Abstract
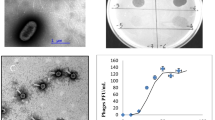
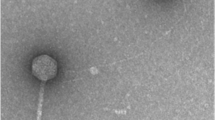
The use of phage and phage-based products for the prevention and treatment of bee disease is one of the promising natural alternatives to chemical or antibiotic treatments in beekeeping. A novel lysogenic bacteriophage, phage Pd22F (vB_PlaM_Pd22F), was isolated from Paenibacillus dendritiformis by the prophage induction method. This phage, which is capable of infecting Paenibacillus larvae and P. dendritiformis strains, was characterized by microbiological and comparative genomic analysis. Transmission electron microscopy images showed that phage Pd22F had the morphology of a myovirus. Whole-genome sequencing results showed that vB_Pla M_Pd22F has an 86,388-bp linear dsDNA genome with a GC content of 50.68%. This genome has 124 coding sequences (CDSs), 53% of which encode functionally unknown proteins and 57 of which encode proteins that show similarity to known proteins. In addition, one tRNA gene was found. The phage Pd22F genome does not contain any antimicrobial resistance genes. The similarity between the genome sequence of phage Pd22F and the whole genome sequences of other Paenibacillus phages available in the NCBI Virus Database was found to be below 50% (42%), indicating that phage Pd22F differs greatly from previously characterized phages at the DNA level. The results of comparative genomics and phylogenetic analysis revealed that Pd22F is a new phage belonging to the family Myoviridae, order Caudovirales. This is the first report of genomic and morphological characterization of a Paenibacillus dendritiformis prophage.







Similar content being viewed by others
References
Genersch E (2010) American Foulbrood in honeybees and its causative agent, Paenibacillus larvae. J Invertebr Pathol 103:S10–S19. https://doi.org/10.1016/j.jip.2009.06.015
Ingham CJ, Ben Jacob E (2008) Swarming and complex pattern formation in Paenibacillus vortex studied by imaging and tracking cells. BMC Microbiol 8:36. https://doi.org/10.1186/1471-2180-8-36
Be’er A, Strain SK, Hernandez RA, Ben-Jacob E, Florin EL (2013) Periodic reversals in Paenibacillus dendritiformis swarming. J Bacteriol 195:2709–2717. https://doi.org/10.1128/JB.00080-13
Forsgren E (2010) European foulbrood in honey bees. J Invertebr Pathol 103(Suppl 1):S5-9. https://doi.org/10.1016/j.jip.2009.06.016
Bailey L, Ball BV (1991) Honey bee pathology, 2nd edn. Academic Press, London
White G (1912) The cause of European foulbrood. US Department of Agriculture Bureau of Entomology; Circular no. 157
Gaggìa F, Baffoni L, Stenico V, Alberoni D, Buglione E, Lilli A, Di Gioia D, Porrini C (2015) Microbial investigation on honey bee larvae showing atypical symptoms of European foulbrood. Bull Insectol 68(2):321–327
Erler S, Lewkowski O, Poehlein A, Forsgren E (2018) The curious case of Achromobacter eurydice, a Gram-variable pleomorphic bacterium associated with European foulbrood disease in honeybees. Microb Ecol 75(1):1–6. https://doi.org/10.1007/s00248-017-1007-x
Lapidot D, Dror R, Vered E, Mishli O, Levy D, Helman Y (2015) Disease protection and growth promotion of potatoes (Solanum tuberosum L.) by Paenibacillus dendritiformis. Plant Pathol 64:545–551. https://doi.org/10.1111/ppa.12285
Hubenova Y, Hubenova E, Mitov M (2020) Electroactivity of the Gram-positive bacterium Paenibacillus dendritiformis MA-72. Bioelectrochemistry 136:107632. https://doi.org/10.1016/j.bioelechem.2020.107632
