Abstract
Magnetic resonance spectroscopic imaging (MRSI) is an imaging modality used for generating metabolic maps of the tissue in-vivo. These maps show the concentration of metabolites in the sample being investigated and their accurate quantification is important to diagnose diseases. However, the major roadblocks in accurate metabolite quantification are: low spatial resolution, long scanning times, poor signal-to-noise ratio (SNR) and the subsequent noise-sensitive non-linear model fitting. In this work, we propose a frequency-phase spectral denoising method based on the concept of non-local means (NLM) that improves the robustness of data analysis and scanning times while potentially increasing spatial resolution. We evaluate our method on simulated data sets as well as on human in-vivo MRSI data. Our denoising method improves the SNR while maintaining the spatial resolution of the spectra.
You have full access to this open access chapter, Download conference paper PDF
Similar content being viewed by others
1 Introduction
Magnetic Resonance Spectroscopic imaging (MRSI), also known as chemical shift imaging, is a clinical imaging modality for studying tissues in-vivo to investigate and diagnose neurological diseases. More specifically, this modality can be used in non-invasive diagnosis and characterization of patho-physiological changes by measuring specific tissue metabolites in the brain. Accurate metabolite quantification is a crucial requirement for effectively using MRSI for diagnostic purposes. However, a major challenge with MRSI is the long scanning time required to obtain spatially resolved spectra due to abundance of metabolites that have a concentration which is approximately 10,000 times smaller than water. Current acquisition techniques such as Parallel Imaging [13] and Echo-Planar Spectroscopic Imaging [9] focus on accelerated scanning times combined with reconstruction techniques to improve the SNR of the spectral signal. Despite this, further accelerated acquistions are desirable. Furthermore, an improved SNR is needed as the non-linear voxel-wise fitting to noisy data leads to a high amount of local minima and noise amplification resulting in poor spatial resolution [7].
The SNR of the signal can be improved by post-processing methods such as denoising algorithms [8], apodization (Gaussian/Lorentzian), filter-based smoothing and transform-based methods [3]. However, these methods reduce resolution and remove important quantifiable information by averaging out the lower-concentration metabolites. Recently, data-dependent approaches such as the Non-local Means (NLM), which use the redundancy inherent in periodic images, are being used extensively for denoising [3]. In the case of MRS, this periodicity implies that the spectra in any voxel may have similar spectra in other voxels in the frequency-phase space. Therefore, it carries out a weighted average of the voxels in this space, depending on the similarity of the spectral information of their neighborhoods to the neighborhood of the voxel to be denoised.
Our Contribution. In this work, we propose a method for spectrally adaptive denoising of MRSI spectra in the frequency-phase space based on the concept of Non-local Means. Our method compensates for the lack of phase-information in the acquired spectra by implementing a dephasing approach on the spectral data. In the next section, we introduce the experimental methods beginning with the concept of NLM in the frequency-phase space followed by the spectral dephasing and rephasing approach. Our proposed method is then validated quantitatively and qualitatively using simulated brain data and human in-vivo MRSI data sets to show the improvements in SNR and spatial-spectral resolution of MRSI data.
2 Methods
MR Spectroscopy. Magnetic resonance spectroscopy is based on the concept of nuclear magnetic resonance (NMR). It exploits the resonance frequency of a molecule, which depends on its chemical structure, to obtain information about the concentration of a particular metabolite [12]. The time-domain complex signal of a nuclei is given by: \(S(t) = \int \mathrm {p}(\omega )\mathrm {exp}(-i\varPhi )\mathrm {exp}(-t/T^{*}_{2})dw\). The frequency-domain signal is given by \(S(\omega )\), \(T^{*}_{2}\) is the magnetization decay in the transverse plane due to magnetic field inhomogeneity and \(\mathrm {p}(\omega )\) comprises of Lorentzian absorption and dispersion line-shapes function having the spectroscopic information about the sample. \(\varPhi \) represents the phase, \((\omega t + \omega _{0})\), of the acquired signal where \(\omega t\) is the time-varying phase change and \(\omega _{0}\) is the initial phase. For the acquired MRSI data, I, \(\varPhi \) is unknown. This process allows generation of metabolic maps through non-linear fitting to estimate concentration of metabolites such as N-acetyl-aspartate (NAA), Creatine (Cr) and Choline (Cho).
