Abstract
Key to sound validation studies is the formalization and harmonization of procedures for design of experiment and interpretation of results. International guidelines (ISO 5725, ENGL) are available for the validation of GMO detection methods, and ad-hoc validation statistics (e.g. per cent bias, repeatability and reproducibility) are used for in-house and inter-laboratory testing and decision-making. Acceptability criteria have been set but not every situation can be covered by a preset rule; the interpretation of results in validation largely depends on expert judgement being a matter of professional judgment and expertise. Fuzzy logic-based techniques may be used to summarize the information obtained by independent validation statistics and are helpful in such respect. A comprehensive indicator of method performance permits direct comparison between methods and facilitates the evaluation of multiple, yet contradictory statistics. The European Union Reference Laboratory for GM Food and Feed has already proposed the fuzzy principle in the context of method validation. Other studies have also proved its applicability in other areas of GMO analysis, but the application has been limited hitherto. In this article, we review the fuzzy logic principle and its potential to support the continuous progress of GMO science and routine laboratory analyses.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Conform to the EU legislation, a traceability system for genetically modified organisms (GMOs) and GMO-derived products in the food and feed chains has been put into place [1, 2] In previous submissions to Accreditation and Quality Assurance [e.g. 3, 4], some of the challenges raising from implementing and enforcing the EU regulations for labelling of food and feed products containing GMOs were already highlighted. Especially, the need for highly performing, adequate GMO test methods represents a major issue.
GMO detection methods are mainly based on the quantitative polymerase chain reaction (qPCR) [5]. Minimal acceptance criteria for acceptable GMO qPCR method performance have been proposed at the International Organization for Standardization [6] level and by the European Network of GMO Laboratories (http://gmo-crl.jrc.ec.europa.eu/doc/Min_Perf_Requir_Analyt_methods_131008.pdf). In general, a method is considered to be valid when various performance statistics (per cent values of bias, repeatability and reproducibility standard deviations, etc.), relevant for a certain ‘purpose of use’ of the method, are matching the method acceptance thresholds, the latter set by expert consensus for each statistic.
Although the commercialization of a GMO necessitates in most parts of the world providing a suitable detection method, no harmonized approaches have been established as to the development of these methods. As a consequence, the detection methods for the different GMOs may vary in their relative performance, and in some cases even different methods with distinct performance have been developed to target the same GMO (see e.g. GMO events GA21, BT11 through http://gmo-crl.jrc.ec.europa.eu). Such situation is most inconvenient and does not facilitate an efficient management of the traceability of GMO along the food and feed production chain. To date, the validation of quantitative GMO detection systems developed in support to applications for GMO use as food or feed (Reg. (EC) No 1829/2003) applies so-called crisp acceptance thresholds set at a single value (e.g. bias = 25%). In the current set-up, the performance of each GMO detection method is described through inter-laboratory validation studies quantifying samples at several GMO concentrations (at http://gmo-crl.jrc.ec.europa.eu). Enforcement laboratories carrying out official controls to verify compliance of GM food and feed to traceability requirements act under ISO 17025 accreditation schemes [7]. This implies meeting accepted standards of validation practice and carefully documenting that they can work out methods in accordance with performance characteristics as reported in the validation exercises—primarily method bias and measurement of uncertainty [8]. Guidance documents have been released concerning the approaches required to estimate major sources of measurement uncertainty (MU) in analytical measurements and to describe the relationship between measurement uncertainty and analytical results by means of validation data from both collaborative and in-house studies. They mainly aim at quantifying the uncertainty [9], providing a frame to set up single-laboratory validations of methods of analysis [10] and describing the relationship between analytical results and measurement uncertainty. In such documents, particular focus is placed on the provisions of EU legislation concerning contaminants in food and undesirable substances in feed [11]. In the field of GM food and feed, a practical guidance to laboratories was published in the context of the activities of the European Network of GMO Laboratories [12] where, in particular, the concept of measurement uncertainty was detailed. MU is a measure of the dispersion of the values attributed to a measured quantity. It has a probabilistic basis and reflects the limitation in knowledge of the exact quantity (as opposed to precision). All measurements are subject to uncertainty, and a report on a measured value (in our case % GM DNA) is only complete if accompanied by a statement of the associated uncertainty. The associated uncertainty is essentially a confidence interval that gives the range of values that can reasonably be attributed to the measurand [9] (measurand is the specific quantity subject to measurement [9]). This can be estimated by combining measurement variability (RSD r and RSD R), the estimated measurement bias and the uncertainty associated with the measurement bias [9].
