Abstract
Background
Paenibacillus larvae is a Firmicute bacterium that causes American Foulbrood, a lethal disease in honeybees and is a major source of global agricultural losses. Although P. larvae phages were isolated prior to 2013, no full genome sequences of P. larvae bacteriophages were published or analyzed. This report includes an in-depth analysis of the structure, genomes, and relatedness of P. larvae myoviruses Abouo, Davis, Emery, Jimmer1, Jimmer2, and siphovirus phiIBB_Pl23 to each other and to other known phages.
Results
P. larvae phages Abouo, Davies, Emery, Jimmer1, and Jimmer2 are myoviruses with ~50 kbp genomes. The six P. larvae phages form three distinct groups by dotplot analysis. An annotated linear genome map of these six phages displays important identifiable genes and demonstrates the relationship between phages. Sixty phage assembly or structural protein genes and 133 regulatory or other non-structural protein genes were identifiable among the six P. larvae phages. Jimmer1, Jimmer2, and Davies formed stable lysogens resistant to superinfection by genetically similar phages. The correlation between tape measure protein gene length and phage tail length allowed identification of co-isolated phages Emery and Abouo in electron micrographs. A Phamerator database was assembled with the P. larvae phage genomes and 107 genomes of Firmicute-infecting phages, including 71 Bacillus phages. Phamerator identified conserved domains in 1,501 of 6,181 phamilies (only 24.3%) encoded by genes in the database and revealed that P. larvae phage genomes shared at least one phamily with 72 of the 107 other phages. The phamily relationship of large terminase proteins was used to indicate putative DNA packaging strategies. Analyses from CoreGenes, Phamerator, and electron micrograph measurements indicated Jimmer1, Jimmer2, Abouo and Davies were related to phages phiC2, EJ-1, KC5a, and AQ113, which are small-genome myoviruses that infect Streptococcus, Lactobacillus, and Clostridium, respectively.
Conclusions
This paper represents the first comparison of phage genomes in the Paenibacillus genus and the first organization of P. larvae phages based on sequence and structure. This analysis provides an important contribution to the field of bacteriophage genomics by serving as a foundation on which to build an understanding of the natural predators of P. larvae.
Similar content being viewed by others
Background
Paenibacillus larvae (P. larvae) is a sporulating Firmicute. It is the causative agent of American Foulbrood (AFB), a disease that infects and destroys the larvae of honeybees (Apis mellifera). The first eight P. larvae phages were reported between 1955 and 1999 and included BLA [1], L3 [2], BL2 [3], PBL1 [4], PBL0.5 [5], PBL2 [5], PBL3 [6], and PPL1c [7]. These phages have not been sequenced. Most of these phages were isolated from lysogens and were used to characterize different strains of P. larvae [8–10]. Phages that infect P. larvae were originally identified as Bacillus larvae phages; however, the names of these phages were changed to Paenibacillus larvae phages following the reclassification of the bacteria [11, 12].
Recent advances in DNA sequencing technology have made it possible to sequence many bacteriophage genomes. When these sequences are analyzed, putative protein functions can be determined. Other studies have used comparative genomics to organize phages into related clusters [13], correlate phage packaging mechanisms with large terminase protein sequences [14], and study gene transfer, phylogenetic relationships, and impacts on host virulence [15, 16].
Comparative genomics can be accomplished using software specialized for phage genomes such as the computer program, Phamerator [17]. Phamerator incorporates available data for each genome entered into its database, such as bacterial host, annotations of genes from GenBank, and conserved domains [18]. Phamerator compares each gene product in the database to each other using BLASTP [19] and ClustalO [20], the scores of which are used to create phamilies (phams) of related gene products. Phamerator provides visual tools such as full genome comparison maps and can display the relationships between proteins within a pham using a circular diagram (pham circle). Proteins within each pham must meet or exceed user-defined cutoffs for E-values and percent identity for at least one other gene product in the pham. Strict cutoffs result in phams that indicate a shared similar function and predict phylogenetic relationships.
In 2013, six P. larvae phages were isolated. These phages were fully sequenced and their genomes published [21, 22]. P. larvae siphovirus phiIBB_Pl23 was isolated in Portugal [21] and P. larvae myoviruses Abouo, Davies, Emery, Jimmer1, and Jimmer2 were isolated in Utah [22]. In this report we compare the genomes of the six fully-sequenced P. larvae phages, categorize all published P. larvae phages into three groups based on structural morphology, use Phamerator to analyze previously unexplored Paenibacillus phages, and explore genetic relationships of the P. larvae phages with 107 other phages that infect Firmicute hosts. We identify gene products with conserved domains including a putative bacteriocin, serine recombinase, and antirepressor, and investigate their conservation among P. larvae phages. Results from Phamerator and CoreGenes indicate a relationship between four P. larvae phages and four small genome myoviruses that infect Lactobacillus, Clostridium and Streptococcus. These results show that comparisons can be drawn between phages that infect a phylum and provide a basis for analyzing and comparing newly isolated phages that infect P. larvae.
Results
Bacterial identification, phage isolation, and phage sequencing
Bacterial isolates were collected from spores found in honey samples. All characteristic tests for P. larvae were positive: the isolates grew on PLA plates and were catalase negative and Gram-positive. PCR products from 16S rRNA primers were sequenced using BigDye sequencing. BLAST results from nine of the ten 16S rRNA sequences showed more than 99% similarity with Paenibacillus larvae subsp. pulvifaciens strains DSM 8442 and DSM 8443 as well as the related bacteria Brevibacillus laterosporus. Of nine isolates, PL2 and PL6 were used for phage isolation. Phages Abouo, Davies, and Emery were isolated using PL6, while phages Jimmer1 and Jimmer2 were isolated using PL2.
Each P. larvae phage sample was plaque-purified at least three times, sequenced, and published [22]. Prior to genome sequencing and electron microscopy we were unaware that one phage sample still contained two different phages. Plaque purification did not successfully separate these two phages. However, assembly of the genomes revealed two clearly independent genomes that separated with ease with over 100-fold coverage of the genomes. These co-isolated phages were named Emery and Abouo. Results from sequencing and annotation of the six P. larvae phages are found in Table 1. The genomes varied in length from ~40 kb to ~58 kb. Most of the genes in each genome were located on the forward strand (90% ± 3%). The average G + C content for these phages is 39.48% ± 1.41%. BLAST hits for proteins within these phages included both Paenibacillus and Brevibacillus bacterial strains as well as bacteriophages that infected other Firmicutes.
Phage cross-infectivity, lysogeny, and lysogen superinfection
Plaques from the sample containing Emery and Abouo were clear while plaques from the other three phages were hazy. Phages Jimmer1 and Jimmer2 were isolated independently using PL2 and neither of these phages could infect PL6. Phages Davies and Emery/Abouo were isolated using PL6 and these phages could not infect PL2. None of these phages were able to produce plaques on lawns of B. cereus, B. subtilis, L. acidophilus, S. aureus, or S. epidermidis. Of the four phage samples, three formed stable lysogens: Jimmer1 in PL2, Jimmer2 in PL2, and Davies in PL6. Stable lysogens were identified when phages were not able to superinfect bacteria lysogenic for the same phage. The lysate containing Emery and Abouo induced lysis in the PL6 Davies lysogen. No other superinfection or induced lysis was observed in any other lysogen-phage combinations. The sample containing Emery and Abouo did not form a stable lysogen and no superinfection data were obtained.
Electron microscopy reveals myovirus structure for five P. larvaephages
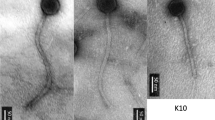
Electron microscopy revealed that these five P. larvae phages were myoviruses, marked by the presence of a contractile tail sheath (Figure 1). Figure 1A shows tail structures separated from the phage capsids. These tail structures were more abundant than intact phages in all samples submitted for electron microscopy. Figure 1B, 1C, 1E, and 1F show intact phages with contracted tail sheaths, while Figure 1D shows an extended tail sheath. Because phages Emery and Abouo were not separated, micrographs for these phages were taken from the same copper grid.
Electron micrographs of P. larvae phages. A) Tails and tail sheaths of P. larvae phages separated from the capsids (Jimmer1). These structures were more abundant than intact phages in electron micrographs. Scale bar represents 0.2 μm. B) Phage Abouo. C) Phage Davies. D) Phage Emery. E) Phage Jimmer1. F) Phage Jimmer2. Scale bars represent 50 nm for panels B-F.