2.1 Non-local Means (NLM) in Frequency-Phase Space
As proposed by Buades et al. [3], the Non-local Means (NLM) method restores the intensity of voxel \(x_{ij}\) by computing a similarity-based weighted average of all the voxels in a given image. In the following, we adapt NLM to the MRSI data: let us suppose that we have complex data, I : \(\varOmega ^3\) \(\longmapsto \) \(\mathbb {C}\) of size \(M\times N\) and noisy spectra \(S_{ij}(\omega )\), where \(({x_{ij} | i\in [1, M], j\in [1, N]})\) and \(\varOmega ^3\) is the frequency-phase grid. Using NLM for denoising, the restored spectra \(\hat{S}_{ij}(\omega )\) is computed as the weighted average of all other spectra in the frequency-phase space defined as:
As a probabilistic interpretation, spectral data \(S_{11}(\omega ),...,S_{MN}(\omega )\) of voxels \(x_{11},.....,x_{MN}\) respectively are considered as MN random variables \(X_{ij}\) and the weighted average estimate \(\hat{S}_{ij}(\omega )\) is the maximum likelihood estimate of \({S_{ij}(\omega )}\).
\(N_{ij} = (2p+1)^3\), \(p \in \mathbb {N}\) is the cubic neighborhood of voxel \(x_{ij}\) within the search volume \(V_{ij} = (2R+1)^3\) around \(x_{ij}\) along the frequency, phase and spatial directions. \(R \in \mathbb {N}\), where R is the radius of search centered at the voxel \(x_{ij}\). The weight \(w(x_{ij}, x_{kl})\) serves as a quantifiable similarity metric between the neighborhoods \(N_{ij}\) and \(N_{kl}\) of the voxels \(x_{ij}\) and \(x_{kl}\) provided \(w(x_{ij}, x_{kl})\) \( \in [0, 1]\) and \(\sum w(x_{ij},x_{kl}) = 1 \). The Gaussian-weighted Euclidean distance is computed between \(S(N_{ij})\) and \(S(N_{kl})\) as shown below:
where \(Z_{ij}\) serves as the normalization constant such that \(\sum _{\j }w(x_{ij},x_{kl}) = 1\), \(S(N_{ij})\) and \(S(N_{kl})\) are vectors containing the spectra of neighborhoods \(N_{ij}\) and \(N_{kl}\) of voxels \(x_{ij}\) and \(x_{kl}\) respectively and h serves as a smoothing parameter [5].
To increase the robustness of our method for MRSI data, in the next section we propose a dephasing approach tailored for use in the frequency-phase NLM.
2.2 Spectral Dephasing
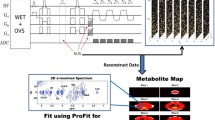
For the acquired data I, as the spectral phase \(\varPhi (I)\) is unknown, the probability of finding a similar neighborhood spectra are very low. To counter this effect, a dephasing step is performed to consider a wide range of possible phase variations in the pattern analysis. For each voxel \(x_{ij}\), the complex time-domain signal \(S_{ij}(t)\) is shifted by a set of phase angles \(\varTheta \). This is given by \(S_{ij}^{\varTheta }(t) = S_{ij}(t).e^{-(i\varTheta )}\), where \(\varTheta \in [-n_{1}\pi , (n_{2}+2)\pi ]\), \(\lbrace n_{1}, n_{2} \in \mathbb {R} \, | \, n_{1}, n_{2} \ge 0 \rbrace \). \(\varTheta \) here is defined to be the range of angles through which the spectrum can be shifted. The dephased signal is transformed into the frequency-domain, \(S_{ij}^{\varTheta }(t)\xrightarrow {\mathscr {F}} S_{ij}^{\varTheta }(\omega )\), following which its real component, \(\mathbb {R}(S_{ij}^{\varTheta }(\omega ))\), is taken to generate a 2D spectral-phase matrix. Note that in this 2D matrix generated, for each voxel \(x_{ij}\), the imaginary part at a given \(\varTheta \) is \(\mathbb {I}(\varTheta ) = \mathbb {R}(\varTheta + \pi /2)\). This approach is illustrated in Fig. 1.
