Abstract
Broad macromolecular baseline (MMBL) is present throughout the magnetic resonance spectroscopy (MRS) spectrum of brain at short echo-time (TE) acquisitions. These variations cause the metabolite peak quantification and processing difficult in diagnostics. MMBL can provide information for specific disease, as an important biomarker. This presents a requirement of an efficient MMBL isolation method in post-acquisition scenario. The volume of medical dataset available are mostly small- or medium-sized along with the constraint of ground truth. The estimation of MM baseline from a noisy spectrum was treated as an ill-conditioned inverse problem. To address both issues, a novel approach of gradient boosted wavelet-feature tree model in a multioutput-regression framework for MRS spectral fitting was adopted to isolate macromolecular baseline from noisy metabolite spectra, where the inverse problem was learned by training over wavelet coefficients of noisy spectral dataset simulated using basis-set of metabolites and macromolecules. The proposed method performed almost perfectly for the simulated dataset with smaller margins of error, compared to an equivalent CNN model. For the simulated test set, RMSE and SSIM of 0.1623 and 0.9571 respectively were obtained and RMSE of 0.2263 was obtained for in-vivo test set. The fitted peak amplitude for individual MM component within ± 4% of error range over the simulated dataset.




Similar content being viewed by others
Data Availability
The in-vivo dataset used during the current study are not publicly available due to ethical issues, but data simulated during the study are available from the corresponding author on reasonable request.
References
C. Cudalbu et al., NMR Biomed. (2021). https://doi.org/10.1002/nbm.4393
M. Hoefemann, C.S. Bolliger, D.G. Chong, J.W. van der Veen, R. Kreis, NMR Biomed. (2020). https://doi.org/10.1002/nbm.4328
A.M. Wright, S. Murali-Manohar, T. Borbath, N.I. Avdievich, A. Henning, Magn. Reson. Med. (2022). https://doi.org/10.1002/mrm.28958
K. Landheer, M. Gajdošík, M. Treacy, C. Juchem, Magn. Reson. Med. (2020). https://doi.org/10.1002/mrm.28282
S. Murali-Manohar, T. Borbath, A.M. Wright, B. Soher, R. Mekle, A. Henning, Magn. Reson. Med. (2020). https://doi.org/10.1002/mrm.28174
D.K. Deelchand, E.J. Auerbach, N. Kobayashi, M. Marjańska, Magn. Reson. Med. (2018). https://doi.org/10.1002/mrm.26826
M. Marjańska, E.J. Auerbach, R. Valabrègue, P.F. Van de Moortele, G. Adriany, M. Garwood, NMR Biomed. (2012). https://doi.org/10.1002/nbm.1754
R.A. de Graaf, P.B. Brown, S. McIntyre, T.W. Nixon, K.L. Behar, D.L. Rothman, Magn. Reson. Med. (2006). https://doi.org/10.1002/mrm.20946
K.L. Behar, T. Ogino, Magn. Reson. Med. (1991). https://doi.org/10.1002/mrm.1910170202
K.L. Behar, D.L. Rothman, D.D. Spencer, O.A. Petroff, Magn. Reson. Med. (1994). https://doi.org/10.1002/mrm.1910320304
D.G. Chong, R. Kreis, C.S. Bolliger, C. Boesch, J. Slotboom, MAGMA (2011). https://doi.org/10.1007/s10334-011-0246-y
A. Henning, in Handbook of Magnetic Resonance Spectroscopy In Vivo: MRS Theory, Practice and Applications. ed. by P.A. Bottomley, J.R. Griffiths (Wiley, 2016), p.303
M. Považan, B. Strasser, G. Hangel, E. Heckova, S. Gruber, S. Trattnig, W. Bogner, Magn. Reson. Med. (2018). https://doi.org/10.1002/mrm.26778
R. Birch, A.C. Peet, H. Dehghani, M. Wilson, Magn. Reson. Med. (2017). https://doi.org/10.1002/mrm.26103
I.A. Giapitzakis, T. Borbath, S. Murali-Manohar, N. Avdievich, A. Henning, Magn. Reson. Med. (2019). https://doi.org/10.1002/mrm.27467
U. Seeger, U. Klose, I. Mader, W. Grodd, T. Nägele, Magn. Reson. Med. (2003). https://doi.org/10.1002/mrm.10332
R. Bartha, D.J. Drost, P.C. Williamson, NMR Biomed. (1999). https://doi.org/10.1002/(SICI)1099-1492(199906)12:4%3c205::AID-NBM558%3e3.0.CO;2-1
W.W.F. Pijnappel, A. Van den Boogaart, R. De Beer, D. Van Ormondt, J. Magn. Reson. (1969). (1992). https://doi.org/10.1016/0022-2364(92)90241-X
T. Laudadio, N. Mastronardi, L. Vanhamme, P. Van Hecke, S. Van Huffel, J. Magn. Reson. (2002). https://doi.org/10.1006/jmre.2002.2593
H. Barkhuijsen, R. De Beer, D. Van Ormondt, J Magn Reson. (1969) (1987). https://doi.org/10.1016/0022-2364(87)90023-0
L. Vanhamme, A. van den Boogaart, S. Van Huffel, J Magn Reson. (1997). https://doi.org/10.1006/jmre.1997.1244
H.H. Lee, H. Kim, Magn. Reson. Med. (2019). https://doi.org/10.1002/mrm.27727
H.H. Lee, H. Kim, Magn. Reson. Med. (2017). https://doi.org/10.1002/mrm.26502
S.S. Gurbani, S. Sheriff, A.A. Maudsley, H. Shim, L.A. Cooper, Magn. Reson. Med. (2019). https://doi.org/10.1002/mrm.27641
M. Marjanska, D. K. Deelchand, R. Kreis, MRS fitting challenge data setup by ISMRM MRS study group. Data Repository for the University of Minnesota. https://experts.umn.edu/en/datasets/mrs-fitting-challenge-data-setup-by-ismrm-mrs-study-group. (2021). Accessed 12 May, 2022.
