Abstract
Spyridium parvifolium is a widespread and morphologically variable shrub from south-eastern Australia. Several varieties have been recognised, and there is disagreement on the accepted taxonomy between Australian states. This study investigated the phylogeography of the species and assessed genetic distinctiveness of its morphological variants. Nuclear ribosomal DNA and complete chloroplast genomes from seventy-two samples of S. parvifolium and seven samples from closely related species were sequenced and analysed using both Bayesian and maximum likelihood phylogenetic methods. The results showed incongruence in the placement of several associated taxa (S. cinereum, S. obcordatum and S. daltonii), plausibly due to long branch attraction, introgression or incomplete lineage sorting. Spyridium parvifolium was resolved as paraphyletic in both phylogenies, with accessions from west of the Murray Darling Depression divergent from those east of the Depression. We found evidence of isolation within S. parvifolium on the inland side of the Great Dividing Range and recent gene flow across Bass Strait. The variants of S. parvifolium were not supported as genetically distinct, and with the prevalence of several variants at single sites and morphological intergrades between variants, we conclude that the taxon is a single, morphologically variable species and that no infraspecific classification is warranted.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Rhamnaceae is a medium-sized family of trees, shrubs, and climbers found throughout the world, excluding Antarctica and the Artic. It includes c. 960 species across c. 60 genera and 11 tribes (Hauenschild et al. 2016; Ladiges et al. 2005; Medan and Schirarend 2004). Diagnostic characters for Rhamnaceae are their small flowers with stamens opposite the (often) hooded petals, which in part cover the anthers (Kellermann et al. 2022). Most Australian Rhamnaceae are from a single tribe, Pomaderreae (c. 240 species), the second largest tribe in the family, consisting of species endemic to Australia and New Zealand. The tribe is easily distinguished within Rhamnaceae by the characteristic stellate hairs found on at least some (if not all) of the plant, which can include the floral parts (Kellermann 2020). Molecular studies focused on the family Rhamnaceae have consistently resolved the tribe as monophyletic (Hauenschild et al. 2016; Onstein et al. 2015; Richardson et al. 2000; van Santen and Linder 2020). One genus of the tribe Pomaderreae, Spyridium, is a group of c. 45 species endemic to semi-arid and temperate regions of southern Australia (Kellermann and Barker 2012). Spyridium species are commonly prostrate–medium shrubs, with tomentose stems and inflorescence of small flowers clustered in cymose heads (Coates 1996). The name Spyridium is derived from the Greek spyridion meaning a small basket, a reference to the inflorescence which is surrounded by leaf-like bracts (Perrin 2018). In many species, these leaf-like bracts (often referred to as floral leaves) are distinct from the stem leaves, covered in velvety hairs and therefore appearing white (Coates 1996). The monophyly of Spyridium is supported in multiple molecular phylogenies (Hauenschild et al. 2016; Kellermann and Udovicic 2007; Kellermann et al. 2005; Richardson et al. 2004).
Spyridium parvifolium is a morphologically variable and widespread shrub from south-eastern Australia (Fig. 1a; Atlas of Living Australia 2020; Canning and Jessop 1986; Curtis and Morris 1993; PlantNET 2021; VicFlora 2021). Found in dry sclerophyll forests and extending to heathlands (VicFlora 2021), S. parvifolium has a disjunct distribution with two main divides: one across Bass Strait between the states of Victoria and Tasmania and the second across the Murray Darling Depression (MDD) between western Victoria and south-central South Australia (SA; Fig. 1a; Atlas of Living Australia 2020). The Bassian Plain underlying the Bass Strait, the body of water separating mainland Australian from Tasmania, has been repeatedly exposed during glacial periods, including the last glacial maximum c. 16–18 ka (Bowler 1982) with the land bridge exposed until c. 13.5–12 ka (Galloway and Kemp 1981). Many other Australian dry forest plant species have disjunct distributions across Bass Strait (Worth et al. 2017). Conversely, the MDD which is currently above sea level has experienced several marine incursions including during both the Miocene, c. 15 mya, and Pliocene, c. 6 mya (Bowler 1982). The most recent shoreline retreat exposed the full extent of the MDD c. 1 mya and gave rise to geomorphology and edaphic features that still impact species distributions in the MDD (Bowler et al. 2006). Many Australian plants have disjunct distributions across the MDD (French et al. 2016), and several studies have found that many of these species show a major genetic divergence across the MDD. This includes Eucalyptus behriana (Fahey et al. 2021), Hardenbergia violacea (Larcombe et al. 2011), Zieria veronicea (Neal et al. 2019), Eucalyptus globulus subsp. bicostata (Jackson et al. 1999) and Themeda triandra (Hayman 1960). Spyridium parvifolium is also discontinuously distributed across Victoria from its western to eastern boundaries and into south-eastern New South Wales.
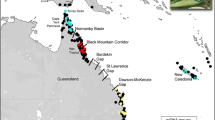
Distribution maps of Spyridium parvifolium (a), S. daltonii (b), S. cinereum (c), and S. obcordatum (d) in south-eastern Australia based on filtered records from Atlas of Living Australia (2020). Australian state abbreviations are as follows: SA South Australia, Vic. Victoria, Tas. Tasmania, NSW New South Wales. Also labelled are the southern section of the MDD, Lowan Sands of the MDD, the NCP, and the southern highlands/alps section of the GDR
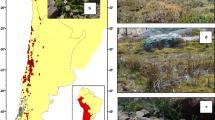
Known commonly as Dusty Miller (VicFlora 2021), S. parvifolium is distinguished from other species of Spyridium by grey–green floral leaves and elliptic–obovate (or orbicular) stem leaves with conspicuous secondary venation and an upper surface that usually has a hispid–sub-velutinous indumentum (VicFlora 2021). Four varieties of the species have been recognised based on morphological features (examples provided in Fig. 2): var. molle with grey–green hairy leaves and described from the Flinders and Cape Barren islands, Tasmania; var. grande with large leaves (> 25 mm long) described from the Dandenong Ranges, Victoria (Fig. 2a); var. hirsutissimum with long hispid hairs on the stems and described from the Grampians, Victoria (Fig. 2b); and the typical var. parvifolium which accommodated all other variation including plants with stem leaves that are “soft and often hairy on the upper side” (Bentham 1863). Many intermediates between the varieties have been noted (VicFlora 2021), and consequently, herbaria in Australian mainland states no longer recognise these varieties as distinct (CHAH 2020). However, in Tasmania two varieties (var. parvifolium and var. molle) are still recognised as separate taxa and both are listed, and protected, as Rare in Tasmanian legislation (Threatened Species Section, 2016a, b). To complicate matters further, additional variants of the species have been informally described in Victoria (Bull 2014; VicFlora 2021). These include a ‘Brisbane Ranges form’, with small (3–5 mm long), notched leaves (Fig. 2c); a ‘Rocky Sites form’, with grey–green hairy leaves (similar to var. molle), found in rocky sites in Gippsland (Fig. 2d; VicFlora 2021); and a small, spreading variant from East Gippsland, marketed in nurseries as ‘Prostrate form’ or ‘Nimbus’ (Bull 2014).
Examples of stem leaf variation in Spyridium parvifolium: a var. grande from Dandenong Ranges National Park; b var. hirsutissimum from Mt Zero Road (north) in the Grampians National Park, c Brisbane Ranges form from Brisbane Ranges National Park, d Rocky Sites form from the same location as Fig. 2a in Dandenong Ranges National Park.
