Abstract
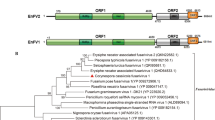
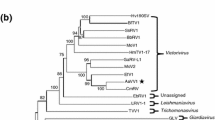
The complete genome sequence of a double-stranded RNA (dsRNA) mycovirus that was isolated from Alternaria solani strain DT-10 causing potato foliar disease was determined. The virus, designated as "Alternaria solani chrysovirus 1" (AsCV1), has four dsRNA segments (dsRNA 1-4) with a length of 3600 bp, 3128 bp, 2996 bp, and 2714 bp, respectively. The RNA-dependent RNA polymerase (RdRp, 1084 amino acids [aa]), putative capsid protein (905 aa), alphachryso-P3 (835 aa), and alphachryso-P4 (729 aa) were encoded by dsRNA1, dsRNA2, dsRNA3, and dsRNA4, respectively, which had the highest sequence identity of 41.77%-72.38% to their counterparts in Helminthosporium victoriae virus 145S (HvV145S) of the genus Alphachrysovirus, family Chrysoviridae. Moreover, the 5′-untranslated regions (UTRs) of AsCV1 dsRNA 1-4, which contained several unique inserts (3-37 bp) and deletions (5-64 bp), shared 51.65%-68.01% identity with those of HvV145S. Phylogenetic analysis based on RdRp sequences showed that AsCV1 clustered the most closely with HvV145S. Considering its distinct host specificity, the low sequence similarity of its encoded proteins to those of other viruses, the unusual features of the 5′-UTRs of its dsRNA 1-4, and the phylogenetic position of its RdRp gene, AsCV1 should be considered a member of a new species in the genus Alphachrysovirus. To the best of our knowledge, this is the first alphachrysovirus identified from phytopathogenic A. solani.


Similar content being viewed by others
References
Xie JT, Jiang DH (2014) New insights into mycoviruses and exploration for the biological control of crop fungal diseases. Annu Rev Phytopathol 52:45–68
Ghabrial SA, Castón JR, Jiang DH, Nibert ML, Suzuki N (2015) 50-plus years of fungal viruses. Virology 479:356–368
Ma GP, Liang ZJ, Hua HH, Zhou T, Wu XH (2019) Complete genome sequence of a new botybirnavirus isolated from a phytopathogenic Alternaria alternata in China. Arch Virol 164:1225–1228
Mahillon M, Decroës A, Liénard C, Bragard C, Legrève A (2019) Full genome sequence of a new polymycovirus infecting Fusarium redolens. Arch Virol 164:2215–2219
Kotta-Loizou I, Coutts RH, Hillman BI, Jiang D, Kim DH, Moriyama H, Suzuki N, ICTV Report Consortium (2020) ICTV Virus Taxonomy Profile: Chrysoviridae. J Gen Virol 101:143–144
Hayashi N, Tsuge T, Kobayashi H, Nishimura S (1988) The presence of double-stranded RNAs in Alternaria alternata Japanese pear pathotype and their participation in AK-toxin productivity. Ann Phytopathol Soc JPN 54:250–252
Aoki N, Moriyama H, Kodama M, Arie T, Teraoka T, Fukuhara T (2009) A novel mycovirus associated with four double-stranded RNAs affects host fungal growth in Alternaria alternata. Virus Res 140:179–187
Okada R, Ichinose S, Takeshita K, Urayama SI, Fukuhara T, Komatsu K, Arie T, Ishihara A, Egusa M, Kodama M, Moriyama H (2018) Molecular characterization of a novel mycovirus in Alternaria alternata manifesting two-sided effects: Down-regulation of host growth and up-regulation of host plant pathogenicity. Virology 519:23–32
Xavier ADS, Barros APO, Godinho MT, Zerbini FM, Souza FO, Bruckner FP, Alfenas-Zerbini P (2018) A novel mycovirus associated to Alternaria alternata comprises a distinct lineage in Partitiviridae. Virus Res 244:21–26
Li H, Bian RL, Liu Q, Yang L, Pang TX, Salaipeth L, Andika IB, Kondo H, Sun LY (2019) Identification of a novel hypovirulence-inducing hypovirus from Alternaria alternata. Front Microbiol 10:1076
Jamal A, Sato Y, Shahi S, Shamsi W, Kondo H, Suzuki N (2019) Novel victorivirus from a Pakistani isolate of Alternaria alternata lacking a typical translational stop/restart sequence signature. Viruses 11:577
Liang ZJ, Wang XY, Hua HH, Cao W, Zou T, Zhao C, Wu XH (2020) Full genome sequence of a new three-segment gammapartitivirus from the phytopathogenic fungus Alternaria tenuissima on cotton in China. Arch Virol 166:973–976
Shang HH, Zhong J, Zhang RJ, Chen CY, Gao BD, Zhu HJ (2015) Genome sequence of a novel endornavirus from the phytopathogenic fungus Alternaria brassicicola. Arch Virol 160:1827–1830
Lin YH, Zhang HL, Zhao CJ, Liu SX, Guo LH (2015) The complete genome sequence of a novel mycovirus from Alternaria longipes strain HN28. Arch Virol 160:577–580
Komatsu K, Katayama Y, Omatsu T, Mizutani T, Fukuhara T, Kodama M, Arie T, Teraoka T, Moriyama H (2016) Genome sequence of a novel victorivirus identified in the phytopathogenic fungus Alternaria arborescens. Arch Virol 161:1701–1704