Wittebole X, De Roock S, Opal SM (2014) A historical overview of bacteriophage therapy as an alternative to antibiotics for the treatment of bacterial pathogens. Virulence 5(1):226–235. https://doi.org/10.4161/viru.25991
Chanishvili N (2016) Bacteriophages as therapeutic and prophylactic means: summary of the Soviet and Post Soviet Experiences. Curr Drug Deliv 13:309–323. https://doi.org/10.2174/156720181303160520193946
Myelnikov D (2018) An alternative cure: the adoption and survival of phage therapy in the USSR, 1922–1955. J Hist Med Allied Sci 73:385–411. https://doi.org/10.1093/jhmas/jry024
Lin DM, Koskella B, Lin HC (2017) Phage therapy: an alternative to antibiotics in the age of multi-drug resistance. World J Gastrointest Pharmacol Therap 8(3):162–173. https://doi.org/10.4292/wjgpt.v8.i3.162
Oliveira A, Melo LDR, Kropinski AM, Azeredo J (2013) Complete genome sequence of the broad-host-range Paenibacillus larvae phage phiIBB_Pl23. Genome Announc 1(5):e00438-e513. https://doi.org/10.1128/genomeA.00438-13
Carson S, Bruff E, DeFoor W, Dums J, Groth A, Hatfield T, Iyer A, Joshi K, McAdams S, Miles D, Miller D, Oufkir A, Raynor B, Riley S, Roland S, Rozier H, Talley S, Miller ES (2015) Genome sequences of six Paenibacillus larvae Siphoviridae phages. Genome Announc 3(3):e00101-e115. https://doi.org/10.1128/genomeA.00101
Beims H, Wittmann J, Bunk B, Spröer C, Rohde C, Günther G, Rohde M, von der Ohe W, Steinert M (2015) Paenibacillus larvae-directed bacteriophage HB10c2 and its application in American foulbrood-affected honey bee larvae. Appl Environ Microbiol 81:5411–5419. https://doi.org/10.1128/AEM.00804-15
Tsourkas PK, Yost D, Krohn A, Leblanc L, Zhang A, Stamereilers C, Amy PS (2015) Complete genome sequences of nine phages capable of infecting Paenibacillus larvae, the causative agent of American foulbrood disease of honeybees. Genome Announc 3(5):e01120-e1215. https://doi.org/10.1128/genomeA.01120-15
Walker JK, Merrill BD, Berg JA, Dhalai A, Dingman DW, Fajardo CP, Graves K, Hill HL, Hilton JA, Imahara C, Knabe BK, Mangohig J, Monk J, Mun H, Payne AM, Salisbury A, Stamereilers C, Velez K, Ward AT, Breakwell DP, Grose JH, Hope S, Tsourkas PK (2018) Complete genome sequences of Paenibacillus larvae phages BN12, Dragolir, Kiel007, Leyra, Likha, Pagassa, PBL1c, and Tadhana. Genome Announc 6:e01602-e1617. https://doi.org/10.1128/genomeA.01602-17
Ribeiro HG, Melo LDR, Oliveira H, Boon M, Lavigne R, Noben JP, Azeredo J, Oliveira A (2019) Characterization of a new Podovirus infecting Paenibacillus larvae. Sci Rep 9:20355. https://doi.org/10.1038/s41598-019-56699-y
Stamereilers C, Fajardo CP, Walker JK, Mendez KN, Castro-Nallar E, Grose JH, Hope S, Tsourkas PK (2018) Genomic Analysis of 48 Paenibacillus larvae Bacteriophages. Viruses 10(7):377. https://doi.org/10.3390/v10070
Tsourkas PK (2020) Paenibacillus larvae bacteriophages: obscure past, promising future. Microbial Genomics 6(2):e000329. https://doi.org/10.1099/mgen.0.000329
Pınarbaş M, Alpay Karaoğlu S (2017) Characterization and antibiotic sensitiveness of gram positive bacteri̇a isolated from American foulbrood suspect bee (Apis mellifera) and bee products, with genetic di̇versity of Paenibacillus larvae isolates. Recep Tayyip Erdogan University, Graduate School of Natural and Applied Sciences, Department of Biology, 117s
Karali M and Alpay Karaoglu S (2019) Investigation of bacteriophage/bacteriocin presence and host width in Paenibacillus dendritiformis isolates. Recep Tayyip Erdogan University, Graduate School of Natural and Applied Sciences, Department of Biology, 61s
Vallat B, Edwards S, O’Neill B (2012) Manuel of diagnostic tests and vaccines for terrestrial animals. OIE 1:365–380