This array of historical varieties and informal variants (both here forth referred to as ‘variants’) and conflicting taxonomy in different Australian states has led to suggestions that a more detailed investigation into the species is required (VicFlora 2021). There has been little molecular research into the species to date, with only a few samples included in broader phylogenetic studies focused on the tribe and family. Kellermann et al. (2005) included one sample of S. parvifolium in a study of the tribe based on nuclear ribosomal DNA (nrDNA) sequences (ITS). That sample was found to be divergent from all other south-east Australian Spyridium species included in the study. Kellermann and Udovicic (2007), extending the previous study by using chloroplast DNA (cpDNA) sequences (trnL-trnF), found S. parvifolium formed an unsupported clade with S. thymifolium (from SA). Hauenschild et al. (2016) included three samples of S. parvifolium in a phylogeny of Rhamnaceae with the combined nrDNA (ITS) and cpDNA (trnL-trnF) analysis placing all three samples in a clade with S. daltonii and S. thymifolium. More recently, Clowes et al. (2022) included eight samples of S. parvifolium in a molecular (nrDNA and chloroplast genomes) phylogeny of Spyridium, with the species found in an unsupported clade (Bayesian posterior probability, 0.73) with S. daltonii, S. obcordatum and S. cinereum in the nrDNA phylogeny and resolved in a strongly supported clade (polytomy; Bayesian posterior probability, 1.0) with S. daltonii in the cpDNA tree. The three most closely related species in the nrDNA tree are narrow range endemics with overlapping (or nearly so) distributions with S. parvifolium (Fig. 1b–d). Spyridium daltonii is also known to hybridise with S. parvifolium, resulting in the named hybrid S. ×-ramosissimum (Kellermann 2006).
The current study aims to extend the work of Clowes et al. (2022), with a specific focus on S. parvifolium. Spyridium parvifolium is sampled from across its geographic range, including all named varieties and known morphological variants (VicFlora 2021), together with representatives of other species of Spyridium. Both nrDNA and complete chloroplast genomes (cpDNA) are used to investigate the phylogeographic history of the species and to compare this to other species of dry sclerophyll forests in south-eastern Australia. The data are also used to assess patterns of genetic variation in S. parvifolium and whether they provide grounds for recognition of distinct species or infraspecific taxa.
Materials and methods
Taxon selection and sampling
In total, 213 individuals representing 49 species of Spyridium and three outgroup genera of tribe Pomaderreae (Pomaderris, Cryptandra and Trymalium) were sampled, including several phrase name taxa. Of these, 66 samples were newly sequenced accessions of S. parvifolium (64 samples) and S. daltonii (two samples) and 145 were previously included in Clowes et al. (2022).
Most samples were collected during fieldwork. Where plants at a site appeared morphologically homogeneous, one voucher specimen (with duplicates) was collected; however, where a site showed morphological variation, more than one voucher was collected and lodged at The University of Melbourne Herbarium (MELU). Based upon State collection and associated permit requirements, duplicates were sent to the National Herbarium of Victoria (MEL), the Tasmanian Herbarium (HO) or the State Herbarium of South Australia (AD). Leaf material was also collected from several individuals at each site (including the vouchered plant/s) and dried in silica gel, with one to four samples from each site included in this study.
Seventy-two samples of S. parvifolium from 36 locations were included in this study (Table 1; Fig. 3), of which eight were also included in Clowes et al. (2022). Samples were collected from across the geographic range and morphological variants of the species. Reference works used to determine the variety or variant of samples are provided in Table 1. For the Brisbane Ranges form, sites identified as this variant were collected from the Brisbane Ranges area and had small, sometimes notched leaves, as described by VicFlora (2021). However, in some cases the leaves were more than 5 mm long, which is longer than the description suggests. Despite this, we have referred to these samples as the Brisbane Ranges form.
Sample locations of Spyridium parvifolium included in this study. Shapes and colours represent morphological variants of S. parvifolium (i.e. var. parvifolium, var. molle etc.) and broad collection regions (e.g. south-east Victoria/NSW, Tasmania including Flinders Island etc.). The approximate location of the MDD shoreline at c 6 mya and c 1 mya is highlighted, and the locations of Scott Creek, Kaiserstuhl and Mt Remarkable are indicated. Other features include: the southern section of the MDD, Lowan Sands of the MDD, the NCP, the southern highlands/alps section of the GDR, Bass Strait and Flinders Island
Three samples of S. daltonii were also included, collected from two different locations within the Grampians ranges: Grampians Road and Mt William (Table 1). One of these accessions was also included in Clowes et al. (2022).
DNA extraction and library preparation
Total genomic DNA was extracted from c. 60 mg of silica dried leaf material following a modified CTAB (cetyl trimethylammonium bromide) DNA extraction protocol (McLay 2017; Shepherd and McLay 2011) based on Doyle and Doyle (1987). Where possible young, dried leaf material from stem tips or dried floral leaves were selected for extraction. DNA quality and quantity were recorded using a Nanodrop 2000 (NanoDrop Products) and Qubit 2.0 fluorometer (Invitrogen). For some samples, in an attempt to improve DNA quality, an additional clean-up was performed using a sodium acetate/ethanol wash following Lamitina Lab Protocols (2007). Genomic DNA was prepared for multiplexed sequencing using the library preparation protocol of Schuster et al. (2018) with modifications per Clowes et al. (2022). Sequencing was performed using an Illumina NextSeq 500 (2 × 150 bp) sequencing platform at the Walter and Eliza Hall Institute of Medical Research Genomics Hub (WEHI).
Sequence assembly and alignment
Quality filtering and base calling were conducted at WEHI with Illumina pipeline software (v.1.7 or later) and pre-processed with custom scripts, as per Schuster et al. (2018). Sequence reads were imported into Geneious 10.0.1 or newer (Kearse et al. 2012) and trimmed, paired and assembled as per Clowes et al. (2022). Sequences were aligned in MAFFT v7.308 (Katoh et al. 2002; Katoh and Standley 2013) using automatic algorithm selection and default settings. Aligned sequences were reviewed in Geneious and manually re-aligned following the protocol described in Clowes et al. (2022). The final nrDNA alignment was 6,386 bp long and the cpDNA alignment totalled 168,343 bp. The nrDNA sequences were partitioned as follows: partial external transcribed spacer (5'ETS), 18S, internal transcribed spacer 1 (ITS1), 5.8S, internal transcribed spacer 2 (ITS2), 26S, and partial non-transcribed spacer (3'ETS + NTS). The 5.8S region was excluded from analyses as it was identical in all samples. The cpDNA alignment was partitioned into four categories: gene coding sequence (CDS); transfer ribonucleic acid (tRNA); ribosomal ribonucleic acid (rRNA); and all remaining sequences, including introns and intergenic spacers (referred to in the partition as spacers). For the cpDNA alignment inverted repeat A (IRA) was excluded from the final analyses. For both the nrDNA and cpDNA alignments, several regions of ambiguous sequence were also excluded (i.e. in repetitive regions of DNA where the number of bases could not be assessed with confidence). Alignments and partition details (including exclusions) are available on Dryad (10.5061/dryad.573n5tb9t).
Phylogenetic analyses
Prior to Bayesian inference (BI) analyses, model testing was performed using MrModeltest 2.3 (Nylander 2004). For the nrDNA dataset, the best fit models selected according to the Akaike information criterion (AIC) were: 5'ETS GTR + G, 18S HKY + I + G, ITS1 SYM + G, ITS2 HKY + G, 26S GTR + I + G and 3'ETS + NTS HKY + G. For the cpDNA data, the best fit models according to AIC were: CDS GTR + I + G, tRNA K80, rRNA HKY and spacers GTR + I + G. For the cpDNA data, the best fit models according to AIC were: CDS GTR + I + G, tRNA K80, rRNA HKY and spacers GTR + I + G. Bayesian inference was undertaken in MrBayes XSEDE (Ronquist and Huelsenbeck 2003) using the CIPRES portal (Miller et al. 2010). For the nrDNA alignment, two independent analyses with four chains (Markov chain Monte Carlo) were run for 10 M generations, sampling every 1,000 steps, with a burnin of 25% and Dirichlet distribution unlinked. For the cpDNA alignment, the analysis was run for 2.5 M generations and sampled every 500 steps. Output files were viewed in Tracer (Rambaut et al. 2014) to check for convergence. The average standard deviation of split frequencies was also reviewed and confirmed to be below 0.01 upon completion of analyses. Branches with values of < 0.95 PP were considered unsupported.