Hu Z, Guo J, Gao BD, Zhong J (2020) A novel mycovirus isolated from the plant-pathogenic fungus Alternaria dianthicola. Arch Virol 165:2105–2109
Gong WJ, Liu H, Zhu XF, Zhao SY, Cheng JL, Zhu HJ, Zhong J, Zhou Q (2021) Molecular characterization of a novel fusarivirus infecting the plant-pathogenic fungus Alternaria solani. Arch Virol 166:2063–2067
Zheng HH, Zhao J, Wang TY, Wu XH (2015) Characterization of Alternaria species associated with potato foliar diseases in China. Plant Pathol 64:425–433
Zhao J, Ma GP, Liu YY, Wu XH (2018) Alternaria species infecting potato in southern China. Can J Plant Pathol 40:312–317
Morris TJ, Dodds JA (1979) Isolation and analysis of double-stranded RNA from virus-infected plant and fungal tissue. Phytopathology 69:854–858
Lyu RL, Zhang Y, Tang Q, Li YY, Cheng JS, Fu YP, Chen T, Jiang DH, Xie JT (2017) Two alphapartitiviruses co-infecting a single isolate of the plant pathogenic fungus Rhizoctonia solani. Arch Virol 163:515–520
Liu LJ, Wang QH, Cheng JS, Fu YP, Jiang DH, Xie JT (2015) Molecular characterization of a bipartite double-stranded RNA virus and its satellite-like RNA co-infecting the phytopathogenic fungus Sclerotinia sclerotiorum. Front Microbiol 6:406
Li SW, Li YT, Hu CH, Han CG, Zou T, Zao C, Wu XH (2020) Full genome sequence of a new mitovirus from the phytopathogenic fungus Rhizoctonia solani. Arch Virol 165:1719–1723
Gong MY, Wang HR, Jia DS, Yan F, Zhang SB (2019) Increased pathogenicity of the pathogen of corn southern leaf blight caused by a novel chrysovirus. Proc Annu Meet Chin Soc Plant Pathol (2019), Beijing, 242
Jiang DH, Ghabrial SA (2004) Molecular characterization of Penicillium chrysogenum virus: reconsideration of the taxonomy of the genus Chrysovirus. J Gen Virol 85:2111–2121
Covelli L, Coutts RHA, Serio FD, Citir A, Açıkgöz S, Hernández C, Ragozzino A, Flores R (2004) Cherry chlorotic rusty spot and Amasya cherry diseases are associated with a complex pattern of mycoviral-like double-stranded RNAs. I. Characterization of a new species in the genus Chrysovirus. J Gen Virol 85:3389–3397
Liu HQ, Fu YP, Xie JT, Cheng JS, Ghabrial SA, Li GQ, Peng YL, Yi XH, Jiang DH (2012) Evolutionary genomics of mycovirus-related dsRNA viruses reveals cross-family horizontal gene transfer and evolution of diverse viral lineages. BMC Evol Biol 12:91
Funding
This work was financially supported by the National Key Research and Development Program of China (2018YFD0200803).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
This study did not include experiments with human participants or animals performed by any of the authors.
Additional information
Handling Editor: Ioly Kotta-Loizou.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
705_2021_5263_MOESM1_ESM.tif
Supplementary file1 (TIF 12467 KB) Fig. S1 (A) Multiple amino acid sequence alignment of the RNA-dependent RNA polymerase (RdRp) domains of AsCV1 and 10 other members of the family Chrysoviridae, using the ClustalX 2.0 program. The eight conserved motifs in RdRp are numbered as I, II, III, IV, V, VI, VII, and VIII, as shown above the amino acid sequence. Numbers in parentheses represent the number of amino acid residues between the motifs. Black stars indicate identical amino acid residues, colons indicate amino acids with high chemical similarity, and dots indicate amino acid residues with low chemical similarity. The red stars indicate AsCV1. Virus names and GenBank accession numbers are as follows: VdCV1, Verticillium dahliae chrysovirus 1 (HM004067); CnCV1, Cryphonectria nitschkei chrysovirus 1 (GQ290649); IjCV1, Isaria javanica chrysovirus 1 (KX898416); AsCV1, Alternaria solani chrysovirus 1 (MW656210); HvV145S-A9, Helminthosporium victoriae virus 145S (AF297176); ACDACV, Amasya cherry disease associated chrysovirus (AJ781166); CgCV1, Colletotrichum gloeosporioides chrysovirus 1 (KT581957); FoCV1, Fusarium oxysporum chrysovirus 1 (EF152346); AaCV1, Alternaria alternata chrysovirus 1 (LC350277); PjCV1, Penicillium janczewskii chrysovirus 1 (KT601115); FgV2, Fusarium graminearum dsRNA mycovirus 2 (HQ343295). (B) Multiple sequence alignment of the 5′-untranslated regions (UTRs) and 3′-UTRs of the four dsRNA segments of AsCV1. Dark blue, pink, and green represent 100%, 75%, and 50% nucleotide sequence identity, respectively
Rights and permissions
About this article
Cite this article
Hu, C., Li, S., Wu, C. et al. Complete genome sequence of the first chrysovirus from the phytopathogenic fungus Alternaria solani on potato in China. Arch Virol 166, 3493–3497 (2021). https://doi.org/10.1007/s00705-021-05263-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00705-021-05263-z