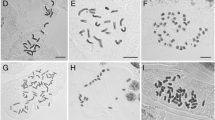
Abstract
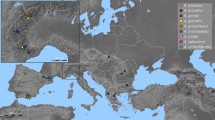
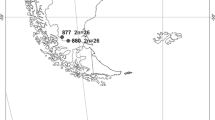
Polyploidy and hybridization are key events in plant evolution. Due to these events, complexes of species can be formed. Dysploidy, a frequent chromosome change in holocentric organisms, may add some difficulties to species delimitation, especially in Cyperaceae. The Eleocharis montana complex is known for its members with overlapping morpho-anatomical features and unclear circumscription. To understand its biological history, several tools were used to investigate different American populations, including morpho-anatomical analysis, genome size estimates, cytogenomic analysis, molecular marker characterization, and genomic in situ hybridization (GISH). Despite overlapping morphological features, it was possible to separate two groups, the first containing E. parodii and E. subarticulata, presenting diploid and dysploid karyotypes, respectively, and small DNA C-values. The second contained E. elegans, E. contracta, and E. montana, with large genomes created by polyploidy. All analyses suggest that E. montana with 2n = 40 is a cytotype of E. contracta with 2n = 20, and both evolved from a natural hybridization involving E. parodii (2n = 10) and a second progenitor that is yet unknown. Furthermore, the GISH results indicated that E. parodii may be an ancestor of E. elegans. All species occur in the probable center of diversification in Austral South America, where the hybridization zone is identified. Fieldwork and information obtained from herbaria indicate that diploid and dysploid species (E. subarticulata and E. parodii) do not occur beyond the center of diversification. However, E. elegans and specially E. montana occur more widely, occupying different flooded environments and landscapes.









Similar content being viewed by others
References
Bennett MD, Leitch IJ (2012) Angiosperm DNA C-values database (release 6.0, Dec. 2012). Available at: https://cvalues.science.kew.org/
Bureš P, Zedek F (2014) Holokinetic drive: centromere drive in chromosomes without centromeres. Evolution 68:2412–2420. https://doi.org/10.1111/evo.12437
Catling PM (1994) Eleocharis compressa × Eleocharis erythropoda, a new natural hybrid spike rush from Ontario. Canad J Bot 72:837–842. https://doi.org/10.1139/b94-108
Catling PM, Reznicek AA, Denford K (1989) Carex lacustris × C. trichocarpa (Cyperaceae), a new natural hybrid. Canad J Bot 67:790–795. https://doi.org/10.1139/b89-106
Chase MW, Knapp S, Cox AV, Clarkson JJ, Butsko Y, Joseph J, Savolainen V, Parokonny AS (2003) Molecular systematics, GISH and the origin of hybrid taxa in Nicotiana (Solanaceae). Ann Bot (Oxford) 92:107–127. https://doi.org/10.1093/aob/mcg087
Da Silva CRM, González-Elizondo MS, Vanzela ALL (2005) Reduction of chromosome number in Eleocharis subarticulata (Cyperaceae) by multiple translocations. Bot J Linn Soc 149:457–464. https://doi.org/10.1111/j.1095-8339.2005.00449.x
Da Silva CRM, Trevisan R, González-Elizondo MS, Ferreira JM, Vanzela ALL (2010) Karyotypic diversification and its contribution to the taxonomy of Eleocharis (Cyperaceae) from Brazil. Austral J Bot 58:49–60. https://doi.org/10.1071/BT09185
Da Silva CRM, Souza TB, Trevisan R, González-Elizondo MS, Torezan JMD, de Souza RF, Vanzela ALL (2017) Genome differentiation, natural hybridisation and taxonomic relationships among Eleocharis viridans, E. niederleinii and E. ramboana (Cyperaceae). Austral Syst Bot 30:183–195. https://doi.org/10.1071/SB17002
Dar TH, Raina SN, Goel S (2017) Cytogenetic and molecular evidences revealing genomic changes after autopolyploidization: a case study of synthetic autotetraploid Phlox drummondii hook. Physiol Molec Biol Pl 23:641–650. https://doi.org/10.1007/s12298-017-0445-8
Del Pozo JC, Ramirez-Parra E (2015) Whole genome duplications in plants: an overview from Arabdopsis. J Exp Bot 66:6991–7003. https://doi.org/10.1093/jxb/erv432