De Graaf DC, Alippi AM, Antúnez K, Aronstein KA, Budge G, De Koker D, De Smet L, Dingman DW, Evans JD, Foster LJ, Fünfhaus A, Garcia-Gonzalez E, Gregorc A, Human H, Murray KD, Nguyen BK, Poppinga L, Spivak M, Van Engelsdorp D, Wilkins S, Genersch E (2013) Standard methods for American foulbrood research. J Apicultural Res. https://doi.org/10.3896/IBRA.1.52.1.11
Bakonyi T, Derakhshifar I, Grabensteiner E, Nowotny N (2003) Development and evaluation of PCR assays for the detection of Paenibacillus larvae in honey samples: comparison with isolation and biochemical characterization. Appl Environ Microbiol 6:1504–1510. https://doi.org/10.1128/AEM.69.3.1504-1510.2003
Halebian S, Harris B, Finegold SM, Rolfe RD (1981) Rapid method that aids in distinguishing Gram-positive from Gram-negative anaerobic bacteria. J Clin Microbiol 13(3):444–448. https://doi.org/10.1128/jcm.13.3.444-448.1981
Rohde M (2011) Microscopy, taxonomy of prokaryotes. Methods Microbiol 38:61–100. https://doi.org/10.1016/B978-0-12-387730-7.00004-8
Sambrook J, Fritsch ER, Maniatis T (1989) Molecular cloning: a laboratory manual, 2nd edn. Cold Spring Harbor Laboratory Press, Cold Spring Harbor
Dobbelaere W, de Graaf DC, Peeters JE (2001) Development of a fast and reliable diagnostic method for American foulbrood disease (Paenibacillus larvae subsp. larvae) using a 16S rRNA gene based PCR. Apidologie 32(4):363–370. https://doi.org/10.1051/apido:2001136
Baker GC, Smith JJ, Cowan DA (2003) Review and re-analysis of domain-specific 16S primers. J Microbiol Methods 55(3):541–555. https://doi.org/10.1016/j.mimet.2003.08.009
Rivas R, Velázquez E, Zurdo-Piñeiro JL, Mateos PF, Martínez Molina E (2004) Identification of microorganisms by PCR amplification and sequencing of a universal amplified ribosomal region present in both prokaryotes and eukaryotes. J Microbiol Methods 56(3):413–426. https://doi.org/10.1016/j.mimet.2003.11.007
Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4(4):406–425. https://doi.org/10.1093/oxfordjournals.molbev.a040454
Kumar S, Stecher G, Li M, Knyaz C, Tamura K (2018) MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol 35:1547–1549. https://doi.org/10.1093/molbev/msy096
Tamura K (1992) Estimation of the number of nucleotide substitutions when there are strong transition-transversion and G + C-content biases. Mol Biol Evol 9:678–687. https://doi.org/10.1093/oxfordjournals.molbev.a040752
Felsenstein J (1985) Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39:783–791. https://doi.org/10.1111/j.1558-5646.1985.tb00420.x
Muniesa M, Jofre J (2007) The contribution of induction of temperate phages to the numbers of free somatic coliphages in waters is not significant. FEMS Microbiol Lett 270:272–276. https://doi.org/10.1111/j.1574-6968.2007.00676.x
Dingman DW, Bakhiet N, Field CC, Stahly DP (1984) Isolation of two bacteriophages from Bacillus larvae, PBL1 and PBL0.5, and partial characterization of PBL1. J Gen Virol 65:1101–1105. https://doi.org/10.1099/0022-1317-65-6-1101
Bradley DE (1967) Ultrastructure of bacteriophage and bacteriocins. Bacteriol Rev 31(4):230–314. https://doi.org/10.1128/br.31.4.230-314.1967
Hershey AD, Dove W (1983) Introduction to lambda. In: Hendrix RW, Roberts JW, Stahl WF, Weisberg RA (eds) Lambda II. Cold Spring Harbor, Cold Spring Harbor Laboratory, pp 3–20
Roberts JW, Devoret R (1983) Lysogenic induction. In: Hendrix RW, Roberts JW, Stahl FW, Weisberg RA (eds) Lambda II. Cold Spring Harbor, Cold Spring Harbor Laboratory, pp 123–144