However, in validation studies it was observed that different test conditions may generate contrasting responses in terms of individual statistics (e.g. Table 1, bias and precision statistics for two GM events).
To obtain a clearer insight into overall method performance and suitability of particular methods for a certain use, the European Union Reference Laboratory for GM Food and Feed (EURL-GMFF) initiated to establish overall method performance assessment of quantitative GMO detection systems based on vague set mathematics in a so-called fuzzy logic approach to decision support [13].
For this, a comprehensive approach aimed at capturing the overall performance of the detection methods in a systematic manner through aggregating the outcomes of the different performance criteria into a single method performance index has been developed. In this respect, the application of fuzzy logic principles has been evaluated at the EURL-GMFF [14].
Validation metrics can be first combined in modules. In a second step, modules are combined in a single performance index according to the inferential rules described for the fuzzy logic. Hereinafter, we describe the modular aggregation indentified by the EURL-GMFF to conflate the parameters typically expressed in method validation for GMO detection.
As explained in [14], three modules can be defined: the qPCR method efficiency (combining the statistics for the efficiencies of the trans- and endo-gene quantitative qPCR method), the method applicability (representing the number of reagents, components in the reaction setup) and the qPCR system accuracy (combining the statistics on bias, repeatability and reproducibility). At a second stage, these three first-level modules are combined into a single GMO detection system performance index. This combination of statistics is supported by an expert weighing expression of the balance of importance of the individual inputs and their aggregation into modules. Modules and their corresponding indicators are dimensionless measures residing between 0 (best response) and 1 (worst response) as calculated via a fuzzy-based procedure [14]. Unlike crisp setting, three so-called membership classes were defined for individual outputs—favourable (F), unfavourable (U), partial membership—using transition curves in the range F to U (Fig. 1).
Example of crisp (top) and fuzzy (bottom) membership functions (x = the value of each individual input). Top graph: a = threshold value; S(x;a) = the crisp membership of the input x. Bottom graph: a = the lower threshold (values of x lower than a have membership to unfavourable class equal to 1 and to favourable class equal to 0) value; b = the upper threshold (values of x greater than b have membership to unfavourable class equal to 0 and to favourable class equal to 1); c = (a + b)/2; S(x;a;b) = the membership of the input x to the favourable set; its complement, 1 − S(x;a;b), gives the degree of membership of the input x to the unfavourable set
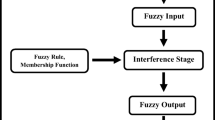
Fuzzy-based indicators permit to construct a flexible model (Fig. 2) in which multiple inputs (x 1 , …, x n , representing basic statistics) are aggregated into a single indicator. Inputs in a module (and, in turn, modules in the indicator) are combined according to decision rules. In particular, expected weights are assigned to each input (and, consequently, to the inputs when they are combined in the fuzzy model), the higher weight being assigned to the preposition that the expert judges farthest from good performance. The expert reasoning runs as follows: if all inputs are F, then the weight is 0 (best performance); if all inputs are U, then the weight is 1 (worst performance), while all the other combinations assume intermediate weights. A single output is ultimately resolved (defuzzification) on a value between 0 (best) and 1 (worst).
Schematic diagram of a two-stage fuzzy inference system for assessment purpose. The operational flow starts with the conversion of basic inputs (x 1 , …, x n ) into fuzzy rules and ends up with the crisp (defuzzified) output (z); aggregation of basic inputs into modules (y 1 , …, y n ) is an intermediate step (w 1 , …, w n are the expert weights applied to basic inputs; ‘w 1 , …, ‘w n are the expert weights applied to modules; U is the unfavourable threshold)
To further document the usefulness of fuzzy logic interpretation of combined statistics in method validation, an analysis of method performance for two cotton GMO MON15985 and LLCotton25 qPCR methods is shown in Table 1 (basic statistics and fuzzy-based indicator).