Doležel J, Greilhuber J, Suda J (2007) Estimation of nuclear DNA content in plants using flow cytometry. Nat Protoc 2:2233–2244. https://doi.org/10.1038/nprot.2007.310
Doyle JJ, Doyle JL (1987) A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull 19:11–15
Eiten LT (1969) The vegetative anatomy of Eleocharis interstincta (Vahl) Roem. and Schult. Arq Bot Estado São Paulo 4:187–228
Escudero M, Hahn M, Hipp AL (2018) RAD-seq linkage mapping and patterns of segregation distortion in sedges: meiosis as a driver of karyotypic evolution in organisms with holocentric chromosomes. J Evol Biol 31:833–843. https://doi.org/10.1111/jeb.13267
Fay MF, Cowan RS, Simpson DA (2003) Hybridisation between Schoenoplectus tabernaemontani and S. triqueter (Cyperaceae) in the British Isles. Watsonia 24:433–442
Fazekas AJ, Kesanakuri PR, Burgess KS, Percy DM, Grahm SW, Barrett SCH, Newmaster SG, Hajibabaei M, Husband BC (2009) Are plant species inherently harder to discriminate than animal species using DNA barcoding markers? Molec Ecol Resources 9:130–139. https://doi.org/10.1111/j.1755-0998.2009.02652.x
Ferreira JPR, Venturi S, Trevisan R (2015) Eleocharis guaglianoniana (Cyperaceae), a new species from southern Brazil. J Torrey Bot Soc 142:186–192. https://doi.org/10.3159/TORREY-D-14-0078
Gerrits PO, Smid L (1983) A new, less toxic polymerization system for the embedding of soft tissues in glycol methacrylate and subsequent preparing of serial sections. J Microscop 132:81–85. https://doi.org/10.1111/j.1365-2818.1983.tb04711.x
Gitaí J, Paule J, Zizka G, Schulte K, Benko-Iseppon AM (2014) Chromosome numbers and DNA content in Bromeliaceae: additional data and critical review. Bot J Linn Soc 176:349–368. https://doi.org/10.1111/boj.12211
Goetghebeur P (1998) Cyperaceae. In: Kubitzki K (ed) Flowering plants monocotyledons, vol. 29. Springer, Berlin, pp 141–190
González-Elizondo MS, Peterson PM (1997) A classification of and key to the supraspecific taxa in Eleocharis (Cyperaceae). Taxon 46:433–449. https://doi.org/10.2307/1224386
González-Elizondo MS, Tena-Flores JA (2000) Eleocharis (Cyperaceae) in the New World. In: Wilson KL, Morrison DA (eds) Monocots: systematics and evolution. CSIRO, Sidney, pp 637–643
Heslop-Harrison JS, Schwarzacher T (2011) Organisation of the plant genome in chromosomes. Pl J 66:18–33. https://doi.org/10.1111/j.1365-313X.2011.04544.x
Hinchliff CE, Lliully Aguilar AE, Carey T, Roalson EH (2010) The origins of Eleocharis (Cyperaceae) and the phylogenetic position of Websteria, Egleria, and Chillania. Taxon 59:709–719
Kalendar R, Schulman AH (2006) IRAP and REMAP for retrotransposon-based genotyping and fingerprinting. Nat Protoc 1:2478–2484. https://doi.org/10.1038/nprot.2006.377
Kerbs B, Ressler J, Kelly JK, Mort ME, Santos-Guerra A, Gibson MJS, Caujapé-Castells J, Crawford DJ (2017) The potential role of hybridization in diversification and speciation in an insular plant lineage: insights from synthetic interspecific hybrids. AoB PLANTS 9:1–12. https://doi.org/10.1093/aobpla/plx043
Košnar J, Košnar J, Herbstová M, Macek P, Rejmánková E, Štech M (2010) Natural hybridization in tropical spikerushes of subgenus (Cyperaceae): Evidence from morphology and DNA markers. Amer J Bot 97:1229–1240
Krähmer H (2016) Aerenchyma within the stem. In: Krähmer H (ed), Atlas of weed mapping. John Wiley & Sons, Hoboken, pp 194–214
Llorens C, Muñoz-Pomer A, Bernad L, Botella H, Moya A (2009) Network dynamics of eukaryotic LTR retroelements beyond phylogenetic trees. Biol Direct 4:41
Luceño M, Guerra M (1996) Numerical variations in species exhibiting holocentric chromosomes: a nomenclatural proposal. Caryologia 49:301–309. https://doi.org/10.1080/00087114.1996.10797374
Marasek A, Hasterok R, Wiejacha K, Orlikowska T (2004) Determination by GISH and FISH of hybrid status in Lilium. Hereditas 140:1–7. https://doi.org/10.1111/j.1601-5223.2004.01721.x
