Abstract
Although a large number of potentially secreted proteins can be predicted on the basis of genomic distribution of signal sequence-bearing proteins, protein secretion in Archaea has barely been studied. A proteomic inventory and comparison of the growth medium proteins in three hyperthermoacidophiles, i.e., Sulfolobus solfataricus, S. acidocaldarius and S. tokodaii, indicates that only few proteins are freely secreted into the growth medium and that the majority originates from cell envelope bound forms. In S. acidocaldarius both cell-associated and secreted α-amylase activities are detected. Inactivation of the amyA gene resulted in a complete loss of activity, suggesting that the same protein is responsible for the a-amylase activity at both locations. It is concluded that protein secretion in Sulfolobus is a limited process, and it is suggested that the S-layer may act as a barrier for the free diffusion of folded proteins into the medium.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Microorganisms utilize various transport systems to secrete proteins from the cell, such as ABC transporters for peptide and toxin excretion (Delepelaire 2004; Kuipers et al. 2004), the universally conserved Sec-system for the translocation of unfolded secretory proteins (Driessen et al. 1998), and the twin-arginine (Tat) system for the export of folded polypeptides (Berks et al. 2005). Most proteins destined to be secreted contain an N-terminal signal peptide, which, in most cases, directs them to the Sec-system. In archaea, the vast majority of the secreted proteins are synthesized as precursors with a N-terminal signal peptide for export via the Sec-system (Saleh et al. 2008), while in haloarchaea the Tat pathway seems to be the predominant route (Rose et al. 2002; Gimenez et al. 2007). The N-terminal signal peptide consists of a universally conserved tripartite domain structure with a positively charged amino-terminal n-domain, a central hydrophobic h-domain, and a polar c-domain which contains the signal peptide cleavage site (von Heijne 1990). The signal peptide is removed by type 1 signal peptidases allowing the extracellular release of the mature protein domain (von Heijne 1990). Archaeal type 1 signal peptidases utilize a Ser-His-Asp triad for catalysis similar to eukaryote type 1 signal peptidases (Bardy et al. 2005). Previously, it was noted that the signal sequences of a variety of archaeal cell surface proteins are processed by specific intracellular peptidases at the n-domain resulting in the removal of the positively charged residues only (Bardy and Jarrell 2002; Albers et al. 2003). This processing activity leaves the hydrophobic h-domain at the new amino-terminus of the secreted protein. These class 3 signal peptides are typically found in bacterial type IV pilins, and may fulfill a structural role in the final assembly of these proteins at the cell surface (Craig et al. 2006). Some archaeal secreted proteins lack a clearly identifiable signal sequence suggesting that they are secreted via another unknown pathway (Cannio et al. 2000).
Currently, the number of experimentally determined signal sequences of archaeal secretory proteins is relatively limited, with only 69 secretory proteins derived from 33 different archaeal species. The h-domain of archaeal signal sequences appears especially rich in alanine, leucine, and isoleucine residues, while valine seems less abundant (Bagos et al. 2009). Based on this limited signal sequence database, an algorithm specifically aimed at predicting signal peptides in archaea has been developed that unlike other prediction methods was claimed to predict archaeal signal peptides with high accuracy (Bagos et al. 2009). A proteomics study of extracellular proteins is a valuable experimental approach to validate such programs and would yield more insights in the mechanisms of protein secretion in archaea. However, the number of secreted proteins identified in archaeal proteomic studies has been limited thus far. In the Antarctic archaeon Methanococcoides burtonii, 7 proteins were identified in the growth medium (Saunders et al. 2006) while a study on the complete Sulfolobus solfataricus proteome resulted in the identification of 32 proteins present in the extracellular medium fraction (Chong and Wright 2005). Since the genes encoding the majority of these proteins lacked a coding sequence for a signal peptide, many of the proteins identified likely originated from cell lysis.
Members of the crenarchaeal genus Sulfolobus are characterized by their lobed appearance and ability to grow both at high temperature (60–85°C) and low pH (2–4). They grow aerobically on a variety of carbon sources. In silico predictions suggested that about 4.2% (124 proteins) of the total proteome of S. solfataricus consists of signal peptide containing secretory proteins (Albers and Driessen 2002), but recently a lower number (~1.6%, 45 proteins) has been reported (Abu-Qarn and Eichler 2007). The number of signal peptide containing proteins of S. acidocaldarius and S. tokodaii amount to 12 (260 proteins) and 11 (322 protein) % of the total proteins, respectively (Saleh et al. 2008). The results of these prediction methods thus vary tremendously necessitating their experimental validation. Hence, we have made an inventory of the proteins present in the growth medium of three Sulfolobus species. In addition, glycosylated cell surface proteins were identified as a subclass of extracellular proteins. Our data suggest that only few proteins are secreted into the growth medium and that the majority of secreted proteins originate from cell-surface associated forms.