Repeating this step for all MN voxels gives us a 3-D dataset on which the NLM is implemented to give the denoised spectra \(\hat{S}_{ij}^{\varTheta }(\omega ) \in \mathbb {R}\). Our approach has 2 key innovations: the denoising method is (i) robust to phase shifts as the range of angles considered varies from 0 to 2\(\pi \) periodically for all spectral signals, and is (ii) adaptive to the imaging sequence as the spectrum is denoised by relying on similar signals in the given data and not on predefined prior assumptions.
MRSI data dephasing shown here for a sample voxel: (A) Changes in spectral pattern as it is shifted by different phase angles. (B) Corresponding 2D frequency-phase image space generated for the voxel. A sample patch (black box) is selected and then denoised by the NLM-based matching in the frequency-phase space.

2.3 Spectral Rephasing and Recombination
Post-NLM, \(\hat{S}_{ij}^{\varTheta }(\omega )\) is rephased in order to generate the denoised complex signal \(\mathbb {C}(\hat{S}_{ij}(\omega ))\). The complex spectral signal \(\mathbb {C}(\hat{S}_{ij}^{\varTheta }(\omega ))\) is re-generated \(\forall \varTheta \) by combining \(\mathbb {R}(\hat{S}_{ij}^{\varTheta }(\omega ))\) and \(\mathbb {I}(\hat{S}_{ij}^{\varTheta }(\omega ))\) (= \(\mathbb {R}(\hat{S}_{ij}^{\varTheta + \pi /2}(\omega ))\)). The equivalent time signal is obtained by \(\mathbb {C}(\hat{S}_{ij}^{\varTheta }(\omega ))\xrightarrow {\mathscr {F}^{-1}} \mathbb {C}(\hat{S}_{ij}^{\varTheta }(t))\). After this, \(\mathbb {C}(\hat{S}_{ij}^{\varTheta }(t))\) undergoes an inverse phase shift by \(-\varTheta \) to remove the dephasing effect as given by \(\mathbb {C}(\hat{S}_{ij}^{-\varTheta }(t)) = \mathbb {C}(\hat{S}_{ij}^{\varTheta }(t)).e^{(i\varTheta )}\). This re-phased signal is transformed back to the spectral domain to obtain \(\mathbb {C}(\hat{S}_{ij}^{-\varTheta }(\omega ))\). Thereafter, the \(\mathbb {C}(\hat{S}_{ij}^{-\varTheta }(\omega ))\) are averaged over all \(\varTheta \) to generate a single complex spectra \(\mathbb {C}(\hat{S}_{ij}(\omega ))\). The entire pipeline for dephasing and rephasing the spectra is shown in Algorithm 1.
3 Experiments and Results
We performed two different experiments to test the improvement in SNR and metabolite quantification using our proposed denoising method. In the first experiment, we evaluate our method on the publicly available BrainWeb database [4], while in the second experiment we use human in-vivo MRSI data. The SNR of a metabolite was calculated by dividing the maximum value of the metabolite peak by the standard deviation of the spectral region having pure noise. For both experiments, we tested with different noise levels against a ground-truth data to assess the improvement in SNR and spatial-spectral resolution.
3.1 Data Acquisition
We used BrainWeb to simulate a brain MRSI image (size: 64\(\,\times \,\)64 voxels, slice thickness = 1 mm, noise level = 3 \(\%\)) with segmented tissue types, namely White Matter (WM), Grey Matter (GM) and Cerebro Spinal Fluid (CSF) as shown in Fig. 2. In order to have a comparable spectrum with the in-vivo data, water was added to the signal and the main metabolites- NAA, Cho and Cr- were simulated using Priorset (Vespa) [1]. Metabolite concentrations for WM, GM and CSF were based on commonly reported literature values [10, 14]. Next, Gaussian noise of levels 2 and 3 times the standard deviation, \(\sigma \), of the original image data were added to the ground-truth signal.