Marques A, Moraes L, Aparecida dos Santos M, Costa I, Costa L, Nunes T, Melo N, Simon MF, Leitch AR, Almeida C, Souza G (2018) Origin and parental genome characterization of the allotetraploid Stylosanthes scabra Vogel (Papilionoideae, Leguminosae), an important legume pasture crop. Ann Bot (Oxford) 122:1143–1159. https://doi.org/10.1093/aob/mcy113
Queiroz K (2007) Species concepts and species delimitation. Syst Biol 56:879–886. https://doi.org/10.1080/10635150701701083
Rambaut A (2007) FigTree, a graphical viewer of phylogenetic trees. Available at: http://tree.bio.ed.ac.uk/software/figtree
Ribeiro T, Buddenhagen CE, Thomas WW, Souza G, Pedrosa-Harand A (2018) Are holocentrics doomed to change? Limited chromosome number variation in Rhynchospora Vahl (Cyperaceae). Protoplasma 255:263–272. https://doi.org/10.1007/s00709-017-1154-4
Roalson EH (2008) A synopsis of chromosome number variation in the Cyperaceae. Bot Rev 74:209–393. https://doi.org/10.1007/s12229-008-9011-y
Roalson EH, Hinchliff CE, Trevisan R, da Silva CRM (2010) Phylogenetic relationships in Eleocharis (Cyperaceae): C4 photosynthesis origins and patterns of diversification in the spikerushes. Syst Bot 35:257–271. https://doi.org/10.1600/036364410791638270
Rocha DC, Martins D (2011) Adaptações morfoanatômicas de Cyperaceae ao ambiente aquático. Pl Danin 29:7–15
Rosen DJ, Reid C (2015) Eleocharis × inaequilatera (Cyperaceae), a new hybrid spikerush from the coastal plain of Louisiana and Texas. Phytoneuron 23:1–5
Soltis PS, Soltis DE (2009) The role of hybridization in plant speciation. Annual Rev Pl Biol 60:561–588. https://doi.org/10.1146/annurev.arplant.043008.092039
Souza TB, Chaluvadi SR, Johnen L, Marques A, González-Elizondo MS, Bennetzen JL, Vanzela ALL (2018) Analysis of retrotransposon abundance, diversity and distribution in holocentric Eleocharis (Cyperaceae) genomes. Ann Bot (Oxford) 122:279–290. https://doi.org/10.1093/aob/mcy066
Strandhede SO (1965) Chromosome studies in Eleocharis, subser. Palustres. III. Observations on Western European taxa. Opera Bot 9:1–86
Strandhede SO (1966) Morphologic variation and taxonomy in European Eleocharis, subser. Palustres. Opera Bot 10:1–187
Tenaillon MI, Hufford MB, Gaut BS, Ross-Ibarra J (2011) Genome size and transposable element content as determined by high-throughput sequencing in maize and Zea luxurians. Genome Biol Evol 3:219–229. https://doi.org/10.1093/gbe/evr008
Trevisan R (2009) Eleocharis (Cyperaceae) na Região Sul do Brasil. PhD Thesis, Federal University of Rio Grande do Sul, Rio Grande do Sul
Ueno O, Samejima M, Koyama T (1989) Distribution and evolution of C4 syndrome in Eleocharis, a sedge group inhabiting wet and aquatic environments, based on culm anatomy and carbon isotope ratios. Ann Bot (Oxford) 64:425–438. https://doi.org/10.1093/oxfordjournals.aob.a087861
WCSP: World Checklist of Selected Plant Families (version Sep 2014). In: Roskov Y, Abucay L, Orrell T, Nicolson D, Kunze T, Flann C. Bailly N, Kirk P, Bourgoin T, DeWalt RE, Decock W, De Wever A (eds) Species 2000 and ITIS Catalogue of Life, 2015 Annual Checklist. Naturalis, Leiden. Available at: www.catalogueoflife.org/col
Whitney KD, Ahern JR, Campbell LG, Albert LP, King MS (2010) Patterns of hybridization in plants. Perspect Pl Ecol 12:175–182. https://doi.org/10.1016/j.ppees.2010.02.002
Yang M, Zhou Y, Zhu Q, Lu F, Wang Y, Chen J, Wu Q, Zhang W (2009) AFLP markers in the detection of Scirpus × mariqueter (CYPERACEAE) hybrid in China. Aquat Bot 91:298–302. https://doi.org/10.1016/j.aquabot.2009.08.005
Yano O, Katsuyama T, Tsubota H, Hoshino T (2004) Molecular phylogeny of Japanese Eleocharis (Cyperaceae) based on ITS sequence data, and chromosomal evolution. Int J Pl Res 117:409–419. https://doi.org/10.1007/s10265-004-0173-3