Experimental methods
Strains and growth conditions
S. acidocaldarius strain DSM639, S. tokodaii strain DSM16993, and S. solfatarcius P2 strain DSM1617 (DSMZ, Germany) were grown in Brocks medium (Brock et al. 1972), supplemented with 0.3% (w/v) glucose, 0.1% (w/v) casamino acids, 0.3% (w/v) starch, and/or 0.1% NZ-amine SA (Sigma) when indicated. With S. acidocaldarius strain MR31 (Reilly and Grogan 2001), 10 μg/ml uracil was added to the medium. Strains were grown at 80°C, except for S. acidocaldarius MR31 which was grown at 78°C. Gelrite (0.64%) plates were supplemented with various nutrients (as indicated below) and solidified with 10 mM MgCl2/3 mM CaCl2. Both liquid and solid media were at pH 3.0. For the isolation of soluble medium proteins, pre-cultures (50 ml) were grown until the late log phase, whereupon 3 ml was transferred into a 800 ml of growth medium in 3-l flasks that were shaking at 160 rpm. Growth was continued until the early log (OD600 of 0.5–0.7) or stationary phase.
Isolation of supernatant proteins
Cultures were cooled on ice for 20 min, followed by low-spin centrifugation at 12,000×g for 10 min at 4°C to collect the cells. Next, membrane vesicles were removed from the supernatant fraction by centrifugation at 125,000×g at 4°C for 45 min. The cleared medium fraction (about 750 ml) was passed through a 0.45 μm cellulose acetate filter, and concentrated to about 2 ml in a 200 ml stirred cell using an YM10 ultrafiltration membrane (10 kD cut-off, Amicon). The sample was further concentrated with a Microcon YM10 centrifugal filter (Millipore), to a final volume of 40–100 μl and 1.5–2.5 ml in the case of early log phase and stationary phase grown cells, respectively.
Isolation of glycosylated membrane proteins
Cells were washed once with 20 mM TrisCl pH 7.5, 150 mM NaCl, 1 mM EDTA, suspended in 20 mM TrisCl pH 7.5, 150 mM NaCl, and frozen in liquid nitrogen. For lysis, cells were thawed and subjected to French press treatment at 14,000 psi. Debris was removed by centrifugation for 10 min at 4°C at 5,500×g, and the membranes were collected by ultracentrifugation at 288,000×g at 4°C for 1 h. Membranes were solubilized for 1 h with 10 ml 1% dodecyl β-d-maltoside (DDM) in 20 mM TrisCl pH 7.5, 150 mM NaCl, and applied to a concanavalin A (ConA) Sepharose column equilibrated with the same buffer. Unbound protein was removed in a washing step with 30 column volumes of 0.1% DDM in 20 mM TrisCl pH 7.5, 150 mM NaCl. The ConA-bound proteins were eluted with 1 column volume of the same buffer containing 100 mM α-methylmannopyranoside.
Protein identification
Proteins were separated on a 12% SDS-PAGE, visualized with Coomassie brilliant blue G250 and protein bands were cut out for identification by mass spectrometry (MS). For LC (liquid chromatography)-MS, 12–15 μg of sample was used. All sample preparations were carried out as described previously (Ellen et al. 2009). Proteins were identified by matching the peptide masses and MS/MS spectra against the individual Sulfolobus species databases as reported previously (Ellen et al. 2009). Data were filtered manually to prevent matches with well-known trypsin autolysis products, human keratin contaminants, or non-peptide masses. False positives were removed manually when the analysis indicated that one spot contained multiple proteins. With LC–MS well-known trypsin autolysis products and human keratin contamination were removed before matching of the spectra. Mass spectra were extracted by the GPS Explorer Software, version 3.5 (Applied Biosystems, Foster City, CA, USA) and combined MS and MS/MS spectra were analyzed using Mascot (Matrix Science, London, UK; version 1.9.05). Mascot results were processed by GPS Explorer and scores reported as percentage confidence intervals (C.I.). Identifications with a C.I. >95% were assigned as significant, provided at least one unique peptide mass was matched.
In silico analysis
To detect the presence of a signal peptide the amino acid sequences of complete proteins were analyzed by SignalP trained on gram-positive bacteria (version 3.0) (Bendtsen et al. 2004, 2005) and PRED-SIGNAL trained on archaea (Bagos et al. 2009). Trans-membrane segment predictions were made with the TMHMM-program. TMHMM and SignalP can be found at: http://www.cbs.dtu.dk/services/. PRED-SIGNAL can be found at: http://bioinformatics.biol.uoa.gr/PRED-SIGNAL/. For the predictions of class 3 signal peptides the Flafind program was used (Szabo et al. 2007) at http://signalfind.org/flafind.html.
α-amylase activity assay
Supernatant fractions (1 ml) of late-exponential phase grown cells were collected by centrifugation (5 min at 16,000×g at room temperature), followed by a second centrifugation step (2 min at 16,000×g). Cell pellets were pooled and suspended to a final volume of 1 ml in demineralized water. A solution of 0.5% (w/v) starch in 250 mM sodium-acetate, 2.5 mM CaCl2, pH 3.4 was boiled for 5 min and diluted fivefold with an aliquot of the cell suspension or supernatant fraction (Haseltine et al. 1996). Samples were incubated overnight at 80°C. The α-amylase activity was determined by measuring the amount of reducing sugar ends (Fuwa 1954). Absorbance of the samples was measured at 500 nm, using solutions of d-maltose in the assay buffer as a standard.