Sequence similarities between phages
Gepard dotplots of the full genome sequences for all six P. larvae phages are shown in Figure 2. Diagonal lines within the dotplots indicated that phages Abouo, Davies, Jimmer1, and Jimmer2 were very similar to each other. Host specificity was also reflected in similarities between these four phages. For instance, the PL2 phages Jimmer1 and Jimmer2 shared 99.8% average nucleotide identity and the PL6 phages Abouo and Davies shared 94.9% identity. However, the average nucleotide identity between PL2 phages Jimmer1 and Jimmer2 and PL6 phages Abouo and Davies was only 80.5%. The lack of diagonal lines in the dotplot indicated that phages Emery and phiIBB_Pl23 were very different from the other P. larvae phages examined. While the Emery and Abouo phages were found in the same sample, sequences assembled independently without conflict with over 100-fold coverage for each genome [22]. No similarities between the sequences of Emery and Abouo were apparent in the dotplot (Figure 2). However, a black line indicates a small section of homology between Emery and Davies that Davies does not share with Abouo. Because Emery and Abouo were co-isolated and sequenced together, individual reads in these sections of Emery, Davies, and Abouo were scrutinized using Consed [23] to ensure the assemblies were correct. The fold coverage before, throughout, and after these sections of Emery, Davies, and Abouo was at least 80.
Distinguishing between phages Emery and Abouo
In addition to the separation of unique DNA sequences for Emery and Abouo in the same sample, two markedly different phages were present in electron micrographs based on measurements of capsid height and tail length. Putative tape measure protein (TMP) genes were identified in each phage genome. The TMP gene found in Emery (gp16) was 3,000 bp long, while the TMP gene in Abouo (gp20) was 2,055 bp long, showing that the TMP gene in Emery was 1.46 times longer than the TMP gene in Abouo. The positive correlation between TMP gene length and tail length [24] was used to suggest a correlation between phages Emery and Abouo in electron micrographs and their respective genome sequences. The average tail length for Emery was 162.2 nm long while the average tail length for Abouo was 113.6 nm, making the tail of phage Emery 1.43 times longer than the tail of phage Abouo (Table 2). In comparison, the TMP gene of Emery was 1.46 times longer than the TMP gene in Abouo. Based on this data, we matched the long TMP gene and tail length with Emery, and the short TMP gene and tail length with Abouo (Figure 1).
Measurements from electron micrographs
P. larvae phages were separated into three distinct groups based on structural morphology. The first group of phages were myoviruses (icosahedral capsids and contractile tails) and included phages Abouo, Davies, Emery, Jimmer1, Jimmer2, PBL0.5 [5], and PBL2 [5]. There were two distinct groups of phages that were siphoviruses. The first siphovirus group contains phages BLA [1], PBL1 [4], and PPL1c [7]. These phages had long, non-contractile tails and elongated capsids. The second siphovirus group contained only PBL3 which had a round capsid [6]. Phage phiIBB_Pl23 was also a siphovirus [21], but could not be categorized into one of the two siphovirus groups because images or measurements were not yet available. There was no apparent correlation between the type of phage and where the phage was isolated. Measurements taken from all published electron micrographs of P. larvae phages are shown in Table 2. Measurements for phages Abouo, Davies, Emery, Jimmer1, and Jimmer2 were taken from at least three different intact phages. Phages grouped into categories by morphology type were found to have similar structural measurements. The structures of the P. larvae myoviruses were similar in size with an average capsid height of 67.2 ± 3.2 nm and an average width of 64.1 ± 2.6 nm. The average tail length was 122.0 ± 27.3 nm and was the most variable of the measured phage features.
Frameshift in P. larvaephage Emery
Phage Emery exhibited a putative ribosomal slippage site in gp4 that encoded for a head morphogenesis protein in the SPP1 gp7 family. This frameshift was identified by the online frameshift finding tool FrameD [25]. The two products in Emery are predicted to be 82.5 kDa following ribosomal slippage and 58.9 kDa if there is no slippage. The presence of both head morphogenesis proteins in the Emery virion has not yet been verified. We were unable to detect a putative frameshift via FrameD in Bacillus phage SPP1 or any protein sequence homology using BLASTP between the morphogenesis proteins in Emery and SPP1.
P. larvaephage genomic comparison using Phamerator software
A database of phage genomes related to the P. larvae phages was assembled for analysis using the phage genome comparison program Phamerator [17]. The finished Phamerator database contained a diverse set of phages that infected Firmicute bacteria including the 6 P. larvae phage genomes, 71 Bacillus phages, 1 Clostridium phage, 3 Enterococcus phages, 2 Geobacillus phages, 7 Lactobacillus phages, 6 Listeria phages, one Paenibacillus glucanolyticus phage, 15 Staphylococcus phages, and 1 Streptococcus phage (Additional file 1). Phages included in the database were selected based on BLAST hits to gene products identified during annotation of the P. larvae phage genomes. Phamerator grouped the 13,697 putative proteins annotated in the 113 phage genomes into 6,181 phamilies (phams). Only 2,233 phams (36.1%) contained two or more members. These 2,233 phams contained 9,749 (71.1%) of the 13,697 putative proteins in the database. The remaining 3,948 (28.8%) putative proteins could not be grouped with other proteins and were designated as "orphams" [17].
The Phamerator database allowed comparison of P. larvae phage genes to each other and to phages infecting other bacteria. A spreadsheet was exported from the Phamerator database to report all phage gene products in the database, the phams to which the gene products are assigned, and the conserved domains found in gene products in those phams (Additional file 2). Table 3 indicates how many putative proteins in each P. larvae phage are orphams, are shared only with P. larvae phages, or are shared with phages infecting other bacterial hosts. Putative proteins from Jimmer1 and Jimmer2 shared phams with 56 non-P. larvae phages, while Abouo shared phams with 52, Davies with 53, Emery with 24, and philBB_P123 with 57 other non-P. larvae phages. Between the six P. larvae phages, there are phams shared with 72 non-P. larvae phages of the 107 (67.3%) included in the database. Of 562 genes in the six P. larvae phages, only 114 (20.3%) encoded proteins grouped into phams with proteins from other types of phages in the database. Of the remaining genes, 300 (53.4%) encoded proteins that were grouped into phams containing only P. larvae phage proteins and 148 (26.3%) were orphams.
Pham groupings reflected the genetic relationships of P. larvae phages. The genomic sequence comparison of Jimmer1 and Jimmer2 using ClustalW identified differences in 80 bp of the 54,312 bp genomes (99.85% similar) [22]. All corresponding genes between Jimmer1 and Jimmer2 shared the same phams. Of the 80 P. larvae phage genes in Jimmer1 and Jimmer2, 31 were unique to these two phages and were not found in any other phages in the database. These 31 genes would be orphams if Jimmer2 were not isolated (Table 3).
Phamerator identified conserved domains in at least one gene in 1,501 phams (24.3%) of the total 6,181 phams in the database. Although many P. larvae phage genes encoded proteins with significant BLAST hits, less than half of the proteins had a known function. Of all P. larvae phage putative proteins, 86% had a BLAST hit with an E-value less than 1 × 10-4 (see Table 3), yet only 48% of the proteins returned BLAST hits listing a function. Conserved domains were identified in only 43% of the P. larvae phage putative proteins (Table 3). Phages Emery and phiIBB_Pl23 contained the most orphams, the fewest BLAST hits, and the most putative proteins with no identifiable conserved domains.
P. larvaephages share structural and regulatory genes with similar functions
Conserved domains and BLAST hits matching phage or bacterial proteins were used to assign functions to 234 gene products in the six P. larvae phages, indicating that these genes were not novel and were characteristically found in other phages. The assembly and structural proteins were grouped according to function in Table 4. The regulatory and non-structural proteins are listed in Table 5. The pham assignment for each gene is shown in parentheses. Pham numbers are specific to the Phamerator database used for this analysis.
Most of the functions listed in Tables 4 and 5 describe proteins found in more than one phage. For example, all five of the P. larvae myoviruses contained seven proteins that belonged to the same family or superfamily but not always to the same pham. The function of these proteins includes head morphogenesis, tape measure, baseplate (see Table 4), LysM peptidoglycan binding, peptidoglycan hydrolase, PBSX, and bacteriocin (see Table 5).
Few proteins with known functions were identified as putative virulence factors. BLAST results indicate that gp26 in P. larvae phage phiIBB_Pl23 is a protein that is toxic to insect larvae. No toxin genes were identified in the P. larvae myoviruses. Other host-related proteins include an ABC transporter-like protein found in P. larvae phages Abouo, Davies, Jimmer1, Jimmer2, and phiIBB_Pl23 as well as an XRE-family transcriptional regulator found in all P. larvae phages. The five myoviruses contained between five and ten of these regulators per genome compared to only two in the siphovirus phiIBB_Pl23. Abouo gp51, Jimmer1 gp58, Jimmer2 gp58 and Emery gp40, gp64, and gp65 (Table 5) are the only transcriptional regulators that share a pham with a non-P. larvae phage. All others are only found in P. larvae phages. It is not known what effects these transcriptional regulators have on the host, but they do contain a canonical helix-turn-helix (HTH) domain. Very few of the regulatory genes in these phages have known functions.