Kiliç AO, Pavlova SI, Alpay S, Kiliç SS, Tao L (2001) Comparative study of vaginal Lactobacillus phages isolated from women in the United States and Turkey: prevalence, morphology, host range, and DNA homology. Clin Diagn Lab Immunol 8(1):31–39. https://doi.org/10.1128/CDLI.8.1.31-39.2001
Hurst CJ, Reynolds KA (2002) Chapter 48: sampling viruses from soil. In: Hurst CJ, Crawford RL, Knudsen GR, McInerney MJ, Stetzenbach DL (eds) Manual of environmental microbiology, 2nd edn. American Society for Microbiology Press, Washington DC, pp 527–534
Santos SB, Carvalho CM, Sillankorva S, Nicolau A, Ferreira EC, Azeredo J (2009) The use of antibiotics to improve phage detection and enumeration by the double-layer agar technique. BMC Microbiol 9:148. https://doi.org/10.1186/1471-2180-9-148
Kim M, Ryu S (2011) Characterization of a T5-like coliphage, SPC35, and differential development of resistance to SPC35 in Salmonella enterica serovar typhimurium and Escherichia coli. Appl Environ Microbiol 77(6):2042–2050. https://doi.org/10.1128/AEM.02504-10
Fauquet C, Mayo MA, Maniloff J, Desselberger U, Ball LA (2005) Virus taxonomy. In: Eighth report of the international committee on the taxonomy of viruses. Elsevier Academic Press, San Diego, CA
Laemmli UK (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680–685. https://doi.org/10.1038/227680a0
Pickard DJ (2009) Preparation of bacteriophage lysates and pure DNA. Methods Mol Biol 502:3–9. https://doi.org/10.1007/978-1-60327-565-1_1
Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, Pyshkin AV, Sirotkin AV, Vyahhi N, Tesler G, Alekseyev MA, Pevzner PA (2012) SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 19(5):455–477. https://doi.org/10.1089/cmb.2012.0021
Wick RR, Judd LM, Gorrie CL, Holt KE (2017) Unicycler: resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput Biol 13(6):e1005595. https://doi.org/10.1371/journal.pcbi.1005595
Overbeek R, Olson R, Pusch GD, Olsen GJ, Davis JJ, Disz T, Edwards RA, Gerdes S, Parrello B, Shukla M, Vonstein V, Wattam AR, Xia F, Stevens R (2014) The SEED and the Rapid Annotation of microbial genomes using Subsystems Technology (RAST). Nucleic Acids Res 42(Database issue):D206–D214. https://doi.org/10.1093/nar/gkt1226
Delcher AL, Bratke KA, Powers EC, Salzberg SL (2007) Identifying bacterial genes and endosymbiont DNA with Glimmer. Bioinformatics 23:673–679. https://doi.org/10.1093/bioinformatics/btm009
Hyatt D, Chen GL, LoCascio PF, Land ML, Larimer FW, Hauser LJ (2010) Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinform 11:119. https://doi.org/10.1186/1471-2105-11-119
Borodovsky M, Lomsadze A (2011) Gene identification in prokaryotic genomes, phages, metagenomes, and EST sequences with GeneMarkS suite. Curr Protoc Bioinform 4(5):1–17. https://doi.org/10.1002/0471250953.bi0405
Brettin T, Davis JJ, Disz T, Edwards RA, Gerdes S, Olsen GJ, Olson R, Overbeek R, Parrello B, Pusch GD, Shukla M, Thomason JA, Stevens R, Vonstein V, Wattam AR, Xia F (2015) RASTtk: a modular and extensible implementation of the RAST algorithm for building custom annotation pipelines and annotating batches of genomes. Sci Rep 5:8365. https://doi.org/10.1038/srep08365
Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25(17):3389–3402. https://doi.org/10.1093/nar/25.17.3389
Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215:403–410. https://doi.org/10.1016/S0022-2836(05)80360-2
Laslett D, Canback B (2004) ARAGORN, A program to detect tRNA genes and tmRNA genes in nucleotide sequences. Nucleic Acids Res 32(1):11–16. https://doi.org/10.1093/nar/gkh152
