Abstract
Cucumber mosaic virus (CMV) was consistently recovered from symptomatic snap bean plants during surveys conducted in 2007 and 2008 in central Wisconsin. A large proportion of these CMV-infected plants contained a single-stranded linear RNA molecule consisting of 339 nucleotides and sharing 90–94% sequence identity with other satellite (sat) RNAs of CMV. Comparison of this satRNA sequence with currently available CMV satRNA sequences suggests this to be a novel satRNA.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Wisconsin remains a domestic leader in the production of processing snap bean (Phaseolus vulgaris L.), averaging over 28,125 hectares from 2000 through 2006 (National Agricultural Statistics Service, 2006). Viral diseases are common in this crop, and a range of symptoms are observed annually. Cucumber mosaic virus (CMV) has been the most prevalent virus among the characterized viruses in snap bean [6, 13, 15].
CMV isolates have been classified into two main strains or subgroups, designated as subgroups I and II based on serology, peptide mapping of the coat protein (CP), nucleic acid hybridization, RT-PCR followed by RFLP and nucleotide sequence identity [16, 18]. The two subgroups show 75% nucleotide identity [18]. Subgroup I has been further divided into IA and IB on the basis of the nucleotide sequences of the 5’ non-translated region of the RNA 3 and the CP gene, with 92–95% nucleotide identity [18].
The genome of CMV contains three positive-sense single-stranded RNAs, packaged in separate particles [16]. Some isolates of CMV, in addition to viral genomic RNAs, contain a small linear, single-stranded RNA molecule known as satellite RNA (satRNA), ranging in size from 332 to 405 nucleotides. These small RNAs are dependent on CMV for replication, encapsidation and vector transmission. Interacting with the CMV helper virus and plant host, satRNAs can influence symptom expression, ranging from attenuation to increased severity [3, 7, 9–12, 19, 20]. These interactions often depend upon the particular CMV genotype and host plant. In this report, we demonstrate the association of a new type of satRNA with field isolates of CMV in snap bean.
A total of eighty symptomatic snap bean plants were collected from among ten different field locations in Wisconsin between August and October in 2007, 2008 and 2010. Specifically, eight samples per field were collected from four fields in 2007, three fields in 2008 and three fields in 2010. An additional subset of CMV isolates (six isolates) collected in New York snap bean fields (in 2009) was used in this study. To initially confirm infection of CMV in these field-collected samples, they were analyzed by triple antibody sandwich (TAS) ELISA using subgroup-specific CMV antisera (CMV-DTL for subgroup I and CMV-ToRS for subgroup II) according to the manufacturer’s specifications (Agdia Inc. Elkhart, IN).
Total RNAs were extracted from leaf samples by using an RNeasy Mini Kit (QIAGEN, CA, USA) according to manufacturer’s instructions. The CMV CP gene was targeted for amplification using a specific primer pair (CP F: 5′-TTGAGTCGAGTCATGGACAAATC-3′ and CP R: 5′-AACACGGAATCAGACTGGGAG-3′) [14] and superscript III (Invitrogen, Carlsbad, USA) as reverse transcriptase during the RT-PCR. Positive CMV samples from field-based ELISA assessment and subsequent RT-PCR CP sequence amplification were later examined for the co-occurrence of satRNA by RT-PCR with a specific primer pair (CMV sat F: 5’GGGAATTCATTTAGGTGACACTATAGTTTTGTTTG 3’ and CMV sat R: 5’GGGGTCTAGACCCGGGTCCTG 3’) [8]. The PCR program was initiated at 53°C for 45 min, followed by 30 cycles at 94°C for 30 s, 54°C for 30 s, 72°C for 1 min and finished with 72°C for 7 min. Products were separated on a 2% agarose gel to initially screen for the presence or absence of satRNA. Non-inoculated, healthy plant foliage from an aphid-free greenhouse and a Kentucky (Ky) isolate of CMV carrying satellite RNA [8] were used as negative and positive controls, respectively. Gel purification was performed using a QIAquick Gel Purification Kit (QIAGEN, CA, USA).