Yano O, Tanaka N, Ito Y (2016) Molecular evidence for a natural hybrid between Isolepis crassiuscula and Isolepis lenticularis (Cyperaceae) in New Zealand. New Zealand J Bot 54:433–445. https://doi.org/10.1080/0028825X.2016.1205106
Zedek F, Šmerda J, Šmarda P, Bureš P (2010) Correlated evolution of LTR retrotransposons and genome size in the genus Eleocharis. BMC Pl Biol 10:265. https://doi.org/10.1186/1471-2229-10-265
Acknowledgements
The authors thank the Brazilian agencies FINEP, Fundação Araucária, CNPq, CAPES, and ProPPG-UEL for their financial support. We thank also the Department of Structural Biology, Molecular, and Genetics from Universidade Estadual de Ponta Grossa for technical support. The authors declare no conflict of interest within this article. A.L.L. Vanzela and R. Trevisan thank CNPq for the productivity scholarship (numbers 309902/2018-5 and 313306/2018-4, respectively).
Author information
Authors and Affiliations
Contributions
L. Johnen, T.B. de Souza, and D. M. Rocha performed all the experiments, wrote and corrected the manuscript. M. S. González-Elizondo and R. Trevisan were responsible for species identifications, support for morphological analyses and for manuscript corrections. S.R. Chaluvadi and J.L. Bennetzen performed the genomic sequencing, supported the bioinformatic analysis, and edited the manuscript. A.L.L. Vanzela designed the study, checked the data analyses, and organized, wrote, and corrected the manuscript.
Corresponding author
Additional information
Handling Editor: Martin A. Lysak.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Information on Electronic Supplementary Material
Information on Electronic Supplementary Material
Supplementary materials include list of species with samples localities and maps, images of achenes morphological analysis, anatomical sections of culms, dendrograms and DNA sequences, and comparative tables and graphs of composition of the repetitive DNA families. All these information complements the results presented in the main text.
Online Resource 1. Data for the Eleocharis montana complex sampled in this study.
Online Resource 2. Geographic localization of the collected populations of Eleocharis parodii, E. contracta, E. elegans, E. montana, and E. subarticulata from Brazilian regions.
Online Resource 3. Sequence alignment used in genetic relationship analysis.
Online Resource 4. Species differentiation using achene shapes and borders.
Online Resource 5. Comparison of morphological features using a distance matrix by the UPGMA method.
Online Resource 6. Measurements of diagnostic morphological structures of Eleocharis montana.
Online Resource 7. Culm anatomies of Eleocharis montana, E. elegans, E. contracta, E. subarticulata, and E. parodii, stained with 0.05% toluidine blue.
Online Resource 8. DNA C-value estimates for the Eleocharis montana complex.
Online Resource 9. Distribution of repetitive DNA elements on the assembled genomes of Eleocharis parodii (2n = 10), E. elegans (2n = 20) and E. montana (2n = 40).
Online Resource 10. In silico microsatellite scanning in three Eleocharis datasets.
Online Resource 11. Phylogeny based on Roalson et al. (2010), using the Brazilian Eleocharis montana complex species (Subgenus Eleocharis), plus E. acutangula (subg. Limnochloa) as an outgroup.
Online Resource 12. Comparison of trees using partial 35S rDNA and chloroplast (petN-psbM and trnC-ycf6) sequences.
Online Resource 13. Dendrogram concatenating data of partial 35S rDNA and chloroplast genes, molecular markers and morphological features using the Mesquite tool.
Rights and permissions
About this article
Cite this article
Johnen, L., de Souza, T.B., Rocha, D.M. et al. Allopolyploidy and genomic differentiation in holocentric species of the Eleocharis montana complex (Cyperaceae). Plant Syst Evol 306, 39 (2020). https://doi.org/10.1007/s00606-020-01666-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00606-020-01666-8