Maximum likelihood (ML) analysis was completed with IQ-Tree using default settings (Nguyen et al. 2015). Model testing was automated in IQ-Tree, with ModelFinder (Kalyaanamoorthy et al. 2017) used to select models for each partition using Bayesian information criterion (BIC). Best fit models for the nrDNA data were: 5'ETS TPM2u + F + G4, 18S K2P + I + G4, ITS1 TIM2e + G4, ITS2 TNe + G4, 26S TIM3 + F + I and 3'ETS + NTS TN + F + G4. Best fit models for the cpDNA sequences were: CDS TVM + F + I, tRNA K2P, rRNA HKY + F and spacers TVM + F + I + G4. The nrDNA analysis contained 274 parsimony-informative characters and the cpDNA analysis included a total of 3,468 parsimony-informative characters. Branches with values of < 95% ultrafast bootstrap support (UFBS) were considered unsupported. Branches that were supported in one analysis (for example BI) but unsupported in the other analysis (for example ML) were considered unsupported overall.
Long branch attraction tests
Preliminary nrDNA phylogenies placed samples of two species (S. cinereum and S. obcordatum) together on comparatively long branches and in positions incongruent with those on cpDNA trees. To assess whether long branch attraction (LBA) could be influencing the positions of these species in the nrDNA trees, BI and ML analyses were re-run (as described above), with each species separately removed from the dataset, following an approach suggested by Bergsten (2005). The resulting consensus trees, as viewed in FigTree (Rambaut and Drummond 2012), are provided in the Electronic Supplementary Material (Online Resources 1–4).
Results
Nuclear rDNA phylogeny
Overall, the topology in the BI (Fig. 4a) and ML (Fig. 4b) nrDNA trees was similar, though there were several unsupported (< 0.95 PP, < 95% UFBS) incongruencies between them. Clades relevant to S. parvifolium are shown in detail in Fig. 4, with all other clades collapsed. The BI tree (Fig. 4a) resolved two sister clades (B and C) that included all S. parvifolium samples, along with samples of S. daltonii (nested in clade B) plus a clade (D) of S. obcordatum and S. cinereum nested within clade C. Those same clades were also present on the ML tree, but differed in their relationships. In the ML tree (Fig. 4b), clade C was not sister to clade B, but rather was sister, with 57% UFBS, to a clade (E) of other Spyridium species mostly endemic to SA. The relationship of S. daltonii samples to other members of clade B, and that of clade D to other members of clade C also differed in the ML tree, but none of the relevant nodes had strong support. Other differences between the BI and ML trees were also unsupported and made no notable difference to the results.
a Bayesian inference (BI) 50% majority rule consensus tree based on analysis of nuclear ribosomal DNA (nrDNA). Only clades relevant to Spyridium parvifolium are shown (A-D) with all other unrelated clades collapsed. b A reduced selection of the ML consensus tree shown only the SA accessions of S. parvifolium and those of S. daltonii, S. cinereum and S. obcordatum. The S. parvifolium clade contains the same samples of the species as shown in Fig. 4a. In both Fig. 4a and 4b only values for unsupported nodes are showing (i.e. < 0.95 PP on the BI tree and < 95% UFBS on the ML tree) with the values of supported nodes not shown (i.e. ≥ 0.95 PP, ≥ 95% UFBS). Coloured shapes next to accession labels indicate variants and geographic regions for S. parvifolium samples, as described in the caption for Fig. 3. The contents of collapsed clades, together with all support values, can be viewed by accessing this phylogeny on Dryad (https://doi.org/10.5061/dryad.573n5tb9t). Clade E contains 70 accessions of Spyridium predominantly endemic to SA and south-eastern Australia more broadly. The NSW/QLD endemics clade contains six samples of Spyridium, the Tasmanian endemics clade contains 19 Spyridium samples, and the predominantly WA endemics clade contains 33 Spyridium samples
Spyridium parvifolium was not resolved as monophyletic in either the BI or ML trees (Fig. 4a, b). Four samples from west of the MDD in SA, Kaiserstuhl and Mt Remarkable, formed a clade (B) with the three S. daltonii samples; this clade (B) was sister to the main S. parvifolium clade (C) in the BI tree. The previously identified morphological variants of S. parvifolium do not form clades in the tree (Fig. 4a). Spyridium obcordatum and S. cinereum were both resolved on long branches within clade D (Fig. 4a, b). When samples of S. cinereum were removed from the alignment and the analyses re-run, the placement of S. obcordatum changed; in both the BI and ML re-analyses, S. obcordatum was placed as sister (0.93 PP; 95% UFBS) to a clade (0.87 PP; 77% UFBS) containing all other samples of clade A (Online Resources 1, 2), congruent with its placement in the cpDNA phylogeny (Fig. 5). When S. obcordatum was removed from the alignment and the analyses re-run, S. cinereum was found in a basal polytomy in clade C in both BI and ML trees (Online Resources 3, 4), congruent with the BI tree Fig. 4a.
Bayesian inference (BI) 50% majority rule consensus tree based on analysis of chloroplast genomes (cpDNA). Posterior probabilities (PP) < 0.95 are shown, followed by ultrafast bootstrap (UFBS) values < 95%; support values are not shown for nodes with ≥ 0.95 PP or ≥ 95% UFBS. Coloured shapes next to accession labels indicate variants and regions of S. parvifolium, described in Fig. 3 caption. Clades relevant to S. parvifolium are labelled F–G and G1-G11. Clades not focused on S. parvifolium that have been collapsed, can be viewed accessing this phylogeny on Dryad (10.5061/dryad.573n5tb9t). Collapsed clades within clade F contain 37 accessions of Spyridium (predominantly endemic to SA and south-eastern Australia more broadly). The predominantly SA endemics clade contains 37 Spyridium samples, the predominantly WA endemics clade contains 28 Spyridium samples, the NSW/QLD endemics clade contains six Spyridium samples, and the Tasmanian endemics clade contains 33 Spyridium samples
Chloroplast genome phylogeny
Overall, the relationships of S. parvifolium samples in the BI and ML analyses of cpDNA genomes were congruent, with differences between the trees unsupported (Fig. 5). Clades relevant to S. parvifolium are shown in detail on the BI tree (Fig. 5), with all other clades collapsed. Despite all S. parvifolium samples being found in a single clade (clade G1), the species was rendered paraphyletic as Spyridium daltonii was nested within clade G2. The three accessions of S. daltonii do not form a monophyletic grouping, all falling near the basal polytomy of this clade (Fig. 5). Two samples of S. daltonii (one from Mt William and one from Grampians Road) were placed together with strong support, while the third sample (also from Mt William) was placed separately on a comparatively long terminal branch. Spyridium obcordatum was resolved as sister to clade G1, within clade G, whereas S. cinereum was resolved in a distant clade (F).
Within clade G1 there was some geographic clustering of S. parvifolium samples (Fig. 5, 6). Where more than one sample of S. parvifolium was included for a site, they mostly grouped together in the tree; exceptions were samples from Jancourt, Wilsons Promontory, Distillery Creek, Croajingolong and Scott Creek sites (Fig. 5). Samples of S. parvifolium collected from west of the MDD (Mt Remarkable, Kaiserstuhl and Scott Creek) formed a grade at the base of clade G1 (Figs. 5, 6a), subtending a clade (G2) including all accessions from east of the MDD (Fig. 5). Within G2, an inland GDR (Great Dividing Range) clade G8 was resolved, as was a south-eastern Victorian (coastal NSW) only clade (G6). Tasmanian and Flinders Island accessions were found in four clades (Figs. 5, 6), and south-eastern Victorian samples were found throughout clade G2, including in three of the four Tasmanian and Flinders Island clades (Fig. 6a–c).