Purified PCR products were cloned into pGEMT Easy Vector (Promega, WI, USA), and these clones were used to transform competent cells of Escherichia coli strain DH5α. Three colonies from each isolate were selected from the plates, and amplicons were sequenced using an automated DNA sequencer (ABI Prism 310 Genetic Analyzer, Applied Biosystems) in the DNA sequencing facility at the Biotechnology Center of the University of Wisconsin-Madison. The satRNAs obtained were designated Wisconsin (IR-WI) and New York (IR-NY) satRNA. The sequences of the IR-WI and IR-NY satellite RNAs were compared to the other available CMV satRNA sequences in the GenBank database.
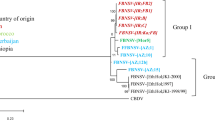
Initial TAS-ELISA results suggested that sixty samples (75%) from Wisconsin and six samples from New York were positive for CMV and belonged to CMV subgroup I (data not shown). Amplification of the CP gene using subgroup-I-specific primers confirmed the ELISA results for the 60 serologically positive field samples. When total nucleic acid extracts were used as a template for CMV satRNA RT-PCR, an approximately 380-bp fragment was obtained for 47 out of the 60 collected CMV isolates. Three out of six CMV isolates collected in New York were also positive for satRNA. Further, sequence analysis showed that IR-WI and IR-NY satRNA sequences are identical. Sequence comparisons with other published CMV satRNAs available in GenBank illustrated that IR-WI and IR-NY satRNAs sequences shared the highest nucleotide identity (ca. 94%) with a Spanish satRNA [1]. Both sequences were differentiated by 17 nucleotide substitutions, two deletions and one insertion (Fig. 1). IR-WI satRNA sequence shared 92% identity with CARNA5 satRNA (GenBank accession number J02061), an attenuated satRNA, originally isolated from cucumber (Cucumis sativus) in Wisconsin and published in 1983 [2]. IR-NY satRNA sequence shared 91% identity with CMV-WL satRNA (GenBank accession number M30590), previously reported in New York, reportedly resulting in white leaf disease on tomato [7]. Furthermore, IR-NY satRNA had 90% identity with B1- satRNA (GenBank accession number M16586) and B2- satRNA (GenBank accession number M16587), other satRNAs [4] from a CMV isolate from red kidney bean in New York [17]. B1 and B2- satRNAs induce chlorosis in tomato and attenuate disease, respectively [5]. These results suggest that the satRNAs found associated with CMV isolates in snap bean in Wisconsin and New York represent a novel type of CMV satRNA. Future studies to address the role of these satRNAs in CMV pathogenicity on snap bean and the variation among satRNAs can provide a better understanding of satRNA dynamic in snap bean fields.
Alignment of IR-WI, IR-NY and Spanish satellite RNA sequences. GenBank accession numbers: IR-WI, JF834526 [present work]; IR-NY, JN029953 [present work]; Spanish-satRNA, Z75870 [1]. Nucleotide variations are boxed
References
Aranda MA, Fraile A, Dopazo J, Malpica JM, Garcia-Arenal F (1997) Contribution of mutation and RNA recombination to the evolution of a plant pathogenic RNA. J Mol Evol 44:81–88
Collmer CW, Tousignant ME, Kaper JM (1983) Cucumber mosaic virus-associated RNA 5. X. The complete nucleotide sequence of a CARNA5 incapable of inducing tomato necrosis. Virology 127:230–234
Crescenzi A, Barbarossa L, Cillo F, Di Franco A, Vovlas N, Gallitelli D (1993) Role of cucumber mosaic virus and its satellite RNA in the etiology of tomato fruit necrosis in Italy. Arch Virol 131:321–333
Edwards MC, Gonsalves D, Provvidenti R (1983) Genetic analysis of cucumber mosaic virus in relation to host resistance: location of determinants for pathogenicity to certain legumes and Lactuca saligna. Phytopathology 73:269–273