Cloning
Genomic DNA of S. acidocaldarius strain DSM639 was isolated using the QuickPick™ SML gDNA kit from BioNobile (Finland) according to the manual. By PCR, a fragment of 841 bp of the upstream region of the α-amylase gene Saci_1162 was amplified using the forward primer 5′-CCCCCCCCCCGCGGCCCCTCATTAGCTTCTGTGAG, which introduces a SacII restriction site, and the reverse primer 5′-CCCCCCCCGGATCCACGAATATACTTAGCACTAGGAC, which introduces a BamH1 restriction site. Of the downstream region, a DNA fragment of 1147 bp was amplified using the forward primer 5′-CCCCCCCCGGTACCGTGTTAGGTCTAACCGTAAGGAGG, which introduces a Kpn1 restriction site, and reverse primer 5′-CCCCCCCCCTCGAGTTCTAACAGAGCTTTTATATATAG, which introduces a Xho1 restriction site. Using the introduced restriction sites the amplified up- and downstream regions were cloned up- and downstream, respectively, of a pyrEF cassette in the pBluescript vector p∆2pyrEF (Wagner et al. 2009) which resulted in the gene inactivation construct pKO1162.
Isolation of competent S. acidocaldarius cells
Competent cells were isolated according to “procedure K” (Kurosawa and Grogan 2005). S. acidocaldarius strain MR31 grown on minimal medium containing 0.1% NZ-amine was used to inoculate 50 ml of minimal medium containing 0.1% NZ-amine and 0.2% d-xylose. At an OD600 between 0.1 and 0.3, the culture was cooled on ice for 20 min. Next, cells were harvested at 2,000×g at 4°C for 20 min, suspended in 50 ml of cold 20 mM sucrose (in demineralized water), whereupon the cells were collected by centrifugation, resuspended in 10 ml of 20 mM sucrose solution, and collected again by centrifugation at 2,000×g at 4°C for 20 min. Cells were suspended in 1 ml of the cold 20 mM sucrose solution, quickly collected by centrifugation in a Eppendorf tabletop centrifuge at 4°C for 5 min at 4,000 rpm, and finally suspended in 20 mM sucrose to an OD600 of 5.0. Cells were stored in 50 μl aliquots at −80°C.
Construction of the α-amylase deletion strain of S. acidocaldarius
Electroporation was performed essentially as described before (Berkner et al. 2007). A 50-μl aliquot of competent S. acidocaldarius cells was mixed on ice with 100–300 ng of unmethylated pKO1162 DNA and the cells were immediately transferred to a 1-mm path length electroporation cuvette. An electroporation pulse was given at 1,250 V, 1,000 Ω, 25 μF using a Biorad gene pulser II (Biorad, USA), whereupon 50 μl of a 2× concentrated recovery solution (1% sucrose, 20 mM β-alanine, 20 mM malate buffer pH 4.5, 10 mM MgSO4) was added. Cells were incubated for 30 min at 75°C under mild shaking conditions (300 rpm), and transferred to gelrite plates containing minimal medium supplemented with 0.1% NZ-amine and 0.2% d-xylose. Plates were incubated for 4–5 days at 78°C, whereupon cells from large clear brown colonies were collected and used to inoculate 50 ml of minimal medium containing 0.1% NZ-amine. Cultures were grown for 2–3 days. To ensure that the culture consisted solely of transformed cells, aliquots were transferred to 0.1% NZ-amine gelrite plate to inspect colonies for uniformity in size and color. Single colonies from this second plating were then screened for the absence or presence of the α-amylase gene by PCR. PCR results were confirmed by sequencing and α-amylase activity assays.
Results
Supernatant proteins of three Sulfolobus species
In order to identify extracellular proteins of Sulfolobus, a comparative study of growth medium proteins was conducted with three different Sulfolobus species, i.e., S. solfataricus, S. acidocaldarius, and S. tokodaii. Cells were grown in minimal medium supplemented with glucose and casamino acids until the early log and stationary phase. To minimize contamination by cell lysis, the growth medium was harvested at the early log phase (OD600 of 0.5–0.7). The maximum OD600 at stationary phase was 3.2 for S. solfataricus, 2.5 for S. tokodaii and 2.0 for S. acidocaldarius. Soluble proteins were recovered from the supernatant after high speed centrifugation that cleared the sample of intact cells and membrane vesicles (Ellen et al. 2009). With all three species, the amount of supernatant protein appeared very small based on total protein determination. Therefore, an extensive ultra filtration concentration step was necessary for further analysis. In early log-grown cells, the total protein content of the growth medium was about 83 μg/l for S. tokodaii, 215 μg/l for S. acidocaldarius, and 625 μg/l for S. solfataricus. These values increased in the stationary phase to about 0.84, 2.7, and 2.1 mg/l, respectively.
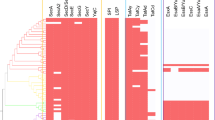
Protein profiles were analyzed by SDS-PAGE (Fig. 1a) and mass spectrometry after gel excision to identify the supernatant proteins. This analysis led to the identification of 13 unique proteins (See Supplementary Table 1 for peptide identification, number of peptides and significance). Only one of the proteins, i.e., SSO2894, a putative transposase, appears to lack a signal peptide, while all other identified proteins are encoded by genes that specify proteins with a signal sequence (see below). Overall, these data suggest that only little cell lysis must have occurred during growth and sample preparation. This was further confirmed by an enzymatic assay of β-galactosidase (LacS), a cytosolic enzyme in S. solfataricus (Cubellis et al. 1990). LacS was choosen as a lysis marker because of its relative high stability at low pH (Pisani et al. 1990). The LacS activity was easily detected in cells disrupted by French pressure treatment, while no significant LacS activity (below the detection limit, <1%) was detected in the concentrated supernatant of the exponential and stationary grown cells. Furthermore, by mass spectrometry, LacS was never identified in the growth medium supernatant fraction.