Distributions of samples from selected cpDNA lineages that include samples across Bass Strait. Clade G3 a, G4 b, G5 c and G9 d. Victoria (Vic.), Tasmanian (Tas.), Flinders Island, Bass Strait, Wilsons Promontory and Cape Liptrap are also highlighted. Other features include the southern section of the MDD, Lowan Sands of the MDD, the NCP and the southern highlands/alps section of the GDR
Of the morphological variants of S. parvifolium, only one, var. hirsutissimum was limited to a single clade (G11, Fig. 5). All samples of the Brisbane Ranges form were found in clade F3, although this clade also contained some samples of var. parvifolium and var. molle from Tasmania. All other variants were found scattered throughout the tree within clade G2.
Discussion
In this study we have presented a molecular analysis of S. parvifolium, across the species range and incorporating all morphological variants previously discussed in the literature. These results provide information about the phylogeographic history of the species and enable an interpretation of the classification of the variants. However, several differences between the nrDNA and cpDNA phylogenies related to the placement of associated taxa (S. cinereum, S. obcordatum and S. daltonii) warrant discussion first.
Incongruent placement of Spyridium cinereum and S. obcordatum in nrDNA and cpDNA phylogenies
The placement of S. cinereum and S. obcordatum differed markedly between the nrDNA and cpDNA phylogenies (Figs. 4, 5), as previously found in the analyses of Clowes et al. (2022). The causes of this incongruence could be biological (e.g. resulting from incomplete lineage sorting or introgression between taxa), as commonly seen in many plant groups (Barrett et al. 2018; Rieseberg and Soltis 1991; Wiley and Lieberman 2011), or could be an artefact of analysis resulting from LBA (Bergsten 2005) or, in the case of nrDNA sequences, from combining different paralogues in the analysis (Bailey et al. 2003; Bayly and Ladiges 2007). The positions of these species in the cpDNA tree (Fig. 5) are more in line with current species-level taxonomy, and morphological features with S. obcordatum resembling, but are distinguished from the Prostrate form of S. parvifolium, and with S. cinereum being considerably more distinctive, in line with placement in clade F with a number of other species (several with similar notched leaves like S. cinereum, for example).
It is the position of S. cinereum and S. obcordatum in the nrDNA tree (Fig. 4a) that seems most anomalous, especially given the very long branches on which they sit relative to surrounding ones. The fact that placement of S. obcordatum moves, and matches that seen in the cpDNA, when S. cinereum is excluded from the nrDNA dataset (Online Resources 1, 2) could be considered evidence that LBA is at play (Bergsten 2005). Other explanations are less plausible, but cannot be ruled out. Of those explanations, incomplete lineage sorting, especially in nrDNA arrays which have small effective population sizes (Buckler and Holtsford 1996) and, therefore, short coalescence times as a result of concerted evolution (Arnheim 1983), seems unlikely and would not account for the unusual branch lengths of S. cinereum and S. obcordatum. Confusion of nrDNA paralogues also seems unlikely, as we scrutinised sequence reads and assemblies closely and saw no evidence of distinctive rDNA paralogues within samples, and especially in S. cinereum and S. obcordatum. Introgression, on the other hand, which can have variable outcomes for sequences of nrDNA arrays (Álvarez and Wendel 2003) could conceivably result in distinctive sequences that combine features of different parental lineages. Although putative hybrids between S. parvifolium and S. cinereum or S. obcordatum have not been reported, S. parvifolium does grow in close proximity to the other two (Atlas of Living Australia 2020), making introgression plausible. The example of S. ×ramosissimum shows that S. parvifolium can hybridise with relatives when they co-occur; however, in the case of S. cinereum, it would have to have been historical rather than recent hybridisation to account for the monophyly of the two highly disjunct samples of S. cinereum (Fig. 1c) in the nrDNA tree.
Broader population sampling and additional nuclear DNA data, preferably from a broad range of loci, are needed to more fully understand the relationships of S. cinereum and S. obcordatum to S. parvifolium and other species in the genus. They could confirm or refute the nrDNA relationships resolved here and could be used to test for evidence of introgression. Target sequence capture methods (e.g. Hyb-Seq; Dodsworth et al. 2019) could be ideal for this purpose.
Placement of Spyridium daltonii among samples of S. parvifolium
Spyridium daltonii, endemic to the Grampians National Park in western Victoria, was nested within S. parvifolium in both the cpDNA and nrDNA trees (Figs. 4a, 5). Although there is uncertainty about its exact placement because nodes at the base of clades B and C in the nrDNA tree are very weakly supported (allowing the possibility that S. daltonii could be sister to S. parvifolium), and because it is placed in a large polytomy near the base of clade G2 in the cpDNA tree, its nested position in the cpDNA tree is strongly supported.
Spyridium daltonii co-occurs with S. parvifolium at several locations in the Grampians (Atlas of Living Australia 2020) and the two species are also known to hybridise, producing the hybrid taxon S. ×ramosissimum (Kellermann 2006). Given this, the nesting of S. daltonii in the S. parvifolium clade, especially in the cpDNA phylogeny, could be attributed to introgression resulting in chloroplast capture. Although the molecular data presented here might suggest that S. daltonii could be synonymised with S. parvifolium, the two are readily distinguished on the basis of morphology to the extent that treating them as conspecific would be difficult to justify. As such, further sampling of S. daltonii individuals and of additional nuclear DNA markers, is needed to assess whether there is any incongruence between nuclear and cpDNA relationships and the extent to which there is evidence of nuclear differentiation or introgression between the two species.
Genetic divergence in Spyridium parvifolium across the Murray Darling Depression
This study has revealed genetic divergence associated with the disjunct distribution of S. parvifolium across the MDD. The SA populations (Mt Remarkable, Kaiserstuhl and Scott Creek; Fig. 3) show genetic differences from more easterly populations based on both cpDNA and nrDNA sequences, although their relationships are resolved differently in the two datasets (Figs. 4a, b, 5). Such divergence is not unexpected as other plant species are disjunct and genetically divergent across the MDD, for example: Eucalyptus behriana (Fahey et al. 2021), Hardenbergia violacea (Larcombe et al. 2011), Eucalyptus globulus subsp. bicostata (Jackson et al. 1999) and Themeda triandra (Hayman 1960).
French et al. (2016) highlighted three factors that have created barriers to dispersal across the MDD, potentially leading to geographic disjunctions in this area: past marine incursions, subsequent edaphic factors and hostile climates associated with Quaternary glacial periods. The MDD and the Naracoorte Coastal Plain (NCP) have experienced multiple marine incursions including during the Miocene (c. 15 mya) and Pliocene (c. 6 mya; Fig. 3; Bowler 1982). Subsequent to these incursions, geomorphology and edaphic factors that remained were also likely a barrier to recolonisation by some plants. During marine incursions in SA, elevated areas including the Lofty Ranges (e.g. Scott Creek and Kaiserstuhl) may have provided refugia for flora species (Byrne 2008), such as S. parvifolium, which is found scattered throughout the southern Lofty Ranges today (Atlas of Living Australia 2020). By c. 1 mya, the shoreline had retreated to the inland margin of the NCP (Fig. 3; Bowler et al. 2006) with the final retreat resulting in additional edaphic barriers (Specht 1972) including calcareous sands and limestone of the NCP. Compounding these incursions and retreat ‘barriers’, were glacial periods of the Quaternary, which resulted in cooler, arid climates in many places including the MDD that inhibited the establishment of plants in areas such as mobile dunes. Given the history of potential barriers to plant dispersal across the MDD, the geographic disjunctions in species’ ranges across this area could be explained by either vicariance (e.g. species predates barrier) or dispersal (i.e. species younger than barrier and disperses across it).