Garneau JR, Depardieu F, Fortier LC, Bikard D, Monot M (2017) PhageTerm: a tool for fast and accurate determination of phage termini and packaging mechanism using next-generation sequencing data. Sci Rep 7(1):8292. https://doi.org/10.1038/s41598-017-07910-5
Stothard P, Wishart DS (2005) Circular genome visualization and exploration using CGView. Bioinformatics 21(4):537–539. https://doi.org/10.1093/bioinformatics/bti054
Krogh A, Larsson B, von Heijne G, Sonnhammer EL (2001) Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J Mol Biol 305(3):567–580. https://doi.org/10.1006/jmbi.2000.4315
Nishimura Y, Yoshida T, Kuronishi M, Uehara H, Ogata H, Goto S (2017) ViPTree: the viral proteomic tree server. Bioinformatics 33(15):2379–2380. https://doi.org/10.1093/bioinformatics/btx157
Lopes A, Tavares P, Petit MA, Guérois R, Zinn-Justin S (2014) Automated classification of tailed bacteriophages according to their neck organization. BMC Genom 15:1027. https://doi.org/10.1186/1471-2164-15-1027
Nylander JAA (2004) MrModeltest v2. Program distributed by the author. Evolutionary Biology Centre, Uppsala University
Ronquist F, Huelsenbeck JP (2003) Mrbayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19(12):1572–1574. https://doi.org/10.1093/bioinformatics/btg180
Rambaut A, Drummond AJ, Xie D, Baele G, Suchard MA (2018) Posterior summarisation in Bayesian phylogenetics using Tracer 1.7. Syst Biol 67(5):901–904. https://doi.org/10.1093/sysbio/syy032
Stamatakis A (2014) RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30(9):1312–1313. https://doi.org/10.1093/bioinformatics/btu033
Rambaut A (2012) FigTree 1.4.0 software. Institute of Evolutionary Biology, University of Edinburgh, Edinburgh
Kropinski AM, Prangishvili D, Lavigne R (2009) Position paper: the creation of a rational scheme for the nomenclature of viruses of Bacteria and Archaea. Environ Microbiol 11:2775–2777. https://doi.org/10.1111/j.1462-2920.2009.01970.x
Gasteiger E, Hoogland C, Gattiker A, Duvaud S, Wilkins MR, Appel RD, Bairoch A (2005) Protein identification and analysis tools on the ExPASy Server. In: Walker JM (ed) The proteomics protocols handbook. Humana Press, New Jersey, pp 571–607
Wu Y (2012) Unwinding and rewinding: double faces of helicase? J Nucleic Acids 140601:1–15. https://doi.org/10.1155/2012/140601
Fuller-Pace FV, Ali S (2008) The DEAD box RNA helicases p68 (Ddx5) and p72 (Ddx17): novel transcriptional co-regulators. Biochem Soc Trans 36(Pt 4):609–612. https://doi.org/10.1042/BST0360609
Schütz P, Karlberg T, van den Berg S, Collins R, Lehtiö L, Högbom M, Holmberg-Schiavone L, Tempel W, Park HW, Hammarström M, Moche M, Thorsell AG, Schüler H (2010) Comparative structural analysis of human DEAD-box RNA helicases. PLoS ONE 5(9):e12791. https://doi.org/10.1371/journal.pone.0012791
Edgell DR, Belfort M, Shub DA (2000) Barriers to intron promiscuity in bacteria. J Bacteriol 182(19):5281–5289. https://doi.org/10.1128/JB.182.19.5281-5289.2000
Chapot-Chartier MP (2014) Interactions of the cell-wall glycopolymers of lactic acid bacteria with their bacteriophages. Front Microbiol. https://doi.org/10.3389/fmicb.2014.00236
Wang IN, Smith DL, Young R (2002) Holins: the protein clocks of bacteriophage infections. Annu Rev Microbiol 54:799–825. https://doi.org/10.1146/annurev.micro.54.1.799
Stark W, Boocock MR, Sherratt DJ (1992) Catalysis by site-specific recombinases. Trends Genet 8:432–439
Hatfull GF, Hendrix RW (2011) Bacteriophages and their genomes. Curr Opin Virol 1:298–303. https://doi.org/10.1016/j.coviro.2011.06.009