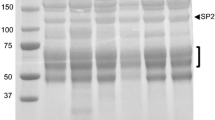
Most of the supernatant proteins identified in S. solfataricus and S. acidocaldarius are substrate-binding proteins that function in conjunction with ABC transporters (Table 1). Although the number of different binding proteins appears to be relatively high in the medium, their amounts are low. This is exemplified by the glucose-binding protein of S. solfataricus which is visible as a very faint band in coomassie blue stained SDS-PAGE of the extracellular medium fraction (Fig. 1a), whereas the binding protein is a major constituent of the membrane fraction (Fig. 1b). The most abundant supernatant protein of S. solfataricus is the large Surface layer protein SlaA (SSO0389) (Veith et al. 2009). On SDS-PAGE, it migrates as a 150-kDa protein and as two closely spaced ~31 kDa proteins. Because of the predicted size of 132 kDa, the latter proteins are likely degradation products. The in-gel method also allowed for the detection of the large S-layer protein of S. acidocaldarius, but not of S. tokodaii. When instead of early log phase, supernatant fractions of stationary phase cultures were analyzed essentially similar results were obtained, except for an archaeal ESCRT-III protein (SSO0881) that was present in S. solfataricus medium fraction, only. The latter was previously identified to be associated with extracellular membrane vesicles (Ellen et al. 2009). The medium fraction of stationary grown S. tokodaii cells contains a cytochromeb558/566 subunit A and an α-amylase (Table 1). Although the in-gel identification method was performed independently for at least three times, this did not lead to the identification of further proteins. To improve the sensitivity of detection, supernatant proteins of stationary phase grown cells were subjected to LC–MS analysis. This resulted in a total of 49 unique proteins (Tables 1, 2). Five of these growth medium proteins were found in all three species, i.e., the large S-layer protein, cytochromeb558/566 subunit A, two substrate binding proteins, and homologs of the S. solfataricus ESCRT-III protein.
Glycosylated membrane protein subproteome
The functional annotation of many of the supernatant proteins of the various Sulfolobus species suggests that they function at the cell-surface. To further determine their localization, a cell envelope subproteome of glycosylated proteins was obtained. Previous studies with S. solfataricus have shown that this fraction is enriched with substrate binding proteins (Elferink et al. 2001). Proteins were separated by SDS-PAGE, excised, and identified by mass spectrometry (See Fig. 1b and Supplementary Table 1 for peptide identification, number of peptides and significance). A total of 28 proteins were found (Tables 1, 2). Of the 28 identified proteins 16 appear to be proteins containing an N-terminal signal sequence (see below). A considerable overlap (13 proteins) was evident with the proteins identified in the glycosylated membrane. The majority (10) of these overlapping proteins corresponded to substrate-binding proteins.
Signal sequence prediction
All proteins
The combined data of the supernatant and the glycosylated fractions show a total of 64 proteins identified either in the medium supernatant, the cell envelope glycosylated fraction, or in both (see Table 3 for summary). The predicted amino acid sequences of these proteins were analyzed with SignalP and PRED-SIGNAL to detect potential signal sequences. For SignalP, the Gram-positive mode was used which has been reported to yield the best results for archaea (Bagos et al. 2009). The archaea-specific PRED-SIGNAL program predicted a signal sequence in only 29 out of the 64 proteins, many of which also contain a transmembrane segment (TMS). In addition, 13 proteins are predicted to contain an N-terminal TMS. The latter could in principle also function as a signal sequence, and indeed the majority of these proteins (10) are found in the medium supernatant fraction. Therefore, from here on all proteins that are predicted to contain either a signal peptide or an N-terminal TM helix are considered to contain a signal peptide regardless whether the signal peptide functions as a TM helix or not. SignalP predicts that 36 proteins are synthesized as precursors with a signal peptide.
Supernatant proteins
Among the 49 proteins identified in the supernatant fraction, the majority (38) is predicted to contain a signal peptide. This high yield of secretory proteins is consistent with the notion that the contamination of the medium supernatant fraction with cytosolic proteins is very low. In S. solfataricus only two proteins were identified that lacked a signal peptide, one of which is an ESCRT-III protein that was previously found to be associated with released membrane vesicles. In both S. acidocaldarius and S. tokodaii four proteins were found that seem to lack a signal peptide. In addition, some proteins contained an unusual signal peptide. The gene encoding the hypothetical protein ST1074 of S. tokodaii specifies a signal peptide that lacks the canonical positively charged amino acids of the N-domain (see Fig. 2), while the S. solfataricus endoglucanase (SSO1354) contains a typical class 1 signal sequence followed by a serine/threonine-rich domain (ST-linker) that previously had only been observed with class 3 signal peptide bearing binding proteins (Elferink et al. 2001).
