibitors. Chem Pharm Bull 2:112–117. https://doi.org/10.1248/cpb.c13-00623
Jia Y, Li P, Song W (2016) Rational design of a profluorescent substrate for S-adenosylhomocysteine hydrolase and its applications in bioimaging and inhibitor screening. ACS Appl Mater Interfaces 8:25818–25824. https://doi.org/10.1021/acsami.6b09190
Lu W, Zhang R, Jiang H, Zhang H, Luo C (2018) Computer-aided drug design in epigenetics. Front Chem 6:57. https://doi.org/10.3389/fchem.2018.00057
Glaab E (2016) Building a virtual ligand screening pipeline using free software: a survey. Brief Bioinform 17:352–366. https://doi.org/10.1093/bib/bbv037
Forli S (2015) Charting a path to success in virtual screening. Molecules 20:18732–18758. https://doi.org/10.3390/molecules201018732
Li Q, Shah S (2017) Structure-based virtual screening. Methods Mol Biol 1558:111–124. https://doi.org/10.1007/978-1-4939-6783-4_5
Kontoyianni M (2017) Docking and virtual screening in drug discovery. Methods Mol Biol 1647:255–266. https://doi.org/10.1007/978-1-4939-7201-2_18
Liu X, Shi D, Zhou S, Liu H, Liu H, Yao X (2018) Molecular dynamics simulations and novel drug discovery. Expert Opin Drug Discov 13:23–37. https://doi.org/10.1080/17460441.2018.1403419
Aci-Sèche S, Ziada S, Braka A, Arora R, Bonnet P (2016) Advanced molecular dynamics simulation methods for kinase drug discovery. Future Med Chem 8:545–566. https://doi.org/10.4155/fmc.16.9
Wang E, Sun H, Wang J, Wang Z, Liu H, Zhang JZH, Hou T (2019) End-point binding free energy calculation with MM/PBSA and MM/GBSA: strategies and applications in drug design. Chem Rev 119:9478–9508. https://doi.org/10.1021/acs.chemrev.9b00055
Clercq DE (1987) S-adenosylhomocysteine hydrolase inhibitors as broad-spectrum antiviral agents. Biochem Pharmacol 36:2567–2575. https://doi.org/10.1016/0006-2952(87)90533-8
Yuan CS, Saso Y, Lazarides E, Borchardt RT, Robins MJ (1999) Recent advances in S-adenosyl-L-homocysteine hydrolase inhibitors and their potential clinical applications. Expert Opin Ther Pat 9:1197–1206. https://doi.org/10.1517/13543776.9.9.1197
Seidel T, Schuetz DA, Garon A, Langer T (2019) The pharmacophore concept and its applications in computer-aided drug design. Prog Chem Org Nat Prod 110:99–141. https://doi.org/10.1007/978-3-030-14632-0_4
Rao SN, Head MS, Kulkarni A, LaLonde JM (2007) Validation studies of the site-directed docking program LibDock. J Chem Inf Model 47:2159–2171. https://doi.org/10.1021/ci6004299
Wu H, Liu Y, Guo M, Xie J, Jiang X (2014) A virtual screening method for inhibitory peptides of angiotensin I-converting enzyme. J Food Sci 79:1635–1642. https://doi.org/10.1111/1750-3841.12559
Wu G, Robertson DH, Brooks CL, Vieth M (2003) Detailed analysis of grid-based molecular docking: a case study of CDOCKER-A CHARMm-based MD docking algorithm. J Comput Chem 24:1549–1562. https://doi.org/10.1002/jcc.10306
Jaghoori MM, Bleijlevens B, Olabarriaga SD (2016) 1001 Ways to run AutoDock Vina for virtual screening. J Comput Aided Mol Des 30:237–249. https://doi.org/10.1007/s10822-016-9900-9
Collier TA, Piggot TJ, Allison JR (2020) Molecular dynamics simulation of proteins. Methods Mol Biol 2073:311–327. https://doi.org/10.1007/978-1-4939-9869-2_17
Hollingsworth SA, Dror RO (2018) Molecular dynamics simulation for all. Neuron 99:1129–1143. https://doi.org/10.1016/j.neuron.2018.08.011
Warren GL, Andrews CW, Capelli AM, Clarke B, LaLonde J, Lambert MH, Lindvall M, Nevins N, Semus SF, Senger S, Tedesco G, Wall ID, Woolven JM, Peishoff CE, Head MS (2006) A critical assessment of docking programs and scoring functions. J Med Chem 49:5912–5931. https://doi.org/10.1021/jm050362n
Kontoyianni M, McClellan LM, Sokol GS (2004) Evaluation of docking performance: comparative data on docking algorithms. J Med Chem 47:558–565. https://doi.org/10.1021/jm0302997
Jain AN (2006) Scoring functions for protein-ligand docking. Curr Protein Pept Sci 7:407–420. https://doi.org/10.2174/138920306778559395
Turner MA, Yuan CS, Borchardt RT, Hershfield MS, Smith GD, Howell PL (1998) Structure determination of selenomethionyl S-adenosylhomocysteine hydrolase using data at a single wavelength. Nat Struct Biol 5:369–376. https://doi.org/10.1038/nsb0598-369
Joshi T, Joshi T, Sharma P, Chandra S, Pande V (2021) Molecular docking and molecular dynamics simulation approach to screen natural compounds for inhibition of Xanthomonas oryzae pv. Oryzae by targeting peptide deformylase. J Biomol Struct Dyn 39:823–840. https://doi.org/10.1080/07391102.2020.1719200
Şahİn K, DurdaĞi S (2020) Combined ligand and structure-based virtual screening approaches for identification of novel AChE inhibitors. Turk J Chem 44:574–588. https://doi.org/10.3906/kim-1911-57
Tirado-Rives J, Jorgensen WL (2006) Contribution of conformer focusing to the uncertainty in predicting free energies for protein-ligand binding. J Med Chem 49:5880–5884. https://doi.org/10.1021/jm060763i
Acknowledgements
We are grateful to the reviewers for their invaluable suggestions.
Funding
This work was supported by the National Natural Science Foundation of China (No. 82060627), Guangxi Natural Science Foundation of China (No. 2020GXNSFAA159149, No. 2018GXNSFAA281114), Innovation Project of Guangxi Graduate Education (YCSW2022367).
Author information
Authors and Affiliations
Contributions
Cong Chen: methodology, software, writing—original draft. Xiang-Hui Zhou: data analysis, manuscript editing. Wa Cheng: data analysis, software. Yan-Fen Peng: investigation, validation. Qi-Ming Yu: investigation, visualization, validation. Xiang-Duan Tan: conceptualization, writing—review and editing, project management.
Corresponding authors
Ethics declarations
Consent to participate
All authors have seen the manuscript and approved to submit the manuscript.
Consent to publish
All authors consent to the publication of the manuscript.
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Chen, C., Zhou, XH., Cheng, W. et al. Identification of novel inhibitors of S-adenosyl-L-homocysteine hydrolase via structure-based virtual screening and molecular dynamics simulations. J Mol Model 28, 336 (2022). https://doi.org/10.1007/s00894-022-05298-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00894-022-05298-2