For S. parvifolium, the genetic pattern in the cpDNA phylogeny (Fig. 5) is consistent with either vicariance (with early differentiation in the west and a single monophyletic group in the east) or with dispersal from west to east. The Spyridium phylogeny (Clowes et al. 2022) implied an early divergence of WA taxa during the Miocene; therefore, the age of the clade including S. parvifolium is such that the Pliocene marine incursion, edaphic barriers or aridification are plausible explanations for the observed SA divergence. Spyridium parvifolium is absent from the Lowan Sands and adjoining regions in SA (Fig. 1a), which could either be associated with current edaphic conditions, or due to previous climatic conditions (e.g. with dune mobilisation in glacial times; Bowler et al. 2006; Conn 1993).
Although S. parvifolium is effectively absent from the MDD, it is found in southern parts of the NCP (Fig. 1a), which suggests that limestone soils are not necessarily a barrier for the species. According to the Australian State of Environment Report, less than 40% of native vegetation remains in the NCP region (Metcalfe and Bui 2017), so it is possible that additional populations of S. parvifolium were present in this area and have been cleared since European settlement. Presence of S. parvifolium on the NCP indicates that some recolonisation of this species has occurred since the sea levels dropped around 1 mya, which could have resulted in the reconnection of previously isolated and divergent eastern and western lineages. Such a process was inferred in the genus Correa (Rutaceae) by French et al. (2016), who identified divergent eastern and western cpDNA clades that overlapped, potentially through reconnection of previously separated lineages, on the Fleurieu Peninsula in SA (i.e. west of the MDD, near the western edge of the NCP).
If S. parvifolium has dispersed across the NCP, e.g. since those plains were exposed above sea level in the last million years, reconnection and intermixing of lineages previously separated across the MDD could potentially explain the contrasting placement of Scott Creek samples in the cpDNA and nrDNA trees. Whereas the cpDNA tree places those samples close to others from west of the MDD, as part of a western grade at the base of Clade F1 (Fig. 5), the nrDNA trees place them separate from the other western samples in a clade (C) of samples from east of the MDD (Fig. 4a). Given the location of Scott Creek near the western edge of the NCP (Fig. 3), it is tempting to speculate that these plants might combine “western” chloroplasts and “eastern” nrDNA because of intermixing of lineages facilitated by dispersal east to west across the NCP. Such a hypothesis could be tested by more intensive sampling of populations from the SA and southwest Victoria, and by using variable genome-wide nuclear markers (e.g. RADseq or similar; Davey and Blaxter 2010), together with cpDNA sequencing, to investigate genetic patterns across this region.
West of the MDD, a north–south pattern of divergence of S. parvifolium was identified, with the most northern population (e.g. Mt Remarkable; Fig. 3) divergent in both phylogenies (Figs. 4a, 5), and especially in the cpDNA tree. Throughout the Lofty Ranges, S. parvifolium has a somewhat continuous distribution with populations separated by relatively short distances (Figs. 1a). North of the Lofty Ranges, S. parvifolium has a discontinuous distribution with only three main localities; Mount Remarkable, Telowie Gorge Conservation Park (both part of the Southern Flinders Ranges) and Tothill Ranges (Atlas of Living Australia 2020). This northern divergence (Mt Remarkable) and discontinued distribution is interesting; however, no other comparative studies with north–south SA samples included have been identified to determine whether this is an isolated finding or pattern also seen in other members of the flora across these areas. The conservation of S. parvifolium across its range in SA is important, because each population (particularly those isolated north of the Lofty Ranges) may be genetically distinct. Further research into SA populations of other taxa found across south-east Australia is warranted, to investigate whether this finding is repeated in other dry sclerophyll forest species.
The Great Dividing Range is a biogeographic barrier in Spyridium parvifolium
The GDR is identified as a barrier to seed-mediated gene flow within S. parvifolium, with samples collected from the inland side of the range forming a clade in the cpDNA tree, distinct from samples collected on the coastal side of the GDR and other parts of Victoria (Fig. 5). This result is interesting because there are relatively few comparable studies of south-east Australian dry sclerophyll forest species with GDR disjunctions, and of those available, the findings were different to those of S. parvifolium. For example, no barrier to gene flow either side of the GDR was detected for the genus Xanthorrhoea (McLay et al. 2021) or the species Eucalyptus melliodora (Broadhurst et al. 2018), both widespread in dry sclerophyll forest of south-eastern Australia. Given the limited number of studies in this area, investigation of more species disjunct and separated by the high parts of the GDR is warranted to see whether there are common patterns. For S. parvifolium in the southern highlands and alps, glacial cycles may have exaggerated the physical barrier, with a snowline that dropped 1,000 m below the range peaks (Frakes et al. 1987). The extension of these alpine and subalpine conditions downward is likely to have significantly fragmented forest vegetation (Byrne 2008), creating even greater potential for isolation of sites on either side of the range. Today, variation in climate and elevation over the GDR may act as a continued physical barrier for species (Milner et al. 2012), such as S. parvifolium.
Recent gene flow in Spyridium parvifolium across Bass Strait
In contrast to the barrier associated with the GDR, there is evidence that seed-mediated gene flow in S. parvifolium has continued until relatively recently across Bass Strait, with multiple close cpDNA relationships identified, predominantly between south-western Victoria, Wilsons Promontory, Tasmania and Flinders Island (Figs. 5, 6). Most clades with Bass Strait connections (G3, G4, G5) also show a degree of sequence divergence between mainland and Tasmanian samples within the clade, except for clade G9 in which there is little differentiation. This sequence divergence suggests the most recent connection across Bass Strait occurred between Wilsons Promontory–Cape Liptrap in south Gippsland (Victoria) and Sisters Beach in north-west Tasmania (Clade G9; Figs. 5, 6d). Other studies have found a similar pattern of recent gene flow between Victoria and Tasmania, for example in Correa (French et al. 2016), Zieria veronicea (Neal et al. 2019), Astroloma humifusum, Epacris impressa, Bursaria spinosa and several other dry forest species (Worth et al. 2017). During glacial maxima, sea levels dropped below those of today exposing the Bassian Plain (Larcombe et al. 2011), with glacial/interglacial oscillations occurring every 100 ka over the last 2.5 M years (Frakes et al. 1987). The last glacial maximum occurred 16–18 ka (Bowler 1982) with the land bridge across Bass Strait exposed until c. 13.5–12 ka (Galloway and Kemp 1981). Evidence suggests that at least some areas of the Bassian Plain were covered in eucalypt woodland (Hope 1978, 1994; Kirkpatrick and Fowler 1998), which may have been suitable habitat for S. parvifolium. If so, this could explain the main points of connection between Victoria and Tasmania identified here and for other dry sclerophyll forest species (Worth et al. (2017): the Otways (e.g. Jancourt, Port Campbell and Distillery Creek), south Gippsland (e.g. Cape Liptrap and Wilson Promontory), north-east Tasmania (e.g. Bay of Fires) and to a lesser extent north-west Tasmania (e.g. Sisters Beach). Over-land dispersal at times of low sea level seems more plausible than multiple chance dispersals over water, as Spyridium species lack obvious features to enable seed dispersal (Coates 1996; Coates et al. 1999).
Spyridium parvifolium is a single, morphologically variable taxon
This study did not resolve the morphological variants of S. parvifolium, including the two currently recognised varieties (var. parvifolium and var. molle) from Tasmania and Flinders Island, as monophyletic or as more genetically differentiated than other populations of the species (Figs. 4a, 5). The cpDNA phylogeny did resolve var. hirsutissimum as monophyletic (Fig. 5); however, both samples were collected from a single site and the cpDNA phylogeny showed strong geographic structuring, such that var. hirsutissimum samples were no more distinct than those from a range of other collecting sites. As discussed above, recent gene flow or close cpDNA relationships are evident between many populations from Tasmania, Flinders Island and Victoria, including between var. parvifolium, var. molle and the Brisbane Ranges form (Figs. 5, 6a, b).