Duffy C, Feiss M (2002) The large subunit of bacteriophage lambda’s terminase plays a role in DNA translocation and packaging termination. J Mol Biol 316(3):547–561. https://doi.org/10.1006/jmbi.2001.5368
Cepko LCS, Garling EE, Dinsdale MJ, Scott WP, Bandy L, Nice T, Faber-Hammond J, Mellies JL (2020) Myoviridae phage PDX kills enteroaggregative Escherichia coli without human microbiome dysbiosis. J Med Microbiol 69(2):309–323. https://doi.org/10.1099/jmm.0.001162
McSpadden GBB (2004) Ecology of Bacillus and Paenibacillus spp. in agricultural systems. Phytopathology 94(11):1252–1258. https://doi.org/10.1094/PHYTO.2004.94.11.1252
Ruiu L (2020) Plant-Growth-Promoting Bacteria (PGPB) against Insects and Other Agricultural Pests. Agronomy 10(6):861. https://doi.org/10.3390/agronomy10060861
Sirota-Madi A, Olender T, Helman Y, Ingham C, Brainis I, Roth D, Hagi E, Brodsky L, Leshkowitz D, Galatenko V, Nikolaev V, Mugasimangalam RC, Bransburg-Zabary S, Gutnick DL, Lancet D, Ben-Jacob E (2010) Genome sequence of the pattern forming Paenibacillus vortex bacterium reveals potential for thriving in complex environments. BMC Genom 11:710. https://doi.org/10.1186/1471-2164-11-710
Dowah ASA, Clokie MRJ (2018) Review of the nature, diversity and structure of bacteriophage receptor binding proteins that target Gram-positive bacteria. Biophys Rev 10:535–542. https://doi.org/10.1007/s12551-017-0382-3
De Jonge PA, Nobrega FL, Brouns SJ, Dutilh BE (2019) Molecular and evolutionary determinants of bacteriophage host range. Trends Microbiol 27:51–63. https://doi.org/10.1016/j.tim.2018.08.006
Merrill BD, Grose JH, Breakwell DP, Burnett SH (2014) Characterization of Paenibacillus larvae bacteriophages and their genomic relationships to firmicute bacteriophages. BMC Genom 15(1):745. https://doi.org/10.1186/1471-2164-15-745
Delesalle VA, Tanke NT, Vill AC, Krukonis GP (2016) Testing hypotheses for the presence of tRNA genes in mycobacteriophage genomes. Bacteriophage 6(3):e1219441. https://doi.org/10.1080/21597081.2016.121944
Sharaf A, Oborník M, Hammad A, El-Afifi S, Marei E (2018) Characterization and comparative genomic analysis of virulent and temperate Bacillus megaterium bacteriophages. Peer J 6:e5687. https://doi.org/10.7717/peerj.5687
Merrill BD, Ward AT, Grose JH, Hope S (2016) Software-based analysis of bacteriophage genomes, physical ends, and packaging strategies. BMC Genom 17(1):679. https://doi.org/10.1186/s12864-016-3018-2
Grose JH, Jensen GL, Burnett SH, Breakwell DP (2014) Correction: genomic comparison of 93 Bacillus phages reveals 12 clusters, 14 singletons and remarkable diversity. BMC Genom 15(1):1184. https://doi.org/10.1186/1471-2164-15-1184
Thorpe HM, Smith MC (1998) In vitro site-specific integration of bacteriophage DNA catalyzed by a recombinase of the resolvase/invertase family. Proc Natl Acad Sci USA 95(10):5505–5510. https://doi.org/10.1073/pnas.95.10.5505
Rezaei Javan R, Ramos-Sevillano E, Akter A, Brown J, Brueggemann AB (2019) Prophages and satellite prophages are widespread in Streptococcus and may play a role in pneumococcal pathogenesis. Nat Commun 10(1):4852. https://doi.org/10.1038/s41467-019-12825-y
Rocker A, Meinhart A (2016) Type II toxin: Antitoxin systems. More than small selfish entities? Curr Genet 62(2):287–290. https://doi.org/10.1007/s00294-015-0541-7
Van Melderen L (2010) Toxin-antitoxin systems: why so many, what for? Curr Opin Microbiol 13(6):781–785. https://doi.org/10.1016/j.mib.2010.10.006
Yamaguchi Y, Inouye M (2011) Regulation of growth and death in Escherichia coli by toxin-antitoxin systems. Nat Rev Microbiol 9(11):779–790. https://doi.org/10.1038/nrmicro2651