Phage genome organization and pham groupings indicate relatedness of four P. larvaephages
A linear genome map of the six P. larvae phages shows that the genes in phages Jimmer1, Jimmer2, Abouo, and Davies are organized similarly (Figure 3). Identically colored genes encode products that share a pham, while white genes encode orpham gene products. There are 58 phams that each contained gene products from phages Abouo, Davies, Jimmer1, and Jimmer2. Proteins in 30 of these phams had identifiable functions based on BLAST hits and are italicized in Tables 4 and 5. Of the 58 conserved phams, 38 did not contain homologs from any other phage in the database. Of the remaining 20 phams that have homologs from other phage types, three of the most populated phams are those containing small terminase (13 members), large terminase (14 members), and portal protein (13 members). Of the 16 other phages that shared one of these phams with the four similar P. larvae myoviruses, only three phages shared all three phams: Staphylococcus phages 37, 88, and PH15.
Linear genome map of the six P. larvae phages. Connecting lines between Jimmer1, Abouo, Davies, and Emery represent nucleotide similarity (E-value less than 1 × 10-4). Genes are color-coded according to phams of the encoded protein. Gene numbers are indicated inside gene boxes. Functions matching gene products described in Tables 4 and 5 are also displayed.
Phamily relationships of large terminase proteins indicate putative DNA packaging strategies
Phage gene products must meet stringent parameters in order be grouped into a pham with other genes that encode similar proteins. Because gene products in a pham are highly similar, phylogenetic analysis indicates that these proteins will be more closely related than others with the same function. A neighbor-joining phylogenetic tree grouped large terminase proteins in the Phamerator database by phamily (Figure 4). Amino acid sequences of large terminase proteins can indicate the DNA packaging strategy [14]. Phages Abouo, Davies, Jimmer1, and Jimmer2 likely use headful packaging and have circularly permuted terminal repeats based on close association with the large terminases of well-characterized headful packaging phages P40 [26], and SPP1 [27] which share a pham. Phage phiIBB_Pl23 likely has 3′ cohesive ends based on close association with phage phiSLT [28]. Further analysis of experimental data indicated that no phams generated by Phamerator contained terminases belonging to phages with different packaging strategies (data not shown). The packaging strategy for phage Emery is still undetermined because its large terminase protein is an orpham.
Neighbor-joining phylogenetic tree of the large terminase gene products from the Phamerator database indicate proposed packaging strategies. Colored boxes indicate proteins belonging to the same pham. Proteins that are not highlighted are orphams. Large terminase proteins grouped into similar phams are closely related on the tree and share a packaging strategy. *Experimentally determined headful packaging, circularly permuted terminal repeats [26, 27], **Experimentally determined 3′ cohesive ends [28]. ***Experimentally determined, long direct terminal repeats [29].
P. larvaephages exhibit genetic and structural similarity with other small genome myoviruses.
The putative proteins encoded in P. larvae phages Abouo, Davies, Jimmer1, and Jimmer2 were often grouped into phams with proteins in Clostridium phage phiC2, Lactobacillus phage KC5a, and Streptococcus phage EJ-1. The similar proteins were mostly structural (Table 6) and included the terminase (small subunit), portal, head morphogenesis, minor structural, tail sheath, and baseplate proteins. All of the gene products listed in the table were grouped into the same pham except for three proteins that narrowly missed the pham cutoff values and are marked by asterisks. The tape measure proteins in Abouo, Davies, Jimmer1, and Jimmer2 were somewhat similar to those found in Streptococcus phage EJ-1 (average E-value < 9 × 10-17, average identity = 24%) and Clostridium phage phiC2 (average E-value < 8 × 10-17, average identity = 22%) but were not near the pham cutoff values of 1 × 10-50.
The electron micrographs of P. larvae phages Abouo, Davies, Jimmer1, and Jimmer2 reveal a structure similar to those reported for Clostridium phages phiC2 [30], phiCD27 [31], and phiCD119 [32], Lactobacillus phages KC5a [33] which is reported as similar to Lactobacillus phages KC21T [33] and phiAQ113 [34], and Streptococcus phage EJ-1 [35]. The capsid of Clostridium phage phiC2 is 65 nm in diameter and the tail is 148 nm long. The capsid of Streptococcus phage EJ-1 is 57 nm in diameter and the tail is 130 nm long. The capsid of Lactobacillus phage KC5a was not measured, however it was reported as similar to KC21T, which had a capsid diameter of 45 nm and a tail length of 160 nm. KC5a was also reported to be genetically similar to phiAQ113, which has a capsid diameter of 55 nm, a tail length of 147 nm, and a tail sheath width of 22 nm. Phamerator grouped all KC5a gene products listed in Table 6 into the same phams as phiAQ113 genes except the small terminase (absent in phiAQ113) and the head morphogenesis protein.
Although P. larvae phage Emery contained gene products with the same functions as those listed in Table 6, the proteins were all orphams. However, the first five gene products encoded in P. larvae phage phiIBB_Pl23 (small terminase, large terminase, portal protein, protease, and major capsid proteins) all shared a pham with similar proteins from five siphovirus Staphylococcus phages (3A, 47, phi12, phiSLT, and tp310-2).
The program CoreGenes 3.5 was used to further compare the genes in the P. larvae phages with small-genome myoviruses. Using the default BLASTP threshold of 75, core proteins were identified in the five P. larvae myoviruses with respect to Clostridium phage phiCD119, Streptococcus phage EJ-1, Lactobacillus phage KC5a, and Lactobacillus phage AQ113. The number of core proteins shared between comparison and reference genomes are listed in Table 7. The percent of core proteins with respect to the reference genome are also reported. Clostridium phage phiCD119 was the only one of these phages that belonged to a genus (phiCD119likevirus); the other three are currently unclassified. Previous analyses of the Podoviridae and Myoviridae families grouped phages together when phages share 40% of core proteins with a reference phage genome [36, 37]. Based on this cutoff value, Abouo, Davies, Jimmer1 and Jimmer2 formed a new group of small genome myoviruses.
Only one pham includes all five myoviruses, and very few phams are shared between unrelated P. larvaephages
Phages Emery and phiIBB_Pl23 are significantly different from each other and from the four similar P. larvae myoviruses, as is evident from the genome maps in Figure 3. However, Tables 4 and 5 demonstrate that some proteins encoded by these phages grouped into similar phams.
Pham 34 is the only pham in the database that included proteins from all five of the new myovirus P. larvae phages. These gene products are Abouo gp34, Davies gp34, Emery gp29, Jimmer1 gp36, and Jimmer2 gp36 and encode a bhlA/bacteriocin protein (Figure 3). No other gene products in the Phamerator database were grouped into this pham. The conserved domain in these proteins was DUF2762, a putative holin-like protein. When comparing amino acid sequences, these five proteins shared > 87% identity and an E-value less than 5 × 10-43.
P. larvae siphovirus phiIBB_Pl23 contained only two proteins that shared a pham with any new myovirus P. larvae phages. The conserved domains in one gene product suggest it encodes a serine recombinase protein (Jimmer1 gp49, Jimmer2 gp49, phiIBB_Pl23 gp33) (Figure 3). The conserved domains in the other gene product suggest it encodes a phage antirepressor protein (Jimmer1 gp19, Jimmer2 gp19, phiIBB_Pl23 gp42) (Figure 3). Antirepressors from 13 other phages are also assigned to this pham (pham 951 in this database), including an antirepressor from Paenibacillus glucanolyticus phage PG1 gp28. A phamily circle links the 16 phages in the database containing a gene product in pham 951 (Figure 5).
Phamily circle connects 16 phages containing a related antirepressor gene. The phamily circle of pham 951 connects all phages with genes encoding proteins in this pham. The phages containing gene products in pham 951 infect Bacillus, Enterococcus, Lactobacillus, Listeria, Clostridium, Paenibacillus, and Staphylococcus bacteria. The antirepressor gene is found in 16 different phages, including Staphylococcus phages (1) phiSLT (gp7), (2) PH15 (gp39), and (3) 55 (gp18); Paenibacillus phages (4) phiIBB_Pl23 (gp42), (5) Jimmer2 (gp19), (6) Jimmer1 (gp19), and (7) PG1 (gp28); Clostridium phage (8) phiC2 (gp52); Listeria phages (9) B054 (gp72), (10) A500 (gp36), and (11) A118 (gp46); Lactobacillus phages (12) phiAT3 (gp23), and (13) A2 (gp28); Enterococcus phage (14) phiEf11 (gp38); and Bacillus phages (15) SPBc2 (gp100) and (16) PM1 (gp26).