Observations made in the field and when reviewing herbarium specimens also suggest that most of the variants of the species are not well supported on morphological or ecological grounds, with instances of several variants recorded at a single site and some morphological intergrades between variants. For example, plants with the morphology of var. parvifolium and var. molle on Flinders Island co-exist at the Hines Road site (and group together based on cpDNA, with no discrimination between them; clade G4, Fig. 5) and plants morphologically matching the Rocky Site form were found side-by-side with var. grande in the Dandenong Ranges (Fig. 2a, d Table 1; and group together in clade G6 based on cpDNA). Several examples of intergrades were observed including examples of var. parvifolium from Flinders Island with a sparse indumentum on the upper surface of the stem leaves (and therefore grading with var. molle), and many examples of the Brisbane Ranges form have small, notched leaves, 1–2 mm longer than the described range for the variant (grading into typical S. parvifolium). Herbarium specimens reveal dozens of collections from throughout the range of S. parvifolium with an indumentum on the upper surface, to varying degrees of density. Hairs on the adaxial surface of the stem leaves appear to be a labile character that randomly occurs in S. parvifolium but might be more common on Flinders Island (hence var. molle is recognised there at the moment).
Given our molecular results, and with consideration to observations made in the field and from herbarium specimens, there is little available evidence to support taxonomic recognition of any of the variants of the species. We therefore recommend S. parvifolium be recognised as a single, morphologically variable and widespread species with no recognition of infraspecific taxa. Our recommendation may impact the Tasmanian legislation (Threatened Species Protection Act 1995), as the varieties currently recognised in Tasmania are both listed as Rare (Threatened Species Section, 2016a, b). A conservation assessment of S. parvifolium as a single taxon may result in the species exceeding the criteria for Rare in Tasmania, and it may therefore require consideration at a different conservation listing level or delisting.
Considering the incongruence of cpDNA and nrDNA phylogenies (Figs. 4a, 5) and the slightly different topologies of the ML and BI trees based on nrDNA (Fig. 4a, b), we do not recommend the merger of the associated species S. cinereum, S. daltonii or S. obcordatum with S. parvifolium. The relationships of all these taxa require further investigation, as our study focused mainly on S. parvifolium. In addition, though the divergence of the Kaiserstuhl and Mt Remarkable was not supported in the nrDNA phylogeny, the morphology of these variants of S. parvifolium should be evaluated. This could be extended to SA variants of the species more generally, given the Scott Creek samples were also unsupported but somewhat divergent in the nrDNA trees and supported and divergent in the cpDNA phylogeny.
Data availability
Individual annotated sequences used in this study are available from GenBank (see Table 1 for accession numbers). Alignments analysed during this study and resulting phylogenetic trees, are available on Dryad (10.5061/dryad.573n5tb9t). Trees resulting from LBA tests are provided in the Electronic Supplementary Material.
References
Álvarez I, Wendel J (2003) Ribosomal ITS sequences and plant phylogenetic inference. Molec Phylogen Evol 29:417–434. https://doi.org/10.1016/S1055-7903(03)00208-2
Arnheim N (1983) Concerted evolution of multigene families. In: Nei M, Koehn RK (eds) Evolution of genes and proteins. Sinauer, Sunderland, pp 38–61
Atlas of Living Australia (2020) Pomaderreae: occurrence download. Available at: https://www.ala.org.au/. Accessed 26 Jun 2020
Bailey CD, Carr TG, Harris SA, Hughes CE (2003) Characterization of angiosperm nrDNA polymorphism, paralogy, and pseudogenes. Molec Phylogen Evol 29:435–455. https://doi.org/10.1016/j.ympev.2003.08.021
Barrett RA, Bayly MJ, Duretto MF, Forster PI, Ladiges PY, Cantrill DJ (2018) Phylogenetic analysis of Zieria (Rutaceae) in Australia and New Caledonia based on nuclear ribosomal DNA shows species polyphyly, divergent paralogues and incongruence with chloroplast DNA. Austral Syst Bot 31:16–47. https://doi.org/10.1071/SB16034
Bayly MJ, Ladiges PY (2007) Divergent paralogues of ribosomal DNA in eucalypts (Myrtaceae). Molec Phylogen Evol 44:346–356. https://doi.org/10.1016/j.ympev.2006.10.027
Bentham G (1863) Spyridium parvifolium. In: Bentham G, von Mueller, F (eds) Flora Australiensis, a description of the plants of the Australian territory, vol. 1. L. Reeve and co, London, p 428
Bowler JM (1982) Aridity in the late tertiary and quaternary of Australia. In: Barker WR, Greenslade PJM (eds) Evolution of the Flora and Fauna of Arid Australia. Peacock Publications in association with the Australian Systematic Botany Society and ANZAAS South Australian Division Inc, Frewville, pp 35–45
Bowler JM, Kotsonis A, Lawrence CR (2006) Environmental evolution of the Mallee region, western Murray Basin. Proc Roy Soc Victoria 118:161–210
Broadhurst LM, Mellick R, Knerr N, Li L, Supple MA (2018) Land availability may be more important than genetic diversity in the range shift response of a widely distributed eucalypt, Eucalyptus melliodora. Forest Ecol Managem 409:38–46. https://doi.org/10.1016/j.foreco.2017.10.024
Buckler ES, Holtsford TP (1996) Zea systematics: ribosomal ITS evidence. Molec Biol Evol 13:612–622
Bull M (2014) Flora of Melbourne: a guide to the indigenous plants of the greater Melbourne area, 4th edn. Hyland House Publishing, Carlton
Byrne M (2008) Evidence for multiple refugia at different time scales during Pleistocene climatic oscillations in southern Australia inferred from phylogeography. Quatern Sci Rev 27:2576–2585. https://doi.org/10.1016/j.quascirev.2008.08.032
Canning EM, Jessop JP (1986) Rhamnaceae. In: Jessop JP, Toelken HR (eds) Flora of South Australia, vol. 2. Government Printer, Adelaide
CHAH (2020) Australian plant census; Spyridium. Available at: https://biodiversity.org.au/nsl/services/search?product=APNI&tree.id=&name=Spyridium&inc._scientific=&inc._cultivar=&inc._other=&max=100&display=&search=true. Accessed 17 Nov 2020
Clowes C, Fowler RR, Fahey PS, Kellermann J, Brown GK, Bayly MJ (2022) Big trees of little baskets: phylogeny of the Australian genus Spyridium (Rhamnaceae: Pomaderreae) focusing on biogeographic patterns and species delimitation. Austral Syst Bot 35:95–119. https://doi.org/10.1071/SB21034