I.W. Selesnick, R.G. Baraniuk, N.C. Kingsbury, IEEE Signal Process. Mag. (2005). https://doi.org/10.1109/MSP.2005.1550194
T. Chen, C. Guestrin, In: Proceedings of the 22nd acm sigkdd International Conference on Knowledge Discovery and Data Mining. (2016), p. 785–794. https://doi.org/10.48550/arXiv.1603.02754.
R.A. De Graaf, In-vivo NMR spectroscopy: principles and techniques, 3rd edn. (Wiley, New York, 2007)
V. Govindaraju, K. Young, A.A. Maudsley, NMR Biomed. (2000). https://doi.org/10.1002/1099-1492(200005)13:3%3c129::AID-NBM619%3e3.0.CO;2-
T.L. Perry, S. Hansen, K. Berry, C. Mok, D. Lesk, J. Neurochem. (1971). https://doi.org/10.1111/j.1471-4159.1971.tb11980.x
C. Sagar, D.K. Singh, N. Sharma, J. Appl. Spectrosc. (2022). https://doi.org/10.1007/s10812-022-01393-7
J. Bergstra, D. Yamins, D. Cox, In PMLR (2013) p. 115–123. https://doi.org/10.48550/arXiv.1209.5111.
D. P. Kingma, J. Ba, arXiv preprint. (2014) https://doi.org/10.48550/arXiv.1412.6980.
R. Kreis, V. Boer, I.Y. Choi, C. Cudalbu et al., NMR Biomed. (2021). https://doi.org/10.1002/nbm.4347
I.A. Giapitzakis, N. Avdievich, A. Henning, Magn Reson Med. (2018). https://doi.org/10.1002/mrm.27070
R. Shwartz-Ziv, A. Armon, Inf Fusion. (2022) https://doi.org/10.48550/arXiv.2106.03253.
S. Elsayed, D. Thyssens, A. Rashed, H. S. Jomaa, L. Schmidt-Thieme, arXiv preprint. (2021) https://doi.org/10.48550/arXiv.2101.02118.
Acknowledgements
The authors express their sincere gratitude to the Indian Institute of Technology (BHU), Varanasi, UP, India, and Apex Super-speciality Hospital and Post Graduate Institute, Varanasi, Uttar Pradesh, India, for supporting the work.
Funding
None.
Author information
Authors and Affiliations
Contributions
CS: principal author, conceptualization, methodology, data curation, software, formal analysis, investigation, writing—original draft. DKS: conceptualization, resources, validation, review & editing, supervision. NS: conceptualization, validation, resources, review & editing, supervision, project administration.
Corresponding author
Ethics declarations
Conflict of Interest
The author(s) declared no potential conflicts of interest concerning the research, authorship, and/or publication of this article.
Ethical Approval
This is a retrospective dataset study. So, the requirement of patient consent was waived off by the institute ethical committee.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Appendix
Appendix
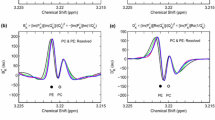
MM spectrum isolation from residual water peak included noisy MR spectra. Top: Noisy spectra with residual waterpeak; Middle: Superimposed individual components contribution of Metabolites, Macromolecular Baseline (GT) and Residual waterpeak, [Blue: Noisy spectrum, Green: GT-MM, Orange: Residual waterpeak]; Bottom: Isolated MM from nosiy spectra with residual waterpeak (GT vs Proposed method) [Blue: GT-MM, Orange: Fitted MM from the proposed method],
Results for MM isolation from spectra with residual waterpeak:
Noisy data with Res. Water content | Res. waterpeak height < = 3 × Highest peak of training spectra | Res. waterpeak height > 3 × Highest peak of training spectra | ||||
|---|---|---|---|---|---|---|
RMSE | SSIM | R2-value | RMSE | SSIM | R2-value | |
Proposed method | 0.2988 | 0.8973 | 0.7836 | > 0.55 | < 0.74 | < 10 |
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Sagar, C., Singh, D.K. & Sharma, N. A Gradient Boosted Regression Tree Ensemble Model Using Wavelet Features for Post-acquisition Macromolecular Baseline Isolation from Brain MR Spectra. Appl Magn Reson 54, 637–655 (2023). https://doi.org/10.1007/s00723-023-01537-8
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00723-023-01537-8