The siphovirus Paenibacillus glucanolyticus phage PG1 also contained three gene products that shared a pham with P. larvae phages Abouo, Davies, Jimmer1, and Jimmer2. Pham 90 encoded an RNA polymerase sigma-70 factor and had 5 members: PG1 gp62, Abouo gp91, Davies gp91, Jimmer1 gp99, Jimmer2 gp99. Pham 75 encoded a recombination protein U and had 9 members, including PG1 gp42, Abouo gp73, Davies gp76, Jimmer1 gp83, and Jimmer2 gp83. Pham 78 encoded a dUTPase and had 13 members, including PG1 gp43, Abouo gp76, Davies gp79, Jimmer1 gp86, and Jimmer2 gp86.
Four P. larvaephages contain duplicated genes
Phages Abouo and phiIBB_Pl23 did not contain any proteins that belonged to the same pham. However, gp52 and gp56 in phages Jimmer1 and Jimmer2 shared 52.1% identity (E-value is 1.39 × 10-36), belonged to pham 50, and encode an XRE family transcriptional regulator that contains a helix-turn-helix DNA binding domain. Additionally, gp62 and gp63 in phages Jimmer1 and Jimmer2 shared 40.3% amino acid identity (E-value is 8.32 × 10-11), belonged to pham 53, and contain an arc-like DNA-binding domain.
Davies gp44 and gp45 and Emery gp53 and gp54 belonged to pham 44 and encoded a putative membrane protein (Figure 6). Comparisons indicated that homologous proteins encoded on the two genomes were more similar than duplicated proteins encoded within one of the genomes. Davies gp44 and Emery gp53 shared 80.1% identity (E-value is 4.69 × 10-107), and Davies gp45 and Emery gp54 shared 82.9% identity (E-value is 3.20 × 10-117). However, Davies gp44 and gp45 shared 31.4% (E-value is 7.29 x 10-28) and Emery gp53 and gp54 shared 35.4% identity (E-value is 3.39 × 10-34). Abouo gp44 also belongs to pham 44 but the nucleotide sequence for this gene is different from the genes encoding the four gene products in Emery and Davies. Abouo gp44-46, Davies gp44-47, and Emery gp53-56 are identified in Figure 2A and 2B by the dark line indicating homology between Emery and Davies and the white gap between Abouo and Davies at the same location.
Davies and Emery share a duplicated gene while Abouo has only one copy. This genome map shows two gene products in both Davies and Emery that belong to pham 44 and encode a putative membrane protein. Davies gp44-47 and Emery gp53-56 are more similar to each other than they are to Abouo gp44-46. Gene product numbers are located inside the colored boxes. The numbers above each gene product indicate the pham number specific to this analysis and the (number of members in the pham). Gene products with the same color share a pham.
Discussion
Prior to this report, nine P. larvae phages were described but were never analyzed collectively or grouped based on similar characteristics. Structural and morphological characteristics are the only published information for grouping the reported P. larvae phages to date. Therefore, for general comparison, P. larvae phages were identified as myovirus, elongated-capsid siphovirus, round-capsid siphovirus, or unknown siphovirus. The five P. larvae myoviruses characterized in this paper are structurally similar to previously isolated P. larvae myoviruses and may also be genetically similar. Since few phages infecting P. larvae have been sequenced, it is useful to compare structural similarity observed in electron micrographs. Now that sequencing data has been published for six P. larvae phage genomes and sequencing of others is sure to follow, genomic grouping will prevail and clusters will likely emerge as occurred with the mycobacteriophages [13].
The five myoviruses were isolated from three soil samples each from a separate location: Jimmer1 and Jimmer2 were isolated independently from the same sample [22], Emery and Abouo were isolated together, and Davies was isolated separately. P. larvae phage PBL2 was isolated from a different sample than BL2, yet all tests indicated no obvious structural or genetic differences between these phages [5]. Similar host properties and selective pressures can result in isolation of similar phages from different locations [13]. More P. larvae siphoviruses need to be sequenced before further correlations between genome and structural morphology can be drawn. As demonstrated in this work and by others, the sequence of the tape measure protein gene may be used to identify individual phages being studied if co-isolation occurs again in the future [38].
Bacteriophages are often unable to superinfect an existing lysogen if the entering and lysogenic phages are genetically similar [39]. The portion of the genome responsible for superinfection immunity has been determined for some phages [40]. Repressor genes involved in superinfection immunity have been characterized and are known to defend the prophage from premature lysis by silencing genes related to lysis [41]. This system does not work against phages that are always lytic or temperate phages that are not sensitive to the prophage repressor genes. Lysogens of Jimmer1, Jimmer2, and Davies displayed superinfection immunity when incubated with the same phage. Jimmer1 and Jimmer2 exhibited nearly identical sequences and were also immune to superinfections of each other.
The host specificity and correlating genome similarity between Jimmer1 and Jimmer2 (infect PL2) and Davies and Abouo (infect PL6) reflect common evolutionary ancestry. The high degree of similarity (over 80% average nucleotide identity) between the four phages may indicate that these phages infect a common host that has not yet been isolated or tested or that two phages recently switched hosts as is common in phages [42]. Jimmer1, Jimmer2, Davies, and Abouo likely coevolved.
Many bacteriophages contain genes that affect the virulence of the bacterial hosts. One toxin gene has been identified in phiIBB_Pl23 (gp26), and no toxin genes have been identified in the five P. larvae myoviruses. P. larvae phages Abouo, Davies, Jimmer1, Jimmer2, and phiIBB_Pl23 encode an ABC transporter-like protein. This was characterized as an extracellular protein produced by P. larvae [43], but it is not known how this protein is involved in host virulence. Future experiments involving the many putative XRE transcriptional regulators encoded by these phages may show a correlation with the virulence of P. larvae. Most of the transcriptional regulators found in the six P. larvae phages do not share phams with phages that infect any other bacterial host, indicating that these regulators are both phage- and host-specific. Two of these transcriptional regulators were duplicated in Jimmer1 and Jimmer2. The differences between these genes indicate that they are ohnologous and arose by gene duplication and subsequent divergence [44]. The duplicated genes in Emery and Davies are putative membrane proteins and likely evolved in a similar fashion. Abouo contains only one copy of this gene (Figure 6). BLAST hits for all six of the sequenced P. larvae phages show similarity to many proteins encoded by Paenibacillus and Brevibacillus bacteria. BLAST hits to these bacteria are not surprising because the genera Paenibacillus and Brevibacillus both belong to the family Paenibacillaceae and are closely related [45].
Analysis of large terminase protein phamilies revealed that Abouo, Davies, Jimmer1, and Jimmer2 likely use the headful packaging mechanism, while phiIBB_Pl23 likely has 3′ cohesive ends. Because of the stringent cutoff values required for inclusion in a pham, these results identify one way experimentally determined properties of a protein can be inferred on others sharing the same phamily.
Several gene products in P. larvae phages have similar functions but do not share phamilies. These include head morphogenesis, tape measure, baseplate, LysM peptidoglycan binding, peptidoglycan hydrolase, PBSX, and bacteriocin proteins. The conserved genes either diverged a long time ago or were acquired via convergent evolution. Additionally, the antirepressor protein in P. larvae phages phiIBB_Pl23, Jimmer1, Jimmer2 shares a pham with antirepressors from 13 other myoviruses and siphoviruses that infect host bacteria in the genera Bacillus, Enterococcus, Geobacillus, Lactobacillus, Listeria, Paenibacillus, and Staphylococcus (Figure 5). The presence of a similar antirepressor among phages of diverse Firmicute hosts may indicate the usefulness of the gene products and their associated conserved domains to regulate production of phage proteins within a diverse set of host bacteria. These data indicate that P. larvae phages have been subjected to multiple evolutionary pressures.
The head morphogenesis protein in phage Emery belongs to the SPP1 gp7 family and contains a ribosomal slippage site that is not found in Bacillus phage SPP1. Although two gene products are produced by the head morphogenesis gene in SPP1 that are 34 kDa and 28 kDa (compared to predicted proteins of 82.5 kDa and 58.9 kDa in Emery), the two SPP1 proteins are thought to be due to an alternative start site, not a frameshift caused by ribosomal slippage [46]. The lack of homology between protein sequences indicates these proteins further illustrates that Emery is not closely related to any other known bacteriophages.
Most of the putative encoded proteins in the P. larvae phages are not grouped into phams containing proteins from other phage types. These data indicate that most P. larvae phage genes are novel among currently identified genes of phages or bacteria. More than half of the P. larvae phage proteins have no identified conserved domains or putative functions, illustrating the diversity of bacteriophages and the vast number of unknown genes yet to be explored.