Non-canonical archaeal signal peptides of secretory proteins found in the medium fraction of Sulfolobus cells. a Signal peptides lacking positively charged residues in the N-domain. The predicted h-domain is underlined. b Class 3 signal peptides of substrate binding proteins. The signal peptidase cleavage recognition site is depicted in bold and underlined, with an arrow indicating the cleavage site. The consensus motifs of the signal cleavage site and of the region connecting the H-domain (underlined) are indicated. The H-domain is followed by a serine/threonine rich region that forms a potential glycosylation site
Of the signal peptide containing medium proteins, 26 are exclusively found in the supernatant. TMHMM predicted that 15 out of these 26 proteins are integral membrane proteins and thus originate from the cell envelope. In S. tokodaii, the fraction also contains the large S-layer protein and a CTP synthase ST0237. The latter catalyses the ATP-dependent amination of UTP to CTP, and this protein is likely of cytosolic origin. This leaves only nine proteins that are potentially secreted directly into the medium: an endogluconase from S. solfataricus, the hypothetical protein Saci_1272 from S. acidocaldarius, and two thermopsins (proteases) plus five hypothetical proteins from S. tokodaii. None of these proteins were identified by the in-gel method suggesting that they are secreted in low quantities. Taken together, these data suggest that Sulfolobus excrete only few proteins directly into the medium.
Glycosylated fraction
The cytoplasmic contamination appeared larger in the glycosylated subproteome with 12 out 28 proteins devoid of an apparent signal peptide. Cytosolic contaminants in this subfraction likely originate as contaminants that associate with the cytoplasmic membranes and that remain in these fraction during the non-stringent washing conditions used during the lectin column chromatography. Remarkably, 12 of the 16 predicted signal peptide-containing proteins were also found in the supernatant fraction further suggesting that they emerged from a cell surface bound protein fraction. One of these proteins, i.e., a putative sugar-binding protein of S. tokodaii (ST1103) is annotated without a signal peptide, whereas the corresponding homologs of S. solfataricus and S. acidocaldarius both contain a class 1 signal peptide. Analysis of the genomic DNA sequence of S. tokodaii revealed the presence of a putative signal peptide in frame with the remainder of the coding sequence, but the gene is preceded by an unusual translation initiation codon, i.e., TTG instead of ATG. Similar observations have been made for other genes in the crenarchaeon Aeropyrum pernix (Yamazaki et al. 2006). The polytopic membrane protein cytochrome b, which was shown to be glycosylated in S. acidocaldarius (Zahringer et al. 2000) was only identified in the glycosylated subproteome of S. tokodaii.
Secretion of α-amylase
Our analysis suggests that the majority of the supernatant proteins originate from cell surface-associated forms. To further examine this hypothesis, the cellular distribution of an enzyme with known function, i.e., α-amylase (Haseltine et al. 1996), was determined in all three analyzed Sulfolobus species. The α-amylase protein contains a putative C-terminal TMS that may function as a membrane anchor. Cells grown into the stationary phase in the presence of starch as a carbon-source were separated from the medium by centrifugation. In S. solfataricus essentially all α-amylase activity was recovered in the medium fraction, while in S. tokodaii and S. acidocaldarius a significant portion of the α-amylase activity was cell-associated, i.e., ~25 and ~30%, respectively. To verify that the cell-associated and soluble α-amylase activity relate to the same protein, the α-amylase gene amyA of S. acidocaldarius strain MR31 was disrupted by an insertion of a pyrEF cassette (see Fig. 3). In contrast to the parental MR31 strain, the ΔamyA strain was unable to grow on minimal medium with starch. However, the strain also failed to grow on minimal medium with glucose or maltose (data not shown), while growth could be recovered when 0.1% NZ amine was included in the growth medium (Fig. 4). At this concentration, NZ amine supported some growth in the absence of any other carbon source, but with wild-type cells, growth was stimulated by the addition of maltose, glucose, or starch. In contrast, growth of the ΔamyA strain was stimulated by glucose or maltose, but not by starch (Fig. 4). Both the medium and cell fraction of the stationary grown ΔamyA strain were devoid of α-amylase activity (Fig. 5). Taken together, these data demonstrate that the cell-bound and medium α-amylase activity in S. acidocaldarius originates from the same protein and suggest that a significant amount of the secreted α-amylase is surface-bound in some Sulfolobus species.
Disruption of the α-amylase gene of S. acidocaldarius. a Schematic representation of the genomic region showing the wild type and ΔamyA mutant in which amyA was been replaced by pyrEF. The PCR primer sets used for the validation of the gene inactivation are indicated with numbers corresponding to the PCR fragments showing in b. PCR primers anneal at either amyA or pyrEF and outside the flanking genomic regions. b PCR validation of the amyA gene disruption. PCR fragments: 1 internal fragment of amyA (500 bp); 2 3′-region of pyrEF (1,251 bp); 3 3′-region of amyA (1,022 bp); 4 5′-region of pyrEF (1,017 bp); and 5 5′-region of amyA (1,023 bp)
Cell-associated (black bars) and medium (white bars) α-amylase activity of S. acidocaldarius MR31 and the derived ΔamyA strain grown to the late log phase on minimal medium with NZ-amine without and with glucose or starch, respectively. The activity expressed in number of hydrolyzed glucosidic bonds in mM/h
Discussion
This study presents an inventory of growth medium proteins and glycosylated cell surface proteins of three species of the hyperthermoacidophilic archaeon Sulfolobus grown on Brocks minimal medium containing glucose and casamino acids. By a combination of in-gel trypsin digestion/MALDI-TOF/TOF mass spectrometry and LC–MS analysis of the concentrated growth medium supernatant fraction, 11 proteins could be identified that found only in the medium supernatant fraction with a predicted signal sequence. These are likely secreted directly into the medium. This number is much lower than predicted by in silico genome analysis for potential secretory proteins (Albers and Driessen 2002; Abu-Qarn and Eichler 2007; Saleh et al. 2008; Bagos et al. 2009). There will be many factors that contribute to this deviation, amongst others the growth conditions used that will result in the expression of a subset of secretory proteins only. Moreover, the in silico analysis also does not distinguish between cell surface and medium localized proteins. Since our focus was on the soluble medium proteins, many of the cell surface localized secreted proteins are not be detected in our analysis.
