Abstract
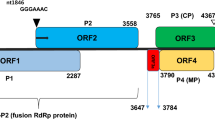
The complete genome sequence of a novel virus found infecting Cnidium officinale, which we have named "cnidium polerovirus 1" (CnPV1), is 6,090 nucleotides in length, similar to those of other poleroviruses. Seven open reading frames (ORF0–5 and ORF3a) were predicted in this genome. CnPV1 shares 32.4%–38.9% full-length nucleotide sequence identity with other known polerovirus genome sequences. The putative P0, P1-2, P3-5, P3, and P4 proteins share 11.3%–19.5%, 37.1%–49.8%, 26.7%–39.5%, 40.8%–49.7%, and 40.8%–49.7% amino acid sequence identity, respectively, with homologous inferred protein sequences from known poleroviruses. Phylogenetic analysis of P1-2 and P3 sequences places CnPV1 with other members of the genus Polerovirus, indicating that it should be classified in a new distinct species.


Similar content being viewed by others
Data availability
The datasets generated during and/or analysed during the current study are available from the corresponding author on reasonable request.
References
Chung BN, Kwon S, Yoon J, Cho I (2022) First report of Cnidium officinale as a natural host plant of apple stem grooving virus in South Korea. Plant Dis 106(1):338
Ramalinagam M, Park Y (2010) Free radical scavenging activities of Cnidium officinale Makino and Ligusticum chuanxiong Hort. Methanolic extracts. Phamacogn Mag 6(24):323–330
Igori D, Kim SE, Kwon S, Moon JS (2022) Complete genome sequence of plantago asiatica virus A, a novel putative member of the genus Polerovirus. Arch Virol 167(1):219–222
ICTV db (2021) https://ictv.global/report/chapter/solemoviridae/solemoviridae/polerovirus. Accessed Jan 2021
Igori D, Lim S, Cho HS, Kim HS, Park JM, Lee HJ, Hong KJ, Kwon SY, Moon JS (2021) Complete genome sequence of artemisia virus B, a new polerovirus infecting Artemisia princeps in South Korea. Arch Virol 166(5):1495–1499
Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215(3):403–410
Filardo FF, Thomas JE, Webb M, Sharman M (2019) Faba bean polerovirus 1 (FBPV-1); a new polerovirus infecting legume crops in Australia. Arch Virol 164(7):1915–1921
Knierim D, Tsai WS, Deng TC, Green SK, Kenyon L (2012) Full-length genome sequences of four polerovirus isolates infecting cucurbits in Taiwan determened from total RNA extracted from field samples. Plant Pathol 62:633–641
Palanga E, Martin AP, Galzi S, Zabre J (2017) Complete genome sequences of cowpea polerovirus 1 and cowpea polerovirus 2 infecting cowpea plants in Burkina Faso. Arch Virol 162:2149–2152
LaTourrette K, Holste NM, Garcia-Ruiz H (2021) Polerovirus genomic variation. Virus Evol 7(2):1–18
Belete MT, Gudeta WF, Kim SE, Igori D, Moon JS (2021) Complete genome sequence of Codonopsis torradovirus A, a novel torradovirus infecting Codonopsis lanceolate in South Korea. Arch Virol 166(12):3473–3476
Kumar S, Stecher G, Li M, Knyaz C, Tamura K (2018) MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol 35:1547–1549
Acknowledgments
This work was supported by IPET (Korea Institute of Planning and Evaluation for Technology in Food, Agriculture, Forestry and Fisheries); through Crop Viruses and Pests Response Industry Technology Development Program (321104-3), funded by the Ministry of Agriculture, Food and Rural Affairs (MAFRA), Republic of Korea. We thank Steven M. Thompson from Edanz (http://www.edanz.com/ac) for editing two drafts of this manuscript.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
This article does not contain any studies involving human participants or animals.
Additional information
Handling Editor: Ralf Georg Dietzgen.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Park, Y.C., Kim, KK., Jun, H.J. et al. Complete genome sequence of a novel member of the genus Polerovirus from Cnidium officinale in South Korea. Arch Virol 168, 104 (2023). https://doi.org/10.1007/s00705-023-05732-7
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00705-023-05732-7