CoreGenes was previously used to verify current taxonomic relationships between phages in the Podoviridae [36] and Myoviridae families [37]. It was also used analyze other "dwarf" myoviruses and group them based on the similarity of core genes [47]. Analysis of core genes and shared phams indicates that P. larvae phages Abouo, Davies, Jimmer1, and Jimmer2 are distantly related to phages in the phiCD119likevirus family as well as phages EJ-1, KC5a, and AQ113. Because proteins grouped into similar phams are phylogenetically related, these proteins likely share a common ancestry. The structural similarities between phages Abouo, Davies, Jimmer1, Jimmer2, phiC2, KC5a, AQ113, and EJ-1 may correlate with their genetic similarities because the conserved core genes include the structural module of each genome. However, the current accepted threshold of 40% for a sufficiently strong CoreGenes percentage prevents any of these phages from being grouped taxonomically (except perhaps KC5a and AQ113, which is not within the scope of this paper). The differences in genome lengths may also prevent the formation of a taxonomic family of these phages as CoreGenes reflects the percentage based on the number of genes, which means that genome length differences and subsequent differences in total gene numbers within a genome can influence the score.
The results of the CoreGenes analysis indicate that P. larvae phages Abouo, Davies, Jimmer1, and Jimmer2 are related phylogenetically. They are also distantly related to phiC2, KC5a, AQ113, and EJ-1 which infect other bacterial hosts. This relationship indicates that these four phages are the closest known phylogenetic relatives to these four P. larvae phages. The conservation of primarily structural genes among the eight small genome myoviruses may indicate that the phages adapted to maintain infectivity as their bacterial hosts diverged, but retained ancestral structural genes that were under less selective pressure.
Conserved genes between different phages may indicate important genes. The bacterial hosts PL2 and PL6 are similar (according to the 16S rRNA sequences and physical properties), and similar BhlA/bacteriocin genes such as found in the shared pham of Jimmer1, Jimmer2, Emery, Abouo, and Davies (pham 34) can likely be used to lyse the bacterial host. It is interesting to note that the only two genes shared between phiIBB_Pl23 and any other P. larvae phage encode a serine recombinase and an antirepressor, shared with the PL2 phages Jimmer1 and Jimmer2. This correlation may indicate similar host interactions, as these genes help regulate the lytic and lysogenic cycles. The PL6 phages do not contain any antirepressor gene products belonging to this pham. Although P. larvae phages Emery and phiIBB_Pl23 do not show significant genetic relatedness to any other sequenced phages, similar genes and phages will likely be discovered in the future. The six newly sequenced genomes of the P. larvae phages compared in this report are an initial foundation for future studies.
Conclusions
This first comparison of P. larvae phage genomes provides insight into the genus Paenibacillus and the important honeybee bacterial pathogen, P. larvae. Although six P. larvae phages show some relatedness to phages that infect other Firmicute bacteria, most P. larvae phage genes do not share phams with non-P. larvae phages and many gene products still have unknown functions. Efforts to characterize these gene products and to isolate, sequence, and analyze new P. larvae phages will help us better understand the genetics of these phages and their bacterial host.
Methods
Identification of field isolates
Paenibacillus larvae spores were extracted from local honey samples using the process described by Hornitzky [48]. Pelleted spores were streaked on PLA [49] plates that contained nalidixic and pipemidic acid and plates were incubated for 48–72 hours at 37°C. Colonies were streaked to purity on PLA plates. Isolates were tested with hydrogen peroxide for the presence of the catalase enzyme [50] and were tested by gram stain [45, 51].
A single colony from each bacterial field isolate was boiled at 98°C for five minutes, and 3 μL of the lysate was used as a PCR template. The 16S rRNA gene region was amplified using universal primers 27 F and 907R [52], and the standard protocol for Taq DNA polymerase (New England Biolabs). Following PCR, amplicon size was checked by agarose gel electrophoresis. Samples producing a ~1 kb band were submitted for BigDye (Applied Biosystems, Life Technologies) sequencing to the BYU DNA Sequencing Center. Resulting 16S sequences were analyzed using BLAST [19].
Superinfection of P. larvaelysogens with phage
Phages described by Sheflo et al. [22] were used in lysogenic superinfection studies using a protocol adapted from [53]. Some agar was removed from the center of an isolated plaque, streaked out on an LB plate, and incubated at 37°C for 24 hours to allow any lysogens to grow. One colony was removed, incubated at 37°C in 1 ml of LB broth for two hours, and then plated using the method described above. When the top agar was solid, 5 μL of each phage lysate was placed on the plate. The plate was incubated agar side down at 37°C for 24 hours. Clearing under the spots indicated superinfection had occurred, while no clearing indicated that the lysogenic bacteria were immune to superinfection.
Electron microscopy of P. larvaephages
Electron microscopy was performed at Brigham Young University in the Life Sciences Microscopy Lab using an FEI Tecnai 12 Spirit transmission electron microscope. To prepare the samples for imaging, 20 μl of high-titer phage lysate was placed on a 200-mesh copper carbon type-B electron microscope grid for one minute. The lysate was wicked away and the grids were stained for two minutes using 2% phosphotungstic acid (pH = 7). Residual liquid was wicked away and the grid was allowed to dry before being imaged. Phage structures in electron micrographs were measured using ImageJ [54]. The average and standard deviation for each measurement was calculated from a minimum of three separate measurements.
Genomic comparison of sequenced phages
The DNA sequences for the six sequenced P. larvae phages were downloaded from GenBank using reported accession numbers [21, 22]. Dotplots of nucleic acid and protein sequences were generated using Gepard [55] and then compared. ClustalW [56] was used to calculate Average Nucleotide Identity (ANI) percentages comparing each of the P. larvae phage genomes. The online tool FrameD [25] was used to search for frameshift mutations. Core genes were identified using the program CoreGenes 3.5 [57, 58] with the default BLASTP threshold of 75.
Phages genes were analyzed using Phamerator [17], an open-source program (GNU general public license) designed to compare phage genes and genomes. For this study, Phamerator was adapted and stored in a GitHub repository (http://github.com/byuphamerator/phamerator-dev) separate from the original version. Phamerator uses BLASTP [19] and ClustalO [20] to compare each protein encoded by the genes in the database. E-values and percent identity scores are used to sort proteins into groups referred to as phamilies (phams) based on user-defined cutoffs for each score. Conserved domains in each protein are then identified. The Phamerator database used in this study was populated with 71 Bacillus phages, one Clostridium phage, 3 Enterococcus phages, 2 Geobacillus phages, 7 Lactobacillus phages, 6 Listeria phages, 6 P. larvae phages, one Paenibacillus glucanolyticus phage, 15 Staphylococcus phages, and one Streptococcus phage. The non-Bacillus phages were included in the database because proteins from these phages appeared in low E-value (<0.0001) BLAST hits for P. larvae phage proteins. In this Phamerator database, genes with E-values smaller than 1 × 10-50 or greater than 32.5% identity with at least one other protein were grouped into phams. These parameters were identical to those used by Cresawn et al. [17]. Conserved domains in proteins were identified using RPS-BLAST [59] to search the Conserved Domain Database (CDD) released by NCBI on 21 March 2013 [18]. The 100 published phages in the Phamerator database are listed below with the respective bacterial hosts and accession numbers. The Bacillus_Draft database can be accessed through Phamerator (see http://phagesdb.org/Phamerator/faq/). The Phamerator database for this study (bphage5) is available at http://phagehunters.byu.edu/BeeProject.aspx.
