Abstract
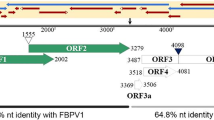
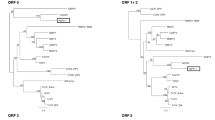
The full-length genome sequences of two novel poleroviruses found infecting cowpea plants, cowpea polerovirus 1 (CPPV1) and cowpea polerovirus 2 (CPPV2), were determined using overlapping RT-PCR and RACE-PCR. Whereas the 5845-nt CPPV1 genome was most similar to chickpea chlorotic stunt virus (73% identity), the 5945-nt CPPV2 genome was most similar to phasey bean mild yellow virus (86% identity). The CPPV1 and CPPV2 genomes both have a typical polerovirus genome organization. Phylogenetic analysis of the inferred P1-P2 and P3 amino acid sequences confirmed that CPPV1 and CPPV2 are indeed poleroviruses. Four apparently unique recombination events were detected within a dataset of 12 full polerovirus genome sequences, including two events in the CPPV2 genome. Based on the current species demarcation criteria for the family Luteoviridae, we tentatively propose that CPPV1 and CPPV2 should be considered members of novel polerovirus species.


Similar content being viewed by others
References
Chen S, Jiang G, Wu J, Liu Y, Qian Y, Zhou X (2016) Characterization of a novel polerovirus infecting maize in China. Viruses 8(120):1–17
Domier LL, D’Arcy CJ (2010) Luteoviruses. In: Mahy BWJ, Van Regenmortel MHV (eds) Desk encyclopedia of plant and fungal virology. Academic, Oxford, pp 197–204
Domier LL (2012) Family Luteoviridae. In: King AMQ, Adams MJ, Carstens EB, Lefkowitz EJ (eds) Virus taxonomy: ninth report of the International Committee on the Taxonomy of Viruses. Elsevier Academic Press, Amsterdam, pp 1045–1053
Guindon S, Dufayard J-F, Lefort V, Anisimova M, Hordijk W, Gascuel O (2010) New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol 59:307–321
Knierim D, Tsai WS, Deng TC, Green SK, Kenyon L (2013) Full-length genome sequences of four polerovirus isolates infecting cucurbits in Taiwan determined from total RNA extracted from field samples. Plant Pathol 62:633–641
Knierim D, Maiss E, Menzel W, Winter S, Kenyon L (2015) Characterization of the complete genome of a novel Polerovirus infecting Sauropus androgynus in Thailand. J Phytopathol 163:695–702
Lotos L, Maliogka VI, Katis NI (2016) New poleroviruses associated with yellowing symptoms in different vegetable crops in Greece. Arch Virol 161:431–436
Martin DP, Murrell B, Golden M, Khoosal A, Muhire B (2015) RDP4: detection and analysis of recombination patterns in virus genomes. Virus Evolution 1(1):1–5
Pagan I, Holmes EC (2010) Long-term evolution of the Luteoviridae: time scale and mode of virus speciation. J Virol 84(12):6177–6187
Pagan I, Holmes EC (2010) Long-term evolution of the Luteoviridae: time scale and mode of virus speciation. J Virol 84:6177–6187
Palanga E, Filloux D, Martin DP, Fernandez E, Gargani D, Ferdinand R, Zabre J, Bouda Z, Neya JB, Sawadogo M, Traore O, Peterschmitt M, Roumagnac P (2016) Metagenomic-based screening and molecular characterization of cowpea-infecting viruses in Burkina Faso. PLoS One 11(10):e0165188
Smirnova E, Firth AE, Miller WA, Scheidecker D, Brault V, Reinbold C, Rakotondrafara AM, Chung BY-W, Ziegler-Graff V (2015) Discovery of a small non-AUG-initiated ORF in poleroviruses and luteoviruses that is required for long-distance movement. PLoS Pathog 11(5):e1004868
Stevens M, Freeman B, Liu HY, Herrbach E, Lemaire O (2005) Beet poleroviruses: close friends or distant relatives? Mol Plant Pathol 6:1–9
Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 30:2725–2729
Theis C, Reeder J, Giegerich R (2008) KNOTINFRAME: prediction of −1 ribosomal frameshift events. Nucleic Acids Res 36:6013–6020
Acknowledgements
EP’s PhD fellowship was funded by the French Embassy of Togo (N°: 808087J; 836014F; 864988K) and International Foundation for Science (N°: C/5489-1). PR is supported by EU grant FP7-PEOPLE-2013-IOF (N° PIOF-GA-2013-622571). We acknowledge Ecole Superieur d’Agronomie of the University of Lomé, which accepted to shelter EP’s IFS funds, and the Laboratoire Mixte International Patho-Bios, which supported sample collection.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
This study was funded by the French Embassy of Togo (N°: 808087J; 836014F; 864988K.), International Foundation for Science (N°: C/5489-1) and EU grant FP7-PEOPLE-2013-IOF (N° PIOF-GA-2013-622571).
Conflict of interest
Author EP has received research grants from the French Embassy of Togo (N°: 808087J; 836014F; 864988K) and the International Foundation for Science (N°: C/5489-1). Author PR has received an EU grant FP7-PEOPLE-2013-IOF (N° PIOF-GA-2013-622571). Author EP and PR declare that they have no conflict of interest.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Palanga, E., Martin, D.P., Galzi, S. et al. Complete genome sequences of cowpea polerovirus 1 and cowpea polerovirus 2 infecting cowpea plants in Burkina Faso. Arch Virol 162, 2149–2152 (2017). https://doi.org/10.1007/s00705-017-3327-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00705-017-3327-z