Remarkably, many of the medium proteins seem to originate from cell surface-associated proteins that are likely released into the extracellular medium. A significant number of these supernatant proteins were also detected in the glycosylated membrane protein fraction. It is possible that cell surface-localized proteins are lost due to “proteolytic shaving” (Tjalsma et al. 2004) as a natural occurring process during growth. An alternative source could be the disintegration of the secretory membrane vesicles that are released into the medium (Ellen et al. 2009). In this respect, vesicle-associated ESCRT-III proteins were also found in the medium supernatant fraction. The exact function of these membrane vesicles is unknown but they may participate in protein secretion. Important examples of released cell-surface proteins are the substrate-binding proteins that function in conjunction with ABC transporters for the uptake of nutrients, the membrane protein cytochromeb558/566 subunit A and the S-layer protein. Substrate-binding proteins are major constituents of the glycosylated membrane protein fraction (Elferink et al. 2001) and in the case of the readily identified glucose-binding protein, only a very small fraction seems to be released into the medium. Release of a substrate-binding protein into the growth medium was previously reported for the crenarcheaon Aeropyrum pernix (Palmieri et al. 2006).
Many of the substrate-binding proteins contain a class 3 signal sequence which is processed in the cytosol such that the N-terminal positively charged residues are removed (Szabo et al. 2007). The remaining stretch of hydrophobic residues at the new N-terminus may function as a scaffold for assembly of the binding proteins into a large oligomeric structure such as the bacterial pilins which assemble into the pilus (Craig et al. 2006). Previous studies have shown that in S. solfataricus the substrate-binding proteins with a class 3 signal peptide require the bas operon for functional assembly in the cell envelope (Zolghadr et al. 2007). The bas operon encodes subunits with homology to bacterial Type II and IV secretion systems, some of which are involved in the assembly of pilin or other cell surface organelles. With the exception of Saci_1038, all the class 3 signal peptide bearing substrate binding-proteins also contain a serine/threonine-rich region (ST-linker) that follows the hydrophobic amino acid stretch (Albers et al. 2004). The ST-linker could serve as a flexible arm between the scaffold assembly region and the substrate-binding domain to allow free movement of this region.
Our data show that only a limited number of proteins are secreted into the medium, and that these proteins are present in low amounts (i.e., only 20–700 μg/ml). This could be a characteristic of hyperthermophiles (Bardy et al. 2003). Possibly, the S-layer of Sulfolobales presents a barrier for the free release of secreted proteins. The pore size estimated by electron microscopy of a self-assembled S. acidocaldarius S-layer is ~5.2 nm at the inner side, but the actual size of the pores is likely much smaller as these S-layers are imaged at a relatively low resolution. These pores may restrict the diffusion of folded proteins through the S-layer (Mark et al. 2006). To investigate this phenomenon, the cellular distribution of α-amylase was analyzed as the activity of this protein can be conveniently assayed. The α-amylase AmyA contains a C-terminal TM domain that may function in anchoring the protein in the cytoplasmic membrane. Except for S. solfataricus (Haseltine et al. 1996) (this study), indeed a significant portion of the α-amylase activity of stationary phase grown S. acidocaldarius and S. tokodaii cells was found to be cell-surface bound. Unfortunately, the α-amylase activities in the exponential phase were too low to allow for an accurate analysis of its distribution. The inactivation of the amyA gene in S. acidocaldarius demonstrates that both supernatant and cell-associated α-amylase activity is due to the same α-amylase. In S. acidocaldarius a thermostable acid protease, termed thermopsin, was found both in the growth medium as well as associated with the cells (Lin and Tang 1990). It is presently unclear why the S. solfataricus α-amylase activity does not occur in a cell-bound form. Secretion might be restricted by the structure and composition of the S-layer that may differ among the three studied species. Alternatively, the S. solfataricus enzyme might be a slower folding variant, which would allow it to pass the S-layer pores more readily. Moreover, the S. solfataricus S-layer protein shares only a low 24 and 14% identity with the S. acidocaldarius and S. tokodaii S-layer proteins, respectively. On the other hand, the S. acidocaldarius and S. tokodaii S-layer proteins share 42% identity. Also the S. solfataricus S-layer protein is almost 200 amino acids shorter than the proteins of the other two species. In addition, the small S-layer protein of S. solfataricus differs significantly in amino acid sequence from the corresponding proteins of S. tokodaii and S. acidocaldarius (Veith et al. 2009). Thus, the S-layer of S. solfataricus may differ in structure from that of the other two species perhaps imposing less of a barrier to the release of α-amylase into the medium. Interestingly, in Bacillus stearothermophilus the S-layer functions as an adhesion site for an extracellular α-amylase (Egelseer et al. 1995).