At different intervals of the tested dynamic range, the method performance resulted in differing values for a number of validation statistics: per cent values of bias, repeatability standard deviation (RSD r) and reproducibility standard deviation (RSD R). In the GM cotton MON15985, for instance, at 0.4 and 2.5% GM level, the method performance resulted in −18 vs. −0.5% per cent values of bias, in 16 vs. 26% of repeatability standard deviation, and 33 vs. 27% reproducibility standard deviation, respectively. In cotton LLCotton25, a lower bias was observed at 0.15% (=12%) than at 0.90% (=20%) GM levels. Opposite performances were observed for the RSD r and the RSD R (23 vs. 18%, and 23 vs. 32%, respectively). As such, these contrasting values for the different statistics make the estimation of the overall performance of a method quite difficult. When aggregated and interpreted in a standard combined way based on fuzzy logic principles and rules, the resulting aggregated index for bias, repeatability and reproducibility measures per GM level, allows estimating more consistently the overall performance of a method. For MON15985, the different performance of this qPCR method at low and high %GM is due to opposing values for different statistics: for bias better at the 2.5% GM level, for repeatability better at the 0.4% GM level. The fuzzy output of the MON15985 values demonstrates, however, acceptable aggregated performances at both GM levels (fuzzy output equal to ~0.50). With LLCotton25, a better performance at low GM level is disclosed by the fuzzy interpretation, which was not obvious from the disaggregated statistics. This example demonstrates that fuzzy outputs on overall method performance may be helpful in supporting a decision as to what method can best be applied in a most efficient way. A more elaborated analysis of the overall performance of a larger set of GMO detection methods applying fuzzy logic (e.g. the EURL validated GMO detection methods) is however, required to enable establishing a Decision Support System for so-called ‘fuzzy logic–balanced GMO Method application’.
The above example shows how combining distinct statistics into a single synthetic number can facilitate a more comprehensive appraisal of the overall performance of a method. The system is based on a balance between operational needs (method validation) and flexibility (hierarchy and aggregation of validation metrics). Fuzzy groups avoid thus crisp decision cut-off values (F and U limits) for basic validation measures, while decision rules provide a rationale to aggregate the distinct validation metrics. The combination of both concepts is attractive because it allows in principle to integrate the interpretation of a large numbers of statistics by a single set of decision rules. Our data presented here and elsewhere on GMO detection method performance provide evidence that in this broad analytical field, effective associations of inference and decision-making can be made by fuzzy logic.
Such fuzzy logic associations were shown useful in a variety of basic aspects in GMO method validation [14]. Thus, fuzzy logic is an innovative concept in GMO testing field which does not substitute for the information provided by the estimation of MU or evaluation of method’s bias. Rather, it offers a valuable tool for stakeholders to look at method performance from another viewpoint: where experts’ consensus on the relative importance of validation metrics (not limited to trueness and precision) is gained for a specific application, a simple indicator can combine and express in a synthetic way the global perception of method behaviour. Given its flexible nature, users’ community can also build up ad-hoc fuzzy inference systems, e.g. for evaluation in comparative fashion of screening methods for detection of GMOs. In such case, by definition, there is no quantification exercise, and no MU but parameters like sensitivity and specificity come into play. Also, fuzzy logic principles were successfully applied in the evaluation of commutability of different types of reference materials as calibrant in quantitative GMO detection [15]. The combinatory power of the fuzzy logic approach was fully exploited as a heuristic decision support in the interpretation of screening results for the presence of particular GMO in a sample [16]. Such approach could be envisaged for other GMO screening approaches that are currently under development and may allow establishing comparative performance criteria for different other screening platforms [e.g. 17–19]. Also, by defining a minimal set of decision criteria, simplified fuzzy-based decision tools could be developed in order to compare performance of in-house validated methods with inter-laboratory tested methods. All the above is, however, inherently dependent on the availability of a set of expert rules that define the applied weights and the transition intervals in the fuzzy logic inferential system. The definition of these settings is crucial in establishing a reliable fuzzy logic method performance evaluation system. Ready-to-use software tools are available to facilitate the implementation [20] and the testing (e.g. sensitivity analysis on the fuzzy settings) [21] of purpose-specific fuzzy-based indicators. For the development of an acceptable, performing system expertise of different fields is essential, including to our feeling at least major input from molecular biologists, bio-mathematicians and legal interpreters (on how non-crisp decisions are to be rendered compatible with legislation and legal obligations).