Bacillus phages vB_BceM_Bc431v3 (B. cereus) [NC_020873], 0305phi8-36 (B. thuringiensis) [NC_009760], 250 (B. cereus) [GU229986], Andromeda (B. pumilus) [KC330684], AP50 (B. anthracis) [NC_011523], B103 (B. subtilis) [NC_004165], B4 (B. cereus) [JN790865], B5S (B. cereus) [JN797796], Bam35c (B. thuringiensis) [NC_005258], Basilisk (B. cereus) [KC595511], Bastille (B. cereus) [NC_018856], BCD7 (B. cereus) [NC_019515], BceA1 (B. cereus) [HE614282], BCJA1c (B. clarkii) [NC_006557], BCP78 (B. cereus) [NC_018860], BCU4 (B. cereus) [JN797798], BMBtp2 (B. thuringiensis) [JX887877], BPS13 (B. cereus) [NC_018857], BtCS33 (B. thuringiensis) [NC_018085], CAM003 (B. thuringiensis) [NC_024216.1], Cherry (B. anthracis) [NC_007457], Curly (B. pumilus) [KC330679], Eoghan (B. pumilus) [KC330680], Evoli, (B. thuringiensis) [NC_024207.1], Fah (B. anthracis) [NC_007814], Finn (B. pumilus) [KC330683], G (B. megaterium) [JN638751], GA-1 (Bacillus sp.) [NC_002649], Gir1 (Bacillus sp.) [Bacillus_Draft], Gamma 51 (B. cereus) [DQ222853], Gamma 53 (B. anthracis) [DQ222855], Gamma isolate d’Herelle (B. cereus) [DQ289556], Gemini (B. pumilus) [KC330681], GIL16c (B. thuringiensis) [NC_006945], Hakuna (B. thuringiensis) [NC_024213.1], Hoody T (B. thuringiensis) [NC_024205], IEBH (B. thuringiensis) [NC_011167], JL (B. cereus) [KC595512], JPB9 (B. thuringiensis) [Bacillus_Draft], Megatron (B thuringiensis) [NC_024211.1], MG-B1 (B. weihenstephanensis) [NC_021336], Nf (B. subtilis) [EU622808], Pappano (B. pumilus) [Bacillus_Draft], PBC1 (B. cereus) [NC_017976], Pegasus (Bacillus sp.) [bacillus.phagesdb.org], pGIL01 (B. thuringiensis) [AJ536073], phi105 (B. subtilis) [NC_004167], phi29 (B. Subtilis) [NC_011048], phiAGATE (B. pumilus) [JX238501], phiNIT1 (B. subtilis) [NC_021856], phIS3501 (B. thuringiensis) [NC_019502], Pleiades (B. pumilus) [Bacillus_Draft], PM1 (B. subtilis) [NC_020883], Polaris (B. pumilus) [Bacillus_Draft], PZA (B. subtilis) [M11813], Riley (B. thuringiensis) [KJ489402], Shanette (B. cereus) [KC595513], SP10 (B. subtilis) [NC_019487], SPbeta (SPBc2) (B. subtilis) [NC_001884], SPO1 (B. subtilis) [NC_011421], SPP1 (B. Subtilis) [NC_004166], Stitch (Bacillus sp.) [Bacillus_Draft], Taylor (B. pumilus) [KC330682], TP21-L (B. cereus) [NC_011645], Troll (B. thuringiensis) [KF208639], W.Ph. (B. cereus) [NC_016563], WBeta (B. cereus) [NC_007734], Wip1 (B. anthracis) [KF188458], phBC6A51 (B. cereus) [NC_004820], phBC6A52 (B. cereus) [NC_004821], Bacillus virus 1 (Bacillus. sp. 6 k512) [NC_009737]; Clostridium phage phiC2 (C. difficile) [NC_009231]; Enterococcus phages phiEf11 (E. faecalis) [NC_013696], phiEF24C (E. faecalis) [NC_009904], phiFL3A (E. faecalis) [NC_013648]; Geobacillus phages GBSV1 (Geobacillus sp. 6 k51) [NC_008376], E2 (Geobacillus) [NC_009552]; Lactobacillus phages A2 (L. casei) [NC_004112], KC5a (L. gasseri) [NC_007924], Lb338-1 (L. paracasei) [NC_012530], Lc-Nu (L. rhamnosus) [NC_007501], LP65 (L. plantarum) [NC_006565], phiAT3 (L. casei) [NC_005893], phig1e (Lactobacillus) [NC_004305]; Listeria phages P100 (L. monocytogenes) [DQ004855], A118 (L. monocytogenes) [NC_003216], A500 (L. monocytogenes) [NC_009810], A511 (L. monocytogenes) [NC_009811], B054 (L. monocytogenes) [NC_009813], P40 (L. monocytogenes) [NC_011308]; Paenibacillus phages Abouo (P. larvae) [KC595517], Davies (P. larvae) [KC595518], Emery (P. larvae) [KC595516], Jimmer1 (P. larvae) [KC595515], Jimmer2 (P. larvae) [KC595514], PG1(P. glucanolyticus) [HQ332138], phiIBB_Pl23 (P. larvae) [KF010834]; Staphylococcus phages phi 12 (S. aureus) [AF424782], 37 (S. aureus) [NC_007055], 3A (S. aureus) [NC_007053], 47 (S. aureus) [NC_007054], 55 (S. aureus) [NC_007060], 88 (S. aureus) [NC_007063], G1 (S. aureus) [NC_007066], K (S. aureus) [NC_005880], PH15 (S. epidermidis) [NC_008723], phiSLT (S. aureus) [NC_002661], SA11-v (S. aureus) [NC_019511], SpaA1 (S. pasteuri) [NC_018277], Twort (S. aureus) [NC_007021], X2 (S. aureus) [NC_007065], tp310-2 (S. aureus) [NC_009762]; Streptococcus prophage EJ-1 (S. pneumoniae) [NC_005294].The neighbor-joining phylogenetic tree (Figure 4) was generated using ClustalO. The phage large terminase proteins included in this tree are listed below with their respective accession numbers.
Bacillus phages 0305phi8-36 gp117 [YP_001429607.1], 250 gp5 [ADB28373.1], Andromeda gp9 [YP_007517474.1], B4 gp4 [YP_006908233.1], B5S gp4 [AEW47238.1], Basilisk gp3 [AGR46580.1], Bastille gp267 [AEQ34197.1], BCD7 gp104 [YP_007005955.1], BceA1 gp4 [CCE73839.1], BCJA1c gp34 [YP_164412.1], BCP78 gp12 [YP_006907847.1], BCU4 gp5 [AEW47511.1], BMBtp2 gp40 [YP_007236398.1], BPS13 gp6 [YP_006907565.1], BtCS33 gp2 [YP_006488672.1], Cherry gp2 [YP_338134.1], Curly gp9 [YP_007517553.1], Eoghan gp9 [YP_007517399.1], Fah gp2 [YP_512312.1], Finn gp9 [YP_007517630.1], G gp1 [YP_009015312.1], Gamma51 gp2 [ABA46445.1], Gamma isolate d’Herelle gp2 [ABC40454.1], Gemini gp9 [AGE60848.1], IEBH gp49 [YP_002154374.1], JL gp4 [AGR46722.1], PBC1 gp2 [YP_006383455.1], phBC6A52 gp2583 [NP_852588.1], phiAGATE gp196 [YP_007349220.1], phiNIT1 gp79 [YP_008318309.1], phIS3501 gp23 [YP_007004362.1], Shanette gp4 [AGR46934.1], SP10 gp119 [YP_007003376.1], SPO1 gp2.11 [YP_002300330.1], Taylor gp9 [AGE60927.1], Troll gp61 [YP_008430845.1], vB_BceM_Bc431v3 gp10 [YP_007676908.1], W.Ph. gp3 [YP_004957018.1], WBeta gp2 [YP_459966.1], and e virus 1 gp18 [YP_001425601.1]; Clostridium phage phiC2 gp2 [YP_001110720.1], Enterococcus phages phiEf11 gp2 [YP_003358792.1], phiEF24C gp5 [YP_001504114.1], and phiFL3A gp43 [YP_003347605.1]; Geobacillus phages GBSV1 gp17 [YP_764473.1] and E2 gp2 [YP_001285808.1]; Lactobacillus phages A2 gp2 [NP_680484.1], KC5a gp35 [YP_529870.1], Lb338-1 gp89 [YP_002790768.1], Lc-Nu gp2 [YP_358760.1], LP65 gp113 [YP_164748.1], phiAT3 gp2 [YP_025027.1], and phig1e gp37 [NP_695170.1]; Listeria phages A118 gp2 [NP_463463.1], A500 gp2 [YP_001468388.1], A511 gp5 [YP_001468454.1], B054 gp2 [YP_001468706.1], P100 gp5 [AAY53308.1], and P40 gp2 [YP_002261418.1]; Paenibacillus phages Abouo gp2 [AGR47449.1], Davies gp2 [YP_008858637.1], Emery gp3 [AGR47349.1], Jimmer1 gp2 [AGR47249.1], Jimmer2 gp2 [AGR47149.1], and phiIBB_Pl23 gp2 [YP_008320338.1]; Staphylococcus phages PH15 gp2 [YP_950664.1], phi 12 gp30 [NP_803336.1], phiSLT gp39 [BAB21732.1], SpaA1 gp4 [YP_006560692.1], and tp310-2 gp45 [ABS87507.1]; Streptococcus phage EJ-1 gp39 [NP_945278.1]; and Mycobacterium phage Nigel gp4 [YP_002003843.1].
Availability of supporting data
The data set supporting the results of this article is included within the article (and its Additional files). The same files are also available at http://phagehunters.byu.edu/BeeProject.aspx.
References
Drobnikova V, Ludvik J: Bacteriophage of Bacillus larvae. J Apic Res. 1982, 21: 53-56.
Popova A, Valerianov T, Toschkoff AS: Bacteriophage of Bacillus larvae. C R Acad Bulg Sci. 1976, 29: 249-252.
Benada O, Ludvik J, Drobnikova V: Morphology of a new bacteriophage isolated from Bacillus larvae. Folia Microbiol (Praha). 1984, 29: 520-521. 10.1007/BF02873162.