In the case of in-vivo data, we acquired a 2D-MRSI data of the brain of a healthy human volunteer using a 3T-HDxt system (GE-Healthcare). PRESS localization [2], CHESS water suppression [6] and EPSI readout [9] were used as part of the sequence. The acquisition parameters were: Field of view (FOV) = \(160\,\times \,160\,\times \,10\,{\text {mm}^{3}}\), voxel size = \(10\,\times \,10\,\times \,10\,{\text {mm}^{3}}\), TE/TR=35/2000 ms and spectral bandwidth = 1 kHz. The dataset was zero-filled and reconstructed to generate a grid of \(32\,\times \,32\) voxels and 256 spectral points. 6 (ground truth), 3 and 1 averages were acquired with a total scan duration of 33 min (5.5 min per average). Figure 4(A) shows the in-vivo data acquired along with the entire field-of-view (white grid) and the corresponding spectra of a voxel (red box).
3.2 Results
Simulated data: In Fig. 3, we show the SNR improvement for NAA for data with noise levels 2\(\sigma \) and 3\(\sigma \) in a \(32\,\times \,32\) region of interest. It is evident that while the spectral SNR improves significantly, the spatial resolution is preserved as the lower concentration metabolite peaks have only a small amount of smoothing and there is no voxel bleeding in the CSF (containing only water).
NLM Denoising results in the simulated data. (From Top) Row 1 (L-R): Full SNR of NAA in – original data, with additive noise 2\(\sigma \), and noise 3\(\sigma \) (\(\sigma \) is the standard deviation of the original data). Row 2 & 3: 32\(\times \)32 Region of Interest (ROI) for applying the frequency-phase NLM: SNR of NAA in the original data, noise level 2\(\sigma \), 3\(\sigma \) and the corresponding spectra of reference WM voxel (red box). Row 4 & 5 (L-R): Denoised SNR for noise level 2\(\sigma \) (SNR improvement = 2.9), for noise level 3\(\sigma \) (SNR improvement = 2.2), and the corresponding denoised spectrum. The SNR improves significantly while retaining the spatial-spectral resolution (seen by no voxel bleeding in the CSF). (Color figure online)
Denoising results for in-vivo data. (A) Original human in-vivo brain MRSI data with the excitation region shown (white grid). (From Top) Row 1 & 2: SNR of NAA and the corresponding WM voxel spectra in: (B) 6-averages data (ground-truth), (C) 3-averages data (original) and (D) denoised, (E) single-scan data (original) and (F) denoised with corresponding spectra. Row 3: LCModel based absolute concentration estimate of NAA in: 6-averages data, 3-averages data (original) and denoised, 1-average data (original) and denoised. The NAA concentration estimate and spectral SNR improve considerably as seen in columns D (SNR = 23.29) and F (SNR = 11.38) against the ground-truth (SNR = 11.44).
In-vivo data: Figure 4 reports the SNR improvement in NAA for the 3-averages and the 1-average data as compared to the ground-truth 6-averages data. The figure also presents the results from the LCModel [11] which is the gold standard quantitation tool in MRS analysis. LCModel fits the spectral signal \(S(\omega )\) using a basis set of spectra of metabolites acquired under identical acquisition conditions as the in-vivo data. As explained earlier for noisy data, the non-linear fitting leads to poor spatial resolution. Therefore, the LCModel can be used to assess the improvement in spatial resolution through a better fit. Due to space constraints, we present the results for NAA only and mention the SNR values for Cho and Cr. LCModel quantification (Fig. 4) shows that the absolute concentration estimation of NAA in the denoised data improves significantly. The Full-Width Half Maximum (FWHM) shows information about the water peak – a narrow peak gives a better spatial resolution. As shown in Table 1, the FWHM of the denoised 1- and 3-averages data is lower than the 6-averages data while the corresponding mean SNR improves considerably. Therefore, we observe here that our method can accelerate MRSI data acquisition by almost 2 times by reducing the number of scans acquired.
4 Conclusion
In this work, we proposed a novel frequency-phase NLM-based denoising method for MRS Imaging to improve the SNR and spatial resolution of the metabolites. A spectral dephasing approach is promoted to compensate for the unknown phase information of the acquired data. To the best of our knowledge, this is a novel application of the concept of NLM and has been validated on both simulated and in-vivo MRSI data.
In particular, we assessed the effect of our method on metabolites such as NAA, Cho and Cr and obtained a visible improvement in SNR while the spatial resolution was preserved which, subsequently, led to a better estimation of the absolute concentration distribution of NAA. This has direct benefits as it would accelerate data acquisition by taking fewer scan averages. Future work would involve using a more robust metabolite-specific search in the given dataset with less smoothing. This can be coupled with optimal computational efficiency and better estimation of the in-vivo metabolites.
