Abstract
Here, we describe the full-length genome sequence of a novel potyvirus, tentatively named “Miscanthus sinensis mosaic virus” (MsiMV), isolated from Miscanthus sinensis (silver grass) held in a post-entry quarantine facility after being imported into Western Australia, Australia. The MsiMV genome is 9604 nucleotides (nt) in length, encoding a 3071-amino-acid (aa) polyprotein with conserved sequence motifs. The MsiMV genome is most closely related to that of sorghum mosaic virus (SrMV), with 74% nt and 78.5% aa sequence identity to the SrMV polyprotein region. Phylogenetic analysis based on the polyprotein grouped MsiMV with SrMV, sugarcane mosaic virus (SCMV), and maize dwarf mosaic virus (MDMV). This is the first report of a novel monopartite ssRNA virus in Miscanthus sinensis related to members of the genus Potyvirus in the family Potyviridae.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Miscanthus sinensis, commonly known as silver grass, is a member of the family Poaceae and is native to eastern Asia, where it is a keystone species in grasslands. In addition to ornamental applications, M. sinensis has been investigated for use as a biofuel crop due to its high biomass yield. Miscanthus has been found to be an alternate host for viruses infecting plants of the family Poaceae, including switchgrass mosaic virus and barley yellow dwarf virus [1].
Potyviruses represent the largest and most economically damaging genus of known plant RNA viruses [2]. Potyviruses are aphid-borne, infecting both monocotyledonous and eudicotyledonous angiosperms [3]. In response to viral infection, plants can disrupt viral transcripts via RNA interference using Dicer-like endonucleases [4]. Infections by potyviruses typically result in silencing responses, predominantly using 21- and 22-nt small RNAs [5].
All potyviruses have a monopartite genome containing a long open reading frame (ORF) [6], which encodes a polyprotein that is proteolytically processed by three viral proteases into 10 functional proteins: P1, HC-Pro, P3, 6K1, cylindrical inclusion (CI) protein, 6K2, viral genome-linked protein (VPg), NIa-Pro, nuclear inclusion b (NIb; also known as RNA-dependent RNA polymerase), and capsid protein (CP) [7, 8].
A Miscanthus sinensis cultivar “Morning Light” plant imported into Western Australia from the USA in 1985 was transferred to the Plant Quarantine Station, Rydalmere, New South Wales, for virus screening, as per new import conditions for clonal grasses at the time [9]. Double-stranded RNA extracted from 38 g of mature leaf tissue electrophoresed in a 0.75% agarose gel revealed bands suspected to be of viral RNA, and subsequent mechanical inoculation of ground grass tissue onto plants of Zea mays cv. Supagold resulted in mosaic symptoms [10]. The infected Morning Light cultivar was never released from post-entry quarantine and was maintained as a positive control for biological indexing (Fig. 1A). Sap from the infected plant contained filamentous particles approximately 600 nm in length (Fig. 1B), further confirming the presence of a potyviral pathogen.
(A) A Miscanthus plant infected with MsiMV, showing leaf mottling symptoms. (B) Morphology of MsiMV virions viewed by transmission electron microscopy. Samples were prepared using a modified ‘quick dip’ method [14]. Freshly cut leaf pieces (2 × 2 mm) were placed into 2% (w/v) phosphotungstic acid stain (pH 7.07) and adsorbed onto a carbon-coated mesh grid (formvar carbon film 400 mesh, copper). Grids were dried at 37 °C for at least 30 min then examined at 26,000x magnification using a Tecnai G2 Spirit transmission electron microscope. (C) Schematic representation of the genome organization of MsiMV. The 5′ (140 nt) and 3′ (251 nt) untranslated regions are represented by solid horizontal bars, while ORFs are depicted as open boxes. Putative sites of polyprotein cleavage mediated by P1, HC-Pro, and NIa-Pro are indicated by red, orange, and blue arrows, respectively. Protein domain sizes in aa are indicated below the protein name. (D) Alignment of small RNAs, ONT, and Illumina ribominus RNA-seq sequences with the MsiMV genome sequence. Scale bars on the y-axis show the total number of mapped reads (x1000).
Collection of RNA samples and in silico assembly of 21- to 22-nt small RNA sequences were performed as described previously [11], yielding a single 9604-nt sequence containing a single long ORF from nt 141 to 9353 and flanking non-coding sequences, including a polyA tail at the 3’ end, which is typical of potyviruses. Mapping of small RNA reads onto the MsiMV genome sequence, allowing no mismatches, resulted in 11,370,140 aligned reads (~25Kx coverage) of which 89.2%, 7.6%, and 0.3% were 21 nt, 22 nt, and 24 nt long, respectively. The high proportion of mapped 21-nt reads is consistent with them being derived from the antiviral silencing response [5].
The completeness of the small-RNA-based genome assembly was confirmed by mapping of both Illumina and long-read Oxford Nanopore Technology (ONT reads > 500 bp) ribosomal-depleted RNA-seq data (Fig.1D). Overall, 7,154,030 Illumina (~74.5Kx coverage) and 21,495 ONT (2140x coverage) reads were mapped onto the MsiMV genome using bowtie [12] and minimap2 [13], respectively. Identical assembled MsiMV genome sequences were obtained using all three technologies. Raw data are available under BioProject PRJNA752836.