Wang X, Lord DM, Cheng HY, Osbourne DO, Hong SH, Sanchez-Torres V, Quiroga C, Zheng K, Herrmann T, Peti W, Benedik MJ, Page R, Wood TK (2012) A new type V toxin-antitoxin system where mRNA for toxin GhoT is cleaved by antitoxin GhoS. Nat Chem Biol 8:855–861. https://doi.org/10.1038/nchembio.1062
Page R, Peti W (2016) Toxin-antitoxin systems in bacterial growth arrest and persistence. Nat Chem Biol 12:208–214. https://doi.org/10.1038/nchembio.2044
Yao J, Guo Y, Wang P, Zeng Z, Li B, Tang K, Liu X, Wang X (2018) Type II toxin/antitoxin system ParESO/CopASO stabilizes prophage CP4So in Shewanella oneidensis. Environ Microbiol 20(3):1224–1239. https://doi.org/10.1111/1462-2920.14068
Breitbart M, Miyake JH, Rohwer F (2004) Global distribution of nearly identical phage-encoded DNA sequences. FEMS Microbiol Lett 236:249–256. https://doi.org/10.1016/j.femsle.2004.05.042
Chow CET, Fuhrman JA (2012) Seasonality and monthly dynamics of marine Myovirus communities. Environ Microbiol 14(8):2171–2183. https://doi.org/10.1111/j.1462-2920.2012.02744.x
Wommack KE, Nasko DJ, Chopyk J, Sakowski EG (2015) Counts and sequences, observations that continue to change our understanding of viruses in nature. J Microbiol 53:181–192. https://doi.org/10.1007/s12275-015-5068-6
Rohwer F, Edwards R (2002) The Phage Proteomic Tree: a genome-based taxonomy for phage. J Bacteriol 184:4529–4535. https://doi.org/10.1128/JB.184.16.4529-4535.2002
Sullivan MB, Coleman M, Weigele P, Rohwer F, Chisholm SW (2005) Three Prochlorococcus cyanophage genomes: signature features and ecological interpretations. PLoS Biol 3:e144. https://doi.org/10.1371/journal.pbio.0030144
Hambly E, Tetart F, Desplats C, Wilson WH, Krisch HM, Mann NH (2001) A conserved genetic module that encodes the major virion components in both the coliphage T4 and the marine cyanophage S-PM2. Proc Natl Acad Sci USA 98(20):11411–11416. https://doi.org/10.1073/pnas.191174498
Hsiao CL, Black LW (1978) Head morphogenesis of bacteriophage T4. III. The role of g20 in DNA packaging. Virology 91(1):15–25. https://doi.org/10.1016/0042-6822(78)90351-3
Acknowledgments
Sincere thanks to Bayram Toraman of Karadeniz Technical University for providing advice on bioinformatics application. Thanks to Dr. Cantekin Dursun of Recep Tayyip Erdoğan University for providing help on the phylogenetic tree of phages. We thank Elif Sevim and Müberra Pinarbaş for their technical comments and assistance in the study.
Funding
This study was financially supported by the RTEU Scientific Research Projects (RTEU-FBA-2017-807).
Author information
Authors and Affiliations
Contributions
All authors contributed to the study's conception and design. Material preparation, experimental studies, data collection, evaluation of results, and interpretation of bioinformatic analysis were performed by Arif Bozdeveci and Şengül Alpay Karaoğlu. The main draft of the article was written by Arif Bozdeveci and Şengül Alpay Karaoğlu, and all authors contributed to and commented on the main draft of the article. All authors read and approved the final version of the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors have no financial conflicts of interest to declare.
Additional information
Handling Editor: Tim Skern.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Bozdeveci, A., Karali, M., Akpinar, R. et al. Isolation, characterization, and comparative genomic analysis of vB_PlaM_Pd22F, a new bacteriophage of the family Myoviridae. Arch Virol 167, 1269–1284 (2022). https://doi.org/10.1007/s00705-022-05429-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00705-022-05429-3