References
European Commission (2003) Official Journal of the European Union, L 268/12
European Commission (2003) Official Journal of the European Union, L 268/24
Popping B (2006) Accred Qual Assur 11:89–93
Žel J, Cankar K, Ravnikar M, Camloh M, Gruden K (2006) Accred Qual Assur 10:531–536
Innis MA, Gelfland DH, Sninsky JJ, White TJ (1990) PCR protocols: a guide to methods for applications. Academic Press, Inc, USA
ISO 5725 (1994) Accuracy (trueness and precision) of measurements methods and results, parts 1–6. International Organization for Standardization, Switzerland
ISO 17025 (2005) General requirements for the competence of testing and calibration laboratories. International Organization for Standardization, Switzerland
Analytical Laboratory Accreditation Criteria Committee (2007) AOAC Intl. http://www.aoac.org/alacc_guide_2008.pdf. Accessed 31 May 2010
EURACHEM/CITAC (2000) Quantifying uncertainty in analytical measurements, 2nd(ed), EURACHEM Secretariat, BAM, Berlin (http://www.eurachem.org/). Accessed 31 May 2010
Thompson M, Ellison SLR, Wood R (2002) Pure Appl Chem 74:835–855
Trapman S, Burns M, Broll H, Macarthur R, Wood R, Zel J (2009) Guidance document on measurement uncertainty for GMO testing laboratories, 2nd (ed), http://irmm.jrc.ec.europa.eu/html/reference_materials_catalogue/user_support/EUR22756EN.pdf13. Accessed 31 May 2010
DG Health and Consumer Protection (2004) http://ec.europa.eu/food/food/chemicalsafety/ contaminants/report-sampling_analysis_2004_en.pdf. Accessed 31 May 2010
Zadeh LA (1965) J Inf Contr 8:338–353
Bellocchi G, Acutis M, Paoletti C, Confalonieri R, Trevisiol P, Grazioli E, Delobel C, Savini C, Mazzara M, Van den Eede G (2008) Food Anal Methods 2:126–135
Lievens A, Bellocchi G, De Bernardi D, Moens W, Savini C, Mazzara M, Van den Eede G, Van den Bulcke M (2010) Anal Bioanal Chem doi:10.1007/s00216-009-3346-z
Bellocchi G, Bertholet V, Hamels S, Moens W, Remacle J, Van den Eede G (2010) Transgenic Res 19:57–65
Van den Bulcke M, Lievens A, Barbau-Piednoir E, MbongoloMbella G, Roosens N, Sneyers M, Casi AL (2009) Anal Bioanal Chem doi:10.1007/s00216-009-3286-7
Waiblinger HU, Grohmann L, Mankertz J, Engelbert D, Pietsch K (2009) Anal Bioanal Chem doi:10.1007/s00216-009-3173-2
Dobnik D, Morisset D, Gruden K (2009) Anal Bioanal Chem doi: 10.1007/s00216-009-3197-7
Acutis M, Trevisiol P, Confalonieri R, Bellocchi G, Grazioli E, Van den Eede G, Paoletti C (2007) J AOAC Int 90:1432–1438
Foscarini F, Bellocchi G, Confalonieri R, Savini C, Van den Eede G (2010) Environ Modell Softw 25:1256–1260
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Author information
Authors and Affiliations
Corresponding author
Additional information
Papers published in this section do not necessarily reflect the opinion of the Editors, the Editorial Board and the Publisher.
A critical and constructive debate in the Discussion Forum or a Letter to the Editor is strongly encouraged!.
Rights and permissions
Open Access This is an open access article distributed under the terms of the Creative Commons Attribution Noncommercial License (https://creativecommons.org/licenses/by-nc/2.0), which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
About this article
Cite this article
Bellocchi, G., Savini, C., Van den Bulcke, M. et al. Fuzzy logic-based procedures for GMO analysis. Accred Qual Assur 15, 637–641 (2010). https://doi.org/10.1007/s00769-010-0690-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00769-010-0690-9