Abstract
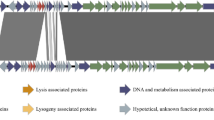
Aeromonas hydrophila is an important finfish pathogen, besides being an opportunistic human pathogen. In the present study, the genomes of three A. hydrophila-specific phages, CF8, PS1, and PS2, were isolated, characterized and sequenced. Transmission electron microscopy showed that all three phages had typical Myoviridae morphology. The linear dsDNA genomes of CF8, PS1, and PS2 were 238,150 bp, 237,367 bp, and 240,447 bp in length, with a GC content of 42.2%, 38.8%, and 38.8%, respectively. The low sequence similarity (67.6% - 69.8% identity with 27.0% - 29.0% query coverage) to other phage genomes in the NCBI database indicated the novel nature of the CF8, PS1, and PS2 genomes. A total of 244, 247, and 250 open reading frames (ORFs) were predicted in the CF8, PS1, and PS2 genome, respectively. During the annotation process, functional predictions were made for 28-31 ORFs, while the rest were classified as “hypothetical proteins” with yet unknown functions. Genes for tRNAs were also detected in all phage genomes. As all three phages in the present study had a very narrow host range with lytic activity against only one strain of A. hydrophila, these phages could be good candidates for phage typing applications. Moreover, the endolysin- and lytic-transglycosylase-encoding genes could be used for recombinant cloning and expression of anti-microbial proteins.


Similar content being viewed by others
Change history
19 May 2020
Authors would like to correct the incorrect version.
References
Janda JM, Abbott SL (2010) The genus aeromonas: taxonomy, pathogenicity, and infection. Clin Microbiol Rev 23:35–73
Fenton M, Ross P, McAuliffe O, O’Mahony J, Coffey A (2010) Recombinant bacteriophage lysins as antibacterials. Bioeng Bugs 1:9–16
Rai S, Tyagi A, Naveen Kumar BT, Kaur S, Singh NK (2019) Isolation, genomic characterization and stability study of narrow-host range Aeromonas hydrophila lytic bacteriophage. J Exp Zool India 22:1075–1082
Fortier LC, Moineau S (2007) Morphological and genetic diversity of temperate phages in Clostridium difficile. Appl Environ Microbiol 73:7358–7366
Bolger AM, Lohse M, Usadel B (2014) Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30:2114–2120
Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, Pyshkin AV, Sirotkin AV, Vyahhi N, Tesler G, Alekseyev MA, Pevzner PA (2012) SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 19:455–477
Cornelissen A, Hardies SC, Shaburova OV, Krylov VN, Mattheus W, Kropinski AM, Lavigne R (2012) Complete genome sequence of the giant virus OBP and comparative genome analysis of the diverse PhiKZ-related phages. J Virol 86:1844–1852
Meier-Kolthoff JP, Goker M (2017) VICTOR: genome-based phylogeny and classification of prokaryotic viruses. Bioinformatics 33:3396–3404
Goker M, Garcia-Blazquez G, Voglmayr H, Telleria MT, Martin MP (2009) Molecular taxonomy of phytopathogenic fungi: a case study in Peronospora. PLoS One 4:e6319
Besemer J, Lomsadze A, Borodovsky M (2001) GeneMarkS: a self-training method for prediction of gene starts in microbial genomes. Implications for finding sequence motifs in regulatory regions. Nucleic Acids Res 29:2607–2618
Jones P, Binns D, Chang HY, Fraser M, Li W, McAnulla C, McWilliam H, Maslen J, Mitchell A, Nuka G, Pesseat S, Quinn AF, Sangrador-Vegas A, Scheremetjew M, Yong SY, Lopez R, Hunter S (2014) InterProScan 5: genome-scale protein function classification. Bioinformatics 30:1236–1240
Zimmermann L, Stephens A, Nam SZ, Rau D, Kubler J, Lozajic M, Gabler F, Soding J, Lupas AN, Alva V (2018) A completely reimplemented MPI bioinformatics toolkit with a new HHpred server at its core. J Mol Biol 430:2237–2243
Lowe TM, Chan PP (2016) tRNAscan-SE On-line: integrating search and context for analysis of transfer RNA genes. Nucleic Acids Res 44:W54–W57
Pastagia M, Schuch R, Fischetti VA, Huang DB (2013) Lysins: the arrival of pathogen-directed anti-infectives. J Med Microbiol 62:1506–1516
Guo M, Feng C, Ren J, Zhuang X, Zhang Y, Zhu Y, Dong K, He P, Guo X, Qin J (2017) A novel antimicrobial endolysin, LysPA26, against Pseudomonas aeruginosa. Front Microbiol 8:293
Larpin Y, Oechslin F, Moreillon P, Resch G, Entenza JM, Mancini S (2018) In-vitro characterization of PlyE146, a novel phage lysin that targets Gram-negative bacteria. PLoS One 13:e0192507
Paradis-Bleau C, Cloutier I, Lemieux L, Sanschagrin F, Laroche J, Auger M, Garnier A, Levesque RC (2007) Peptidoglycan lytic activity of the Pseudomonas aeruginosa phage phiKZ gp144 lytic transglycosylase. FEMS Microbiol Lett 266:201–209
Matamp N, Bhat SG (2019) Phage endolysins as potential antimicrobials against multidrug resistant Vibrio alginolyticus and Vibrio parahaemolyticus: current status of research and challenges ahead. Microorganisms 7:84
Acknowledgements
The authors are grateful to Dr. Ashoo Toor, Assistant Professor, Punjab Agricultural University, Ludhiana, and Dr. Prabjeet Singh, Assistant Professor, Guru Angad Dev Veterinary and Animal Sciences University, Ludhiana, for English and scientific language corrections.
Funding
This work was supported by RKVY Grant RKVY-11:I3 “Development of biotechnological intervention strategies to enhance the safety and shelf life of fishery products”.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
All authors declare that they have no conflict of interest.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Additional information
Handling Editor: Johannes Wittmann.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
The original version of this article was revised: Corrected version of Fig. 2 updated.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Rai, S., Tyagi, A., Kalia, A. et al. Characterization and genome sequencing of three Aeromonas hydrophila-specific phages, CF8, PS1, and PS2. Arch Virol 165, 1675–1678 (2020). https://doi.org/10.1007/s00705-020-04644-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00705-020-04644-0