Dingman DW, Bakhiet N, Field CC, Stahly DP: Isolation of two bacteriophages from Bacillus larvae, PBL1 and PBL0.5, and partial characterization of PBL1. J Gen Virol. 1984, 65: 1101-1105. 10.1099/0022-1317-65-6-1101.
Bakhiet N, Stahly DP: Properties of clear plaque mutants of the Bacillus larvae bacteriophages PBL0.5 AND PBL2. J Invertebr Pathol. 1988, 52: 78-83. 10.1016/0022-2011(88)90105-X.
Campana CF, Bakhiet N, Stahly DP: Morphology of Bacillus larvae bacteriophage PBL3 and physical map of its DNA. J Invertebr Pathol. 1991, 57: 141-143. 10.1016/0022-2011(91)90055-U.
Stahly DP, Alippi AM, Bakhiet N, Campana CF, Novak CC, Cox R: PPL1c, a virulent mutant bacteriophage useful for identification of Paenibacillus larvae subspecies larvae. J Invertebr Pathol. 1999, 74: 295-296. 10.1006/jipa.1999.4893.
Gochnauer T: The isolation of a bacteriophage (bacterial virus) from Bacillus larvae. Bee World. 1955, 36: 101-103.
Gochnauer T: The use of bacteriophages in the analysis of the foulbrood diseases. Proc 10th Intern Congr Entomol Congr Montreal 1956. 1958, 4: 1091-1096.
Gochnauer TA: Some properties of a bacteriophage from Bacillus larvae. J Invertebr Pathol. 1970, 15: 149-156. 10.1016/0022-2011(70)90228-4.
Heyndrickx M, Vandemeulebroecke K, Hoste B, Janssen P, Kersters K, DeVos P, Logan NA, Ali N, Berkeley RCW: Reclassification of Paenibacillus (formerly Bacillus) pulvifaciens (Nakamura 1984) Ash et al. 1994, a later subjective synonym of Paenibacillus (formerly Bacillus) larvae (White 1906) Ash et al. 1994, as a subspecies of P. larvae, with emended descriptions of P. larvae as P. larvae subsp. larvae and P. larvae subsp. pulvifaciens. Int J Syst Bacteriol. 1996, 46: 270-279. 10.1099/00207713-46-1-270.
Genersch E, Forsgren E, Pentikainen J, Ashiralieva A, Rauch S, Kilwinski J, Fries I: Reclassification of Paenibacillus larvae subsp. pulvifaciens and Paenibacillus larvae subsp. larvae as Paenibacillus larvae without subspecies differentiation. Int J Syst Evol Microbiol. 2006, 56: 501-511. 10.1099/ijs.0.63928-0.
Hatfull GF, Jacobs-Sera D, Lawrence JG, Pope WH, Russell DA, Ko CC, Weber RJ, Patel MC, Germane KL, Edgar RH, Hatfull GF, Jacobs-Sera D, Lawrence JG, Pope WH, Russell DA, Ko CC, Weber RJ, Patel MC, Germane KL, Edgar RH, Hoyte NN, Bowman CA, Tantoco AT, Paladin EC, Myers MS, Smith AL, Grace MS, Pham TT, O'Brien MB, Vogelsberger AM, Hryckowian AJ, Wynalek JL, Donis-Keller H, Bogel MW, Peebles CL, Cresawn SG, Hendrix RW: Comparative genomic analysis of 60 Mycobacteriophage genomes: genome clustering, gene acquisition, and gene size. J Mol Biol. 2010, 397: 119-143. 10.1016/j.jmb.2010.01.011.
Casjens SR, Gilcrease EB: Determining DNA packaging strategy by analysis of the termini of the chromosomes in tailed-bacteriophage virions. Methods Mol Biol. 2009, 502: 91-111. 10.1007/978-1-60327-565-1_7.
Wagner PL, Waldor MK: Bacteriophage control of bacterial virulence. Infect Immun. 2002, 70: 3985-3993. 10.1128/IAI.70.8.3985-3993.2002.
Boyd EF: Bacteriophage-encoded bacterial virulence factors and phage-pathogenicity island interactions. Adv Virus Res. 2012, 82: 91-118.
Cresawn SG, Bogel M, Day N, Jacobs-Sera D, Hendrix RW, Hatfull GF: Phamerator: a bioinformatic tool for comparative bacteriophage genomics. BMC Bioinformatics. 2011, 12: 395-10.1186/1471-2105-12-395.
Marchler-Bauer A, Zheng CJ, Chitsaz F, Derbyshire MK, Geer LY, Geer RC, Gonzales NR, Gwadz M, Hurwitz DI, Lanczycki CJ, Lu F, Lu SN, Marchler GH, Song JS, Thanki N, Yamashita RA, Zhang DC, Bryant SH: CDD: conserved domains and protein three-dimensional structure. Nucleic Acids Res. 2013, 41: D348-D352. 10.1093/nar/gks1243.
Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ: Basic local alignment search tool. J Mol Biol. 1990, 215: 403-410. 10.1016/S0022-2836(05)80360-2.
Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li WZ, Lopez R, McWilliam H, Remmert M, Soding J, Thompson JD, Higgins DG: Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol. 2011, 7: 6-
Oliveira A, Melo LDR, Kropinski AM, Azeredo J: Complete genome sequence of the broad-host-range Paenibacillus larvae phage phiIBB_Pl23. Genome Announc. 2013, 1: e00438-00413.
Sheflo MA, Gardner AV, Merrill BD, Fisher JNB, Lunt BL, Breakwell DP, Grose JH, Burnett SH: Complete genome sequences of five Paenibacillus larvae bacteriophages. Genome Announc. 2013, 1: e00668-00613.
Gordon D, Abajian C, Green P: Consed: a graphical tool for sequence finishing. Genome Res. 1998, 8: 195-202. 10.1101/gr.8.3.195.
Belcaid M, Bergeron A, Poisson G: The evolution of the tape measure protein: units, duplications and losses. BMC Bioinformatics. 2011, 12 (Suppl 9): S10-10.1186/1471-2105-12-S9-S10.
Schiex T, Gouzy J, Moisan A, de Oliveira Y: FrameD: a flexible program for quality check and gene prediction in prokaryotic genomes and noisy matured eukaryotic sequences. Nucleic Acids Res. 2003, 31: 3738-3741. 10.1093/nar/gkg610.
Dorscht J, Klumpp J, Bielmann R, Schmelcher M, Born Y, Zimmer M, Calendar R, Loessner MJ: Comparative genome analysis of Listeria bacteriophages reveals extensive mosaicism, programmed translational frameshifting, and a novel prophage insertion site. J Bacteriol. 2009, 191: 7206-7215. 10.1128/JB.01041-09.
Alonso JC, Lüder G, Stiege AC, Chai S, Weise F, Trautner TA: The complete nucleotide sequence and functional organization of Bacillus subtilis bacteriophage SPP1. Gene. 1997, 204: 201-212. 10.1016/S0378-1119(97)00547-7.
Narita S, Kaneko J, Chiba J-i, Piémont Y, Jarraud S, Etienne J, Kamio Y: Phage conversion of Panton-Valentine leukocidin in Staphylococcus aureus: molecular analysis of a PVL-converting phage, φSLT. Gene. 2001, 268: 195-206. 10.1016/S0378-1119(01)00390-0.
Stewart CR, Casjens SR, Cresawn SG, Houtz JM, Smith AL, Ford ME, Peebles CL, Hatfull GF, Hendrix RW, Huang WM, Pedulla ML: The genome of Bacillus subtilis bacteriophage SPO1. J Mol Biol. 2009, 388: 48-70. 10.1016/j.jmb.2009.03.009.
Goh S, Riley TV, Chang BJ: Isolation and characterization of temperate bacteriophages of Clostridium difficile. Appl Environ Microbiol. 2005, 71: 1079-1083. 10.1128/AEM.71.2.1079-1083.2005.
Mayer MJ, Narbad A, Gasson MJ: Molecular characterization of a Clostridium difficile bacteriophage and its cloned biologically active endolysin. J Bacteriol. 2008, 190: 6734-6740. 10.1128/JB.00686-08.
Govind R, Fralick JA, Rolfe RD: Genomic organization and molecular characterization of Clostridium difficile bacteriophage ΦCD119. J Bacteriol. 2006, 188: 2568-2577. 10.1128/JB.188.7.2568-2577.2006.
Kilic AO, Pavlova SI, Alpay S, Kilic SS, Tao L: Comparative study of vaginal Lactobacillus phages isolated from women in the United States and Turkey: prevalence, morphology, host range, and DNA homology. Clin Diagn Lab Immunol. 2001, 8: 31-39.