Taken together, our findings demonstrate that in Sulfolobus the secretion of proteins into the medium is only a marginal process. Most of the proteins observed in the medium also exist as cell surface-bound forms. Future studies should address to what extent the S-layer poses a physical barrier for the free diffusion of folded proteins and their secretion.
References
Abu-Qarn M, Eichler J (2007) An analysis of amino acid sequences surrounding archaeal glycoprotein sequons. Archaea 2:73–81
Albers SV, Driessen AJM (2002) Signal peptides of secreted proteins of the archaeon Sulfolobus solfataricus: a genomic survey. Arch Microbiol 177:209–216
Albers SV, Szabo Z, Driessen AJM (2003) Archaeal homolog of bacterial type IV prepilin signal peptidases with broad substrate specificity. J Bacteriol 185:3918–3925
Albers SV, Koning SM, Konings WN, Driessen AJM (2004) Insights into ABC transport in archaea. J Bioenerg Biomembr 36:5–15
Bagos PG, Tsirigos KD, Plessas SK, Liakopoulos TD, Hamodrakas SJ (2009) Prediction of signal peptides in archaea. Protein Eng Des Sel 22:27–35
Bardy SL, Jarrell KF (2002) FlaK of the archaeon Methanococcus maripaludis possesses preflagellin peptidase activity. FEMS Microbiol Lett 208:53–59
Bardy SL, Eichler J, Jarrell KF (2003) Archaeal signal peptides–a comparative survey at the genome level. Protein Sci 12:1833–1843
Bardy SL, Ng SY, Carnegie DS, Jarrell KF (2005) Site-directed mutagenesis analysis of amino acids critical for activity of the type I signal peptidase of the archaeon Methanococcus voltae. J Bacteriol 187:1188–1191
Bendtsen JD, Nielsen H, von HG, Brunak S (2004) Improved prediction of signal peptides: SignalP 3.0. J Mol Biol 340:783–795
Bendtsen JD, Kiemer L, Fausboll A, Brunak S (2005) Non-classical protein secretion in bacteria. BMC Microbiol 5:58
Berkner S, Grogan D, Albers SV, Lipps G (2007) Small multicopy, non-integrative shuttle vectors based on the plasmid pRN1 for Sulfolobus acidocaldarius and Sulfolobus solfataricus, model organisms of the (cren-)archaea. Nucleic Acids Res 35:e88
Berks BC, Palmer T, Sargent F (2005) Protein targeting by the bacterial twin-arginine translocation (Tat) pathway. Curr Opin Microbiol 8:174–181
Brock TD, Brock KM, Belly RT, Weiss RL (1972) Sulfolobus: a new genus of sulfur-oxidizing bacteria living at low pH and high temperature. Arch Mikrobiol 84:54–68
Cannio R, D’angelo A, Rossi M, Bartolucci S (2000) A superoxide dismutase from the archaeon Sulfolobus solfataricus is an extracellular enzyme and prevents the deactivation by superoxide of cell-bound proteins. Eur J Biochem 267:235–243
Chong PK, Wright PC (2005) Identification and characterization of the Sulfolobus solfataricus P2 proteome. J Proteome Res 4:1789–1798
Craig L, Volkmann N, Arvai AS, Pique ME, Yeager M, Egelman EH, Tainer JA (2006) Type IV pilus structure by cryo-electron microscopy and crystallography: implications for pilus assembly and functions. Mol Cell 23:651–662
Cubellis MV, Rozzo C, Montecucchi P, Rossi M (1990) Isolation and sequencing of a new beta-galactosidase-encoding archaebacterial gene. Gene 94:89–94
Delepelaire P (2004) Type I secretion in gram-negative bacteria. Biochim Biophys Acta 1694:149–161
Driessen AJM, Fekkes P, van der Wolk JP (1998) The Sec system. Curr Opin Microbiol 1:216–222
Egelseer E, Schocher I, Sara M, Sleytr UB (1995) The S-layer from Bacillus stearothermophilus DSM 2358 functions as an adhesion site for a high-molecular-weight amylase. J Bacteriol 177:1444–1451
Elferink MG, Albers SV, Konings WN, Driessen AJ (2001) Sugar transport in Sulfolobus solfataricus is mediated by two families of binding protein-dependent ABC transporters. Mol Microbiol 39:1494–1503
Ellen AF, Albers SV, Huibers W, Pitcher A, Hobel CF, Schwarz H, Folea M, Schouten S, Boekema EJ, Poolman B, Driessen AJ (2009) Proteomic analysis of secreted membrane vesicles of archaeal Sulfolobus species reveals the presence of endosome sorting complex components. Extremophiles 13:67–79
Fuwa H (1954) A new method for microdetermination of amylase activity by the use of amylose as the substrate. J Biochem 41:583–603
Gimenez MI, Dilks K, Pohlschroder M (2007) Haloferax volcanii twin-arginine translocation substrates include secreted soluble, C-terminally anchored and lipoproteins. Mol Microbiol 66:1597–1606
Haseltine C, Rolfsmeier M, Blum P (1996) The glucose effect and regulation of alpha-amylase synthesis in the hyperthermophilic archaeon Sulfolobus solfataricus. J Bacteriol 178:945–950