Coates F (1996) Ecological and biogeographical correlates of rarity in two narrow endemics in Tasmania: Spyridium microphyllum (F. Muell. ex Reisseck) Druce and Spyridium obcordatum (Hook f.) WM Curtis. University of Tasmania, Hobart
Coates F, Kirkpatrick JB, Minchin PR (1999) Towards an explanation of the causes of the rarity of two Tasmanian Spyridium species. Austral J Ecol 24:11–17
Conn BJ (1993) Natural regions and vegetation of Victoria. In: Foreman DB, Walsh NG (eds) Flora of Victoria, vol. 1. Inkata Press, Port Melbourne, pp 79–158
Curtis WM, Morris DI (1993) The student’s flora of Tasmania. St. David’s Park Publishing, Hobart
Davey JW, Blaxter ML (2010) RADSeq: next-generation population genetics. Brief Funct Genomics 9:416–423. https://doi.org/10.1093/bfgp/elq031
Dodsworth S, Pokorny L, Johnson MG, Kim JT, Maurin O, Wickett NJ, Forest F, Baker WJ (2019) Hyb-Seq for flowering plant systematics. Trends Pl Sci 24:887–891. https://doi.org/10.1016/j.tplants.2019.07.011
Doyle J, Doyle J (1987) Genomic plant DNA preparation from fresh tissue-CTAB method. Phytochem Bull 19:11–15
Fahey PS, Fowler RM, McLay TGB, Udovicic F, Cantrill DJ, Bayly MJ (2021) Divergent lineages in a semi-arid mallee species, Eucalyptus behriana, correspond to a major geographic break in southeastern Australia. Ecol Evol 11:664–678. https://doi.org/10.1002/ece3.7099
Frakes LA, McGowran B, Bowler JM (1987) Evolution of Australian environments. In: Dyne GR, Walton DW (eds) Fauna of Australia, vol. 1. Australian Government Printer, Canberra, pp 1–16
French PA, Brown GK, Bayly MJ (2016) Incongruent patterns of nuclear and chloroplast variation in Correa (Rutaceae): introgression and biogeography in south-eastern Australia. Pl Syst Evol 302:447–468. https://doi.org/10.1007/s00606-016-1277-7
Galloway RW, Kemp EM (1981) Late Cainozoic environments of Australia. In: Keast A (ed) Ecological Biogeography of Australia, vol. 1. Junk, The Hague, pp 53–80
Hauenschild F, Matuszak S, Muellner-Riehl AN, Favre A (2016) Phylogenetic relationships within the cosmopolitan buckthorn family (Rhamnaceae) support the resurrection of Sarcomphalus and the description of Pseudoziziphus gen. nov. Taxon 65:47–64. https://doi.org/10.12705/651.4
Hayman DL (1960) The distribution and cytology of the chromosome races of Themeda australis in southern Australia. Austral J Bot 8:58–68
Hope GS (1978) The late Pleistocene and Holocene vegetational history of Hunter Island, north-western Tasmania. Austral J Bot 26:493–514
Hope GS (1994) Quaternary vegetation. In: Hope GS (ed) History of the Australian vegetation: Cretaceous to recent. University of Adelaide Press, Adelaide, pp 368–389
Jackson HD, Steane DA, Potts BM, Vaillancourt RE (1999) Chloroplast DNA evidence for reticulate evolution in Eucalyptus (Myrtaceae). Molec Ecol 8:739–751
Kalyaanamoorthy S, Minh BQ, Wong TKF, von Haeseler A, Jermiin LS (2017) ModelFinder: fast model selection for accurate phylogenetic estimates. Nature Meth 14:587. https://doi.org/10.1038/nmeth.4285
Katoh K, Misawa K, Kuma K, Miyata T (2002) MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucl Acids Res 30:3059–3066. https://doi.org/10.1093/nar/gkf436
Katoh K, Standley DM (2013) MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Molec Biol Evol 30:772–780. https://doi.org/10.1093/molbev/mst010
Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C (2012) Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28:1647–1649. https://doi.org/10.1093/bioinformatics/bts199
Kellermann J (2006) New combinations for two species of Spyridium (Rhamnaceae: Pomaderreae) from the Grampians, Victoria. Muelleria 22:97–104
Kellermann J (2020) A preliminary survey of the leaf-indumentum in the Australian Pomaderreae (Rhamnaceae) using scanning electron microscopy. Swainsona 33:75–102
Kellermann J, Barker WR (2012) Revision of the Spyridium bifidum–S. halmaturinum complex (Rhamnaceae: Pomaderreae) from South Australia and Victoria. Muelleria 30:26–58
Kellermann J, Thiele KR, Udovicic F, Walsh NG (2022) Rhamnaceae. In: Kodela PG (ed) Flora of Australia [partially published]. Australian Biological Resources Study, Canberra. Available at: http://www.ausflora.org.au
Kellermann J, Udovicic F (2007) Large indels obscure phylogeny in analysis of chloroplast DNA (trnL-F) sequence data: Pomaderreae (Rhamnaceae) revisited. Telopea 12:1–22
Kellermann J, Udovicic F, Ladiges PY (2005) Phylogenetic analysis and generic limits of the tribe Pomaderreae (Rhamnaceae) using internal transcribed spacer DNA sequences. Taxon 54:619–631. https://doi.org/10.2307/25065419
Kirkpatrick JB, Fowler M (1998) Locating likely glacial forest refugia in Tasmania using palynological and ecological information to test alternative climatic models. Biol Conservation 85:171–182
Ladiges PY, Kellermann J, Nelson G, Humphries CJ, Udovicic F (2005) Historical biogeography of Australian Rhamnaceae, tribe Pomaderreae. J Biogeogr 32:1909–1919. https://doi.org/10.1111/j.1365-2699.2005.01347.x
Lamitina Lab Protocols (2007) Ethanol Precipitation of DNA. Available at: http://toddlamitina.wixsite.com/lamitina-lab/protocols. Accessed 4 Mar 2016
Larcombe MJ, McKinnon GE, Vaillancourt RE (2011) Genetic evidence for the origins of range disjunctions in the Australian dry sclerophyll plant Hardenbergia violacea. J Biogeogr 38:125–136. https://doi.org/10.1111/j.1365-2699.2010.02391.x
McLay TG (2017) High quality DNA extraction protocol from recalcitrant plant tissue. Available at: https://www.protocols.io/view/high-quality-dna-extraction-protocol-from-recalcit-i8jchun. Accessed 12 Jul 2018
McLay TGB, Ladiges PY, Doyle SR, Bayly MJ (2021) Phylogeographic patterns of the Australian grass trees (Xanthorrhoea Asphodelaceae) shown using targeted amplicon sequencing. Austral Syst Bot 34:206–225. https://doi.org/10.1071/SB20013
Medan D, Schirarend C (2004) Rhamnaceae. In: Kubitzki K (ed) The Families and Genera of Vascular Plants, vol. 6. Springer, Berlin, pp 320–338
Metcalfe DJ, Bui EN (2017) Australia state of the environment 2016: land, independent report to the Australian Government Minister for the Environment and Energy. Australian Government Department of the Environment and Energy, Canberra. https://doi.org/10.26194/6EAM-6G50
Miller M, Pfeiffer W, Schwartz T (2010) Creating the CIPRES science gateway for inference of large phylogenetic trees. LA Gateway Computing Environments Workshop (GCE), New Orleans, pp 1–8. https://doi.org/10.1109/GCE.2010.5676129.