References
Vespa project (Versatile simulation, pulses and analysis). https://scion.duhs.duke.edu/vespa/project
Bottomley, P.A.: Spatial localization in NMR spectroscopy in vivo. Ann. N. Y. Acad. Sci. 508, 333–348 (1987). doi:10.1111/j.1749-6632.1987.tb32915.x
Buades, A., Coll, B.: A non-local algorithm for image denoising. Comput. Vis. Pattern 2(0), 60–65 (2005)
Collins, D.L., Zijdenbos, P., Kollokian, V., Sled, J.G., Kabani, N.J., Holmes, C.J., Evans, C.: Design and construction of a realistic digital brain phantom. IEEE Trans. Med. Imaging 17(3), 463–468 (1998)
Coupé, P., Yger, P., Prima, S., Hellier, P., Kervrann, C.: An optimized blockwise non local means denoising filter for 3D magnetic resonance images. IEEE Trans. Med. Imaging 27(4), 425–441 (2008)
Haase, A., Frahm, J., Hänicke, W., Matthaei, D.: 1H NMR chemical shift selective (CHESS) imaging. Phys. Med. Biol. 30(4), 341–344 (1985). http://stacks.iop.org/0031-9155/30/i=4/a=008
Kelm, B.M., Kaster, F.O., Henning, A., Weber, M.A., Bachert, P., Boesiger, P., Hamprecht, F.A., Menze, B.H.: Using spatial prior knowledge in the spectral fitting of MRS images. NMR Biomed. 25(1), 1–13 (2012)
Nguyen, H.M., Peng, X., Do, M.N., Liang, Z.: Spatiotemporal denoising of MR spectroscopic imaging data by low-rank approximations. IEEE ISBI: From Nano to Macro 0(3), 857–860 (2011)
Posse, S., DeCarli, C., Le Bihan, D.: Three-dimensional echo-planar MR spectroscopic imaging at short echo times in the human brain. Radiology 192(3), 733–738 (1994). http://pubs.rsna.org/doi/abs/10.1148/radiology.192.3.8058941
Pouwels, P.J.W., Frahm, T.: Regional metabolite concentrations in human brain as determined by quantitative localized proton MRS. Magn. Reson. Med. 39(1), 53–60 (1998)
Provencher, S.W.: Estimation of metabolite concentrations from localized in vivo proton NMR spectra. Magn. Reson. Med. 30(6), 672–679 (1993). http://www.ncbi.nlm.nih.gov/pubmed/8139448
de Graaf, R.A.: In Vivo NMR Spectroscopy: Principles and Techniques, 2nd edn. Wiley, Hoboken (2013)
Schulte, R.F., Lange, T., Beck, J., Meier, D., Boesiger, P.: Improved two-dimensional J-resolved spectroscopy. NMR Biomed. 19(2), 264–270 (2006)
Wang, Y., Li, S.: Differentiation of metabolic concentrations between gray matter and white matter of human brain by in vivo ’ h magnetic resonance spectroscopy. Magn. Reson. Med. 39(1), 28–33 (2005)
Acknowledgments
The research leading to these results has received funding from the European Union’s H2020 Framework Programme (H2020-MSCA-ITN-2014) under grant agreement no 642685 MacSeNet.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2016 Springer International Publishing AG
About this paper
Cite this paper
Das, D., Coello, E., Schulte, R.F., Menze, B.H. (2016). Spatially Adaptive Spectral Denoising for MR Spectroscopic Imaging using Frequency-Phase Non-local Means. In: Ourselin, S., Joskowicz, L., Sabuncu, M.R., Unal, G., Wells, W. (eds) Medical Image Computing and Computer-Assisted Intervention - MICCAI 2016. MICCAI 2016. Lecture Notes in Computer Science(), vol 9902. Springer, Cham. https://doi.org/10.1007/978-3-319-46726-9_69
Download citation
DOI: https://doi.org/10.1007/978-3-319-46726-9_69
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-319-46725-2
Online ISBN: 978-3-319-46726-9
eBook Packages: Computer ScienceComputer Science (R0)