Garcia-Arenal F, Zaitlin M, Palukaitis P (1987) Nucleotide sequence analysis of six satellite RNAs of cucumber mosaic virus: Primary sequence and secondary structure alteration Do Not correlate with differences in pathogenicity. Virology 158:339–347
German TL, Thompson A, Wills DK (2004) Statewide distribution of virus problems on processing beans. In: Proceedings for the 2004 Midwest Food Processors Association Processing crops Conference Feb 23–24, La Cross, WI Department of Horticulture, University of Wisconsin
Gonsalves D, Provvidenti R, Edwards MC (1982) Tomato white leaf: the relation of an apparent satellite RNA and cucumber mosaic virus. Phytopathology 72:1533–1538
Hajimorad MR, Ghabrial SA, Roossinck MJ (2009) De novo emergence of a novel satellite RNA of cucumber mosaic virus following serial passages of the virus derived from RNA transcripts. Arch Virol 154:137–140
Jordá C, Alfaro A, Aranda MA, Moriones E, García-Arenal F (1992) Epidemic of cucumber mosaic virus plus satellite RNA in tomatoes in eastern Spain. Plant Dis 76:363–366
Kaper JM, Waterworth HE (1977) Cucumber mosaic virus-associated RNA 5: causal agent for tomato necrosis. Science 196:429–431
Kaper JM, Tousignant ME, Thompson SM (1981) Cucumber mosaic virus associated RNA 5. VIII. Identification and partial characterization of a CARNA 5 incapable of inducing tomato necrosis. Virology 114:526–533
Kosaka Y, Hanada K, Fukunishi T, Tochihara H (1989) Cucumber mosaic virus tomato necrotic disease in Kyoto prefecture. Ann Phytopathol Soc Jpn 55:229–232
Larsen RC, Miklas PN, Eastwell KC, Grau CR, Mondjana A (2002) A virus disease complex devastating late season snap bean production in the Midwest. Annu Rep Bean Improv Coop 45:36–37
Lin HX, Rubio L, Smythe A, Falk BW (2004) Molecular population genetics of cucumber mosaic virus in California: evidence for founder effects and reassortments. J Virol 78:6666–6675
Nault BA (2003) Mid-season update on the snap bean / virus situation in western New York. Cornell Coop Exten Lake Plains Veg News 12:5–6
Palukaitis P, Roossinck MJ, Dietzgen RG, Francki RIB (1992) Cucumber mosaic virus. Adv Virus Res 41:281–348
Provvidenti R (1976) Reaction of Phaseolus and Macroptilium species to a strain of cucumber mosaic virus. Plant Dis Rep 60:289–293
Roossinck MJ (2002) Evolutionary history of cucumber mosaic virus, deduced by phylogenetic analyses. J Virol 76:3382–3387
Simon AE, Roossinck MJ, Havelda Z (2004) Plant virus satellite and defective interfering RNAs: new paradigms for a new century. Ann Rev Phytopathol 42:415–437
Takanami YA (1981) A striking change in symptoms on cucumber mosaic virus—infected tobacco plants induced by a satellite RNA. Virology 109:120–126
Acknowledgments
The authors wish to thank Dr. Keith Perry, Cornell University, and Dr. Said Ghabrial, University of Kentucky, for providing CMV isolates from New York and Kentucky (Ky), respectively.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Author information
Authors and Affiliations
Corresponding author
Additional information
The nucleotide sequence data reported in this article are available in the GenBank databases under the accession numbers JF834526 and JN029953 for IR-WI and IR-NY isolates, respectively.
Rights and permissions
Open Access This is an open access article distributed under the terms of the Creative Commons Attribution Noncommercial License (https://creativecommons.org/licenses/by-nc/2.0), which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
About this article
Cite this article
Nouri, S., Falk, B.W. & Groves, R.L. A new satellite RNA is associated with natural infections of cucumber mosaic virus in succulent snap bean. Arch Virol 157, 375–377 (2012). https://doi.org/10.1007/s00705-011-1154-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00705-011-1154-1