Milner ML, Rossetto M, Crisp MD, Weston PH (2012) The impact of multiple biogeographic barriers and hybridization on species-level differentiation. Amer J Bot 99:2045–2057. https://doi.org/10.3732/ajb.1200327
Neal WC, James EA, Bayly MJ (2019) Phylogeography, classification and conservation of pink zieria (Zieria veronicea; Rutaceae): influence of changes in climate, geology and sea level in south-eastern Australia. Pl Syst Evol 305:503–520. https://doi.org/10.1007/s00606-019-01589-z
New York Botanic Gardens (2020) Index Herbariorum: a global directory of public herbaria and associated staff. New York Botanic Garden, Bronx. Available at: http://sweetgum.nybg.org/science/ih/
Nguyen L, Schmidt HA, Von Haeseler A, Minh BQ (2015) IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Molec Biol Evol 32:268–274. https://doi.org/10.1093/molbev/msu300
Nylander JAA (2004) MrModeltest v2.3. Program distributed by the author. Evolutionary Biology Centre, Uppsala University, Uppsala
Onstein RE, Carter RJ, Xing Y, Richardson JE, Linder HP (2015) Do Mediterranean-type ecosystems have a common history?—Insights from the Buckthorn family (Rhamnaceae). Evolution 69:756–771. https://doi.org/10.1111/evo.12605
Perrin D (2018) Dictionary of botanical names: Australian plant names, meanings, derivation and application. JT Press, Queensland
PlantNET (2021) The NSW plant information network system. Available at: http://plantnet.rbgsyd.nsw.gov.au/cgi-bin/NSWfl.pl?page=nswfl&lvl=sp&name=Spyridium~parvifolium. Accessed 12 Jul 2021
Rambaut A, Drummond AJ (2012) FigTree v1.4.4. Retrieved from http://tree.bio.ed.ac.uk/software/figtree/
Rambaut A, Drummond AJ, Suchard M (2014) Tracer v1. 6. Available at: http://tree.bio.ed.ac.uk/software/tracer/
Richardson JE, Chatrou LW, Mols JB, Erkens RHJ, Pirie MD (2004) Historical biogeography of two cosmopolitan families of flowering plants: Annonaceae and Rhamnaceae. Phil Trans Roy Soc London Ser B Biol Sci 359:1495–1508. https://doi.org/10.1098/rstb.2004.1537
Richardson JE, Fay MF, Cronk QC, Bowman D, Chase MW (2000) A phylogenetic analysis of Rhamnaceae using rbcL and trnL-F plastid DNA sequences. Amer J Bot 87:1309–1324
Rieseberg LH, Soltis DE (1991) Phylogenetic consequences of cytoplasmic gene flow in plants. Evol Trends Pl 5:65–84
Ronquist F, Huelsenbeck JP (2003) MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19:1572–1574. https://doi.org/10.1093/bioinformatics/btg180
Schuster TM, Setaro SD, Tibbits JFG, Batty EL, Fowler RM, McLay TGB, Wilcox S, Ades PK, Bayly MJ (2018) Chloroplast variation is incongruent with classification of the Australian bloodwood eucalypts (genus Corymbia, family Myrtaceae). PLoS ONE 13:e0195034. https://doi.org/10.1371/journal.pone.0195034
Shepherd LD, McLay TGB (2011) Two micro-scale protocols for the isolation of DNA from polysaccharide-rich plant tissue. J Pl Res 124:311–314. https://doi.org/10.1007/s10265-010-0379-5
Specht RL (1972) Origin and migration of the South Australian Flora. In: James AB (ed) The vegetation of south Australia, 2. Government Printer, Adelaide, pp 204–219
Threatened Species Section (2016a) Spyridium parvifolium var. molle (soft dustymiller): species management profile for Tasmania's threatened species link. Available at: http://www.threatenedspecieslink.tas.gov.au/Pages/Spyridium-parvifolium-var-molle.aspx. Accessed 24 Apr 2018
Threatened Species Section (2016b) Spyridium parvifolium var. parvifolium (coast dustymiller): species management profile for Tasmania's threatened species link. Available at: http://www.threatenedspecieslink.tas.gov.au/Pages/Spyridium-parvifolium-var-parvifolium.aspx. Accessed 24 Apr 2018
van Santen M, Linder HP (2020) The assembly of the Cape flora is consistent with an edaphic rather than climatic filter. Molec Phylogen Evol 142:106645. https://doi.org/10.1016/j.ympev.2019.106645
VicFlora (2021) Flora of Victoria: Rhamnaceae. Available at: https://vicflora.rbg.vic.gov.au/flora/taxon/93b292b4-ab2c-4980-926a-6448a297c9e8. Accessed 21 Nov 21
Wiley EO, Lieberman BS (2011) Phylogenetics: theory and practice of phylogenetic systematics. Wiley, Hoboken
Worth JRP, Holland BR, Beeton NJ, Schönfeld B, Rossetto M, Vaillancourt RE, Jordan GJ (2017) Habitat type and dispersal mode underlie the capacity for plant migration across an intermittent seaway. Ann Bot (Oxford) 120:539–549. https://doi.org/10.1093/aob/mcx086
Acknowledgements
We would like to thank The Ecological Society of Australia and the Australasian Systematic Botany Society for their funding support (Holsworth Wildlife Research Endowment and Hansjörg Eichler Scientific Research Fund, respectively) for this research. Catherine Clowes thanks the Botany Foundation (University of Melbourne) for the Megan Klemm Postgraduate Research Award and Sophie Ducker Postgraduate Scholarship and the Soroptimist International of the South West Pacific for the Dame Margaret Blackwood Soroptimist Scholarship. Jürgen Kellerman thanks the Australian Biological Resources Study (ABRS) for their financial support for the broader Rhamnaceae treatment, of which this S. parvifolium project is a component. Thank you to Rob Dabal, Heather Merrylees, Rose Barrett, Todd McLay, Daniel Murphy, Tanja Schuster, Mark Wapstra and Emma Yearwood for collecting samples. Mark Wapstra and the Flinders Island Landcare Group (particularly Kat Hopkins) are thanked for advice on collecting sites. We appreciated the laboratory support provided by Erin Batty and Stephen Wilcox and the herbarium support of Joanne Birch and Margaret Brookes. Thank you to Todd McLay for providing laboratory tips, bioinformatic suggestions and general research troubleshooting guidance. Catherine Clowes would also like to thank Andrew Drinan, Tanja Schuster and Peter Vesk for their support as members of her PhD committee. The Halls Gap Outdoor Community Facebook Group (particularly Yvette Harris and Paul Margetts) are thanked for retrieving a lost field book.
Funding
Open Access funding enabled and organized by CAUL and its Member Institutions. This project was supported by The Holsworth Wildlife Research Endowment administered by The Ecological Society of Australia and the Hansjörg Eichler Scientific Research Fund of the Australasian Systematic Botany Society. Catherine Clowes was supported by the Botany Foundation (University of Melbourne) as a recipient of the Megan Klemm Postgraduate Research Award and the Sophie Ducker Postgraduate Scholarship. Catherine Clowes also received a Dame Margaret Blackwood Soroptimist Scholarship funded by Soroptimist International of the South West Pacific. Jürgen Kellerman was supported by the Australian Biological Resources Study (ABRS).
Author information
Authors and Affiliations
Contributions
The study concept and design were developed by Catherine Clowes with guidance from Michael Bayly, Gillian Brown and Jürgen Kellerman. Sample collection was performed by Catherine Clowes with assistance from Michael Bayly. Laboratory work, bioinformatics and data analysis were performed by Catherine Clowes with support from Patrick Fahey, Rachael Fowler and Michael Bayly. The first draft of the manuscript was written by Catherine Clowes, and all authors commented on versions of the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors have no competing interests to declare that are relevant to the content of this article.
Additional information
Handling Editor: Christoph Oberprieler.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Information on Electronic Supplementary Material
Information on Electronic Supplementary Material
Online Resource 1. Bayesian inference (BI) 50% majority rule consensus tree based on analysis of nuclear ribosomal DNA (nrDNA) with Spyridium cinereum samples removed. Clades not discussed have been collapsed.
Online Resource 2. Maximum likelihood (ML) consensus tree based on analysis of nuclear ribosomal DNA (nrDNA) with Spyridium cinereum samples removed. Clades not discussed have been collapsed.
Online Resource 3. Bayesian inference (BI) 50% majority rule consensus tree based on analysis of nuclear ribosomal DNA (nrDNA) with Spyridium obcordatum samples removed. Clades not discussed have been collapsed.
Online Resource 4. Maximum likelihood (ML) consensus tree based on analysis of nuclear ribosomal DNA (nrDNA) with Spyridium obcordatum samples removed. Clades not discussed have been collapsed.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Clowes, C., Fowler, R., Fahey, P. et al. Phylogeography and classification of Dusty Miller (Spyridium parvifolium; Rhamnaceae): a morphologically variable shrub from south-east Australia. Plant Syst Evol 309, 15 (2023). https://doi.org/10.1007/s00606-023-01851-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00606-023-01851-5