Zago M, Scaltriti E, Rossetti L, Guffanti A, Armiento A, Fornasari ME, Grolli S, Carminati D, Brini E, Pavan P, Felsani A, D'Urzo A, Moles A, Claude J-B, Grandori R, Ramoni R, Giraffa G: Characterization of the genome of the dairy Lactobacillus helveticus bacteriophage ΦAQ113. Appl Environ Microbiol. 2013, 79: 4712-4718. 10.1128/AEM.00620-13.
Diaz E, Lopez R, Garcia JL: EJ-1, a temperate bacteriophage of Streptococcus pneumoniae with a Myoviridae morphotype. J Bacteriol. 1992, 174: 5516-5525.
Lavigne R, Seto D, Mahadevan P, Ackermann HW, Kropinski AM: Unifying classical and molecular taxonomic classification: analysis of the Podoviridae using BLASTP-based tools. Res Microbiol. 2008, 159: 406-414. 10.1016/j.resmic.2008.03.005.
Lavigne R, Darius P, Summer E, Seto D, Mahadevan P, Nilsson A, Ackermann H, Kropinski A: Classification of Myoviridae bacteriophages using protein sequence similarity. BMC Microbiol. 2009, 9: 224-10.1186/1471-2180-9-224.
Smith KC, Castro-Nallar E, Fisher JNB, Breakwell DP, Grose JH, Burnett SH: Phage cluster relationships identified through single gene analysis. BMC Genomics. 2013, 14: 15-10.1186/1471-2164-14-15.
Alatossava T, Forsman P, Ritzenthaler P: Genome homology and superinfection immunity between temperate and virulent Lactobacillus delbrueckii bacteriophages. Arch Virol. 1995, 140: 2261-2268. 10.1007/BF01323245.
Dhaese P, Dobbelaere MR, Van Montagu M: The temperate B. subtilis phage phi 105 genome contains at least two distinct regions encoding superinfection immunity. Mol Gen Genet. 1985, 200: 490-492. 10.1007/BF00425736.
Emond E, Moineau S: Bacteriophages and food fermentations. Bacteriophage: Genetics and Molecular Biology. Edited by: McGrath S, vanSinderen D, Wymondham UK. 2007, Caister Academic Press, 93-123.
Pickard D, Toribio AL, Petty NK, van Tonder A, Yu L, Goulding D, Barrell B, Rance R, Harris D, Wetter M, Wain J, Choudhary J, Thomson N, Dougan G: A conserved acetyl esterase domain targets diverse bacteriophages to the Vi capsular receptor of Salmonella enterica serovar typhi. J Bacteriol. 2010, 192: 5746-5754. 10.1128/JB.00659-10.
Antunez K, Anido M, Evans JD, Zunino P: Secreted and immunogenic proteins produced by the honeybee bacterial pathogen, Paenibacillus larvae. Vet Microbiol. 2010, 141: 385-389. 10.1016/j.vetmic.2009.09.006.
Bergthorsson U, Andersson DI, Roth JR: Ohno’s dilemma: evolution of new genes under continuous selection. Proc Natl Acad Sci U S A. 2007, 104: 17004-17009. 10.1073/pnas.0707158104.
Ludwig W, Schleifer KH, Whitman WB: Revised road map to the phylum Firmicutes. Bergey’s Manual of Systematic Bacteriology, Volume 3. Edited by: De Vos P, Garrity G, Jones D, Krieg NR, Ludwig W, Rainey FA, Schleifer KH, Whitman WB. 2009, New York: Springer-Verlag, 1-13. 2
Becker B, delaFuente N, Gassel M, Gunther D, Tavares P, Lurz R, Trautner TA, Alonso JC: Head morphogenesis genes of the Bacillus subtilis bacteriophage SPP1. J Mol Biol. 1997, 268: 822-839. 10.1006/jmbi.1997.0997.
Comeau AM, Tremblay D, Moineau S, Rattei T, Kushkina AI, Tovkach FI, Krisch HM, Ackermann H-W: Phage morphology recapitulates phylogeny: the comparative genomics of a new group of myoviruses. PLoS One. 2012, 7: e40102-10.1371/journal.pone.0040102.
Hornitzky MAZ, Clark S: Culture of Bacillus larvae from bulk honey samples for the detection of American foulbrood. J Apic Res. 1991, 30: 13-16.
Schuch DMT, Madden RH, Sattler A: An improved method for the detection and presumptive identification of Paenibacillus larvae subsp larvae spores in honey. J Apic Res. 2001, 40: 59-64.
Bakonyi T, Derakhshifar I, Grabensteiner E, Nowotny N: Development and evaluation of PCR assays for the detection of Paenibacillus larvae in honey samples: comparison with isolation and biochemical characterization. Appl Environ Microbiol. 2003, 69: 1504-1510. 10.1128/AEM.69.3.1504-1510.2003.
Halebian S, Harris B, Finegold SM, Rolfe RD: Rapid method that aids in distinguishing Gram-positive from Gram-negative anaerobic bacteria. J Clin Microbiol. 1981, 13: 444-448.
Lane DJ, Stackebrandt E, Goodfellow MC: 16S/23S rRNA sequencing. Nucleic acid techniques in bacterial systematics. Edited by: Stackebrandt E, Goodfellow MC. 1991, England: John Wiley & Sons, Ltd, 115-175.
Sarkis GJ, Hatfull GF: Mycobacteriophages. Methods Mol Biol. 1998, 101: 145-173.
Abràmoff MD, Magalhães PJ, Ram SJ: Image processing with ImageJ. Biophotonics International. 2004, 11: 36-42.
Krumsiek J, Arnold R, Rattei T: Gepard: a rapid and sensitive tool for creating dotplots on genome scale. Bioinformatics. 2007, 23: 1026-1028. 10.1093/bioinformatics/btm039.
Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG: Clustal W and Clustal X version 2.0. Bioinformatics. 2007, 23: 2947-2948. 10.1093/bioinformatics/btm404.
Mahadevan P, King JF, Seto D: Data mining pathogen genomes using GeneOrder and CoreGenes and CGUG: gene order, synteny and in silico proteomes. Int J Comput Biol Drug Des. 2009, 2: 100-114. 10.1504/IJCBDD.2009.027586.
Turner D, Reynolds D, Seto D, Mahadevan P: CoreGenes3.5: a webserver for the determination of core genes from sets of viral and small bacterial genomes. BMC Res Notes. 2013, 6: 1-4. 10.1186/1756-0500-6-1.
Marchler-Bauer A, Bryant SH: CD-Search: protein domain annotations on the fly. Nucleic Acids Res. 2004, 32: W327-W331. 10.1093/nar/gkh454.
Acknowledgements
The authors thank Dr. Steven Cresawn for his assistance in setting up the Phamerator database and the support of Byron Doyle at Brigham Young University for adaptation of the computer code to run Phamerator on our local computers. We appreciate the knowledgeable advice of Dr. Graham Hatfull in preparing this manuscript. We appreciate the assistance of Michael Standing from the BYU Microscopy Lab and Dr. Edward Wilcox from the BYU DNA Sequencing Center. We are grateful for the help of student researchers Parker A. Ayer, Alexander P. Beckstead, Christopher P. Fajardo, Nicholas C. Ferguson, Joshua N. B. Fisher, Adam V. Gardner, Kiel A. Graves, Kaitlin A. Hartmann, Johnathan E. Liu, Bryce L. Lunt, Ashley K. Kennedy, Curtis A. Merrill, Elizabeth Newey, Robert C. Russell, Jonathan E. Schoenhals, Michael A. Sheflo, Braden N. Wake, Kurt R. Williams, and Lauren J. Zimmerman.
This project was funded by the Department of Microbiology and Molecular Biology at Brigham Young University (BYU), a student grant from the BYU Office of Research and Creative Activities, and a BYU Technology Transfer grant.
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
BDM, JHG, DPB and SHB were responsible for the design and coordination of the research. BDM conducted the laboratory research and genomic comparison work. BDM drafted the manuscript and figures, SHB edited extensively. All authors contributed to editing of the manuscript.
Electronic supplementary material
12864_2014_6449_MOESM2_ESM.xlsx
Additional file 2: Spreadsheet exported from the Phamerator database to report all phage gene products in the database, the phams to which the gene products are assigned, and the conserved domains found in gene products in those phams.(XLSX 1 MB)
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
This article is published under an open access license. Please check the 'Copyright Information' section either on this page or in the PDF for details of this license and what re-use is permitted. If your intended use exceeds what is permitted by the license or if you are unable to locate the licence and re-use information, please contact the Rights and Permissions team.
About this article
Cite this article
Merrill, B.D., Grose, J.H., Breakwell, D.P. et al. Characterization of Paenibacillus larvae bacteriophages and their genomic relationships to firmicute bacteriophages. BMC Genomics 15, 745 (2014). https://doi.org/10.1186/1471-2164-15-745
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/1471-2164-15-745