The qualifiers accepted by the International Committee on Taxonomy of Viruses for a new species of potyvirus are < 76% nt and < 82% aa ORF sequence identity [8]. Comparison of the MsiMV polyprotein and coat protein sequences against those of SrMV (KM025045) showed that the new virus met these criteria. Additionally, the polyprotein is predicted to be cleaved into 10 subdomains corresponding to those of other potyviruses (Fig. 1C).
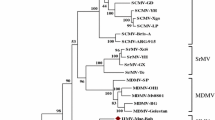
Phylogenetic analysis based on polyprotein sequences placed MsiMV within a clade with SrMV and MDMV, with SCMV as a sister clade (Fig. 2). The clade formation of these viruses may be reflective of the similarity of their plant hosts, which are all in the subfamily Panicoideae of the family Poaceae. The sequence similarity of these viruses could also explain overlaps in host susceptibility seen with SrMV, which can infect Miscanthus, corn, and sugarcane. The phylogenetic position of MsiMV, separate from SrMV, SCMV, and MDMV, further supports the classification of MsiMV as a member of a novel species in the genus Potyvirus.
Neighbor-joining tree of polyprotein aa sequences of MsiMV and selected members of the family Potyviridae. Bootstrap analysis was applied, using 1000 bootstrap replicates in MEGA X [15]. Percent bootstrap values are shown at each node.
References
Bolus S, Malapi-Wight M, Grinstead SC, Fuentes-Bueno I, Hendrickson L, Hammond RW, Mollov D (2020) Identification and characterization of Miscanthus yellow fleck virus, a new polerovirus infecting Miscanthus sinensis. PLoS ONE 15:e0239199
Braidwood L, Muller SY, Baulcombe D (2019) Extensive recombination challenges the utility of Sugarcane mosaic virus phylogeny and strain typing. Sci Rep 9:20067
Gibbs AJ, Hajizadeh M, Ohshima K, Jones RA (2020) The potyviruses: an evolutionary synthesis is emerging. Viruses 12:132
Blevins T, Rajeswaran R, Shivaprasad PV, Beknazariants D, Si-Ammour A, Park H-S, Vazquez F, Robertson D, Meins F Jr, Hohn T (2006) Four plant Dicers mediate viral small RNA biogenesis and DNA virus induced silencing. Nucleic Acids Res 34:6233–6246
Ho T, Wang H, Pallett D, Dalmay T (2007) Evidence for targeting common siRNA hotspots and GC preference by plant Dicer-like proteins. FEBS Lett 581:3267–3272
Cui H, Wang A (2019) The biological impact of the hypervariable N-terminal region of potyviral genomes. Annu Rev Virol 6:255–274
Shen W, Shi Y, Dai Z, Wang A (2020) The RNA-dependent RNA polymerase NIb of potyviruses plays multifunctional, contrasting roles during viral infection. Viruses 12
Wylie SJ, Adams M, Chalam C, Kreuze J, Lopez-Moya JJ, Ohshima K, Praveen S, Rabenstein F, Stenger D, Wang A, Zerbini FM, Ictv Report C (2017) ICTV virus taxonomy profile: Potyviridae. J Gen Virol 98:352–354
Service AQaI (1986) Quarantine Circular Memorandum GM:PG L85/1386. Canberra, Australian Capital Territory.
Davis K, Gillings M (1996) Detection of latent viruses and double-stranded RNAs in clonal grasses during post-entry quarantine. In: ACIAR PROCEEDINGS. Australian Centre for International Agricultural Research, pp 95–99
Barrero RA, Napier KR, Cunnington J, Liefting L, Keenan S, Frampton RA, Szabo T, Bulman S, Hunter A, Ward L (2017) An internet-based bioinformatics toolkit for plant biosecurity diagnosis and surveillance of viruses and viroids. BMC Bioinformatics 18:1–12
Langmead B, Trapnell C, Pop M, Salzberg SL (2009) Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10:R25
Li H (2018) Minimap2: pairwise alignment for nucleotide sequences. Bioinformatics 34:3094–3100
Dijkstra J, de Jager CP (1998) Practical plant virology: protocols and exercises. Springer-Verlag, Berlin Heidelberg New York, pp 291–292
Kumar S, Stecher G, Li M, Knyaz C, Tamura K (2018) MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol 35:1547–1549
Funding
Open Access funding enabled and organized by CAUL and its Member Institutions. Project Improving plant industry access to new genetics through faster and more accurate diagnostics using Next Generation Sequencing. MT18005 is funded by Hort Innovation, using the Hort Innovation Citrus, Grape Tables, Rubus, Potato and Nursery research and development levy, co-investment from Queensland University of Technology and contributions from the Australian Government. Hort Innovation is the grower-owned, not-for-profit research and development corporation for Australian horticulture.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Additional information
Handling Editor: Massimo Turina.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Leblanc, Z., Gauthier, ME., Lelwala, R. et al. Complete genome sequence of a novel potyvirus infecting Miscanthus sinensis (silver grass). Arch Virol 167, 1701–1705 (2022). https://doi.org/10.1007/s00705-022-05445-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00705-022-05445-3