Kuipers A, de BE, Rink R, Fekken S, Kluskens LD, Driessen AJ, Leenhouts K, Kuipers OP, Moll GN (2004) NisT, the transporter of the lantibiotic nisin, can transport fully modified, dehydrated, and unmodified prenisin and fusions of the leader peptide with non-lantibiotic peptides. J Biol Chem 279:22176–22182
Kurosawa N, Grogan DW (2005) Homologous recombination of exogenous DNA with the Sulfolobus acidocaldarius genome: properties and uses. FEMS Microbiol Lett 253:141–149
Lin X, Tang J (1990) Purification, characterization, and gene cloning of thermopsin, a thermostable acid protease from Sulfolobus acidocaldarius. J Biol Chem 265:1490–1495
Mark SS, Bergkvist M, Yang X, Teixeira LM, Bhatnagar P, Angert ER, Batt CA (2006) Bionanofabrication of metallic and semiconductor nanoparticle arrays using S-layer protein lattices with different lateral spacings and geometries. Langmuir 22:3763–3774
Palmieri G, Casbarra A, Fiume I, Catara G, Capasso A, Marino G, Onesti S, Rossi M (2006) Identification of the first archaeal oligopeptide-binding protein from the hyperthermophile Aeropyrum pernix. Extremophiles 10:393–402
Pisani FM, Rella R, Raia CA, Rozzo C, Nucci R, Gambacorta A, De Rosa M, Rossi M (1990) Thermostable beta-galactosidase from the archaebacterium Sulfolobus solfataricus. Purification and properties. Eur J Biochem 187:321–328
Reilly MS, Grogan DW (2001) Characterization of intragenic recombination in a hyperthermophilic archaeon via conjugational DNA exchange. J Bacteriol 183:2943–2946
Rose RW, Bruser T, Kissinger JC, Pohlschroder M (2002) Adaptation of protein secretion to extremely high-salt conditions by extensive use of the twin-arginine translocation pathway. Mol Microbiol 45:943–950
Saleh M, Song C, Nasserulla S, Leduc LG (2008) Indicators from archaeal secretomes. Microbiol Res 03:002
Saunders NF, Ng C, Raftery M, Guilhaus M, Goodchild A, Cavicchioli R (2006) Proteomic and computational analysis of secreted proteins with type I signal peptides from the Antarctic archaeon Methanococcoides burtonii. J Proteome Res 5:2457–2464
Szabo Z, Stahl AO, Albers SV, Kissinger JC, Driessen AJ, Pohlschroder M (2007) Identification of diverse archaeal proteins with class III signal peptides cleaved by distinct archaeal prepilin peptidases. J Bacteriol 189:772–778
Tjalsma H, Antelmann H, Jongbloed JD, Braun PG, Darmon E, Dorenbos R, Dubois JY, Westers H, Zanen G, Quax WJ, Kuipers OP, Bron S, Hecker M, van Dijl JM (2004) Proteomics of protein secretion by Bacillus subtilis: separating the “secrets” of the secretome. Microbiol Mol Biol Rev 68:207–233
Veith A, Klingl A, Zolghadr B, Lauber K, Mentele R, Lottspeich F, Rachel R, Albers SV, Kletzin A (2009) Acidianus, Sulfolobus and Metallosphaera surface layers: structure, composition and gene expression. Mol Microbiol 73:58–72
von Heijne G (1990) The signal peptide. J Membr Biol 115:195–201
Wagner M, Berkner S, Ajon M, Driessen AJ, Lipps G, Albers SV (2009) Expanding and understanding the genetic toolbox of the hyperthermophilic genus Sulfolobus. Biochem Soc Trans 37:97–101
Yamazaki S, Yamazaki J, Nishijima K, Otsuka R, Mise M, Ishikawa H, Sasaki K, Tago S, Isono K (2006) Proteome analysis of an aerobic hyperthermophilic crenarchaeon, Aeropyrum pernix K1. Mol Cell Proteomics 5:811–823
Zahringer U, Moll H, Hettmann T, Knirel YA, Schafer G (2000) Cytochrome b558/566 from the archaeon Sulfolobus acidocaldarius has a unique Asn-linked highly branched hexasaccharide chain containing 6-sulfoquinovose. Eur J Biochem 267:4144–4149
Zolghadr B, Weber S, Szabo Z, Driessen AJM, Albers SV (2007) Identification of a system required for the functional surface localization of sugar binding proteins with class III signal peptides in Sulfolobus solfataricus. Mol Microbiol 64:795–806
Acknowledgments
This work was supported by the Netherlands Proteomics Centre (NPC). We thank Wim Huibers for technical assistance in the mass spectrometry analysis and Bert Poolman for support. S.V.A received a VIDI grant from the Netherlands Science Organization (NWO) and intramural funds of the Max Planck Society.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by L. Huang.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This is an open access article distributed under the terms of the Creative Commons Attribution Noncommercial License (https://creativecommons.org/licenses/by-nc/2.0), which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
About this article
Cite this article
Ellen, A.F., Albers, SV. & Driessen, A.J.M. Comparative study of the extracellular proteome of Sulfolobus species reveals limited secretion. Extremophiles 14, 87–98 (2010). https://doi.org/10.1007/s00792-009-0290-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00792-009-0290-y