Abstract
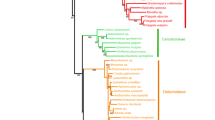
The species phylogeny of Medicago L. (Leguminosae) remains unresolved, as there is significant incongruence between the published gene phylogenies. Here, we compare six of these gene phylogenies of Medicago, inferred from unlinked loci from the nuclear, chloroplast and mitochondrial genomes. Data from all loci were re-analysed, including gap-coding of initial data sets, and dated phylogenies were produced. The patterns of species relationships observed in the six dated phylogenies are compatible with several different biological processes, such as incomplete lineage sorting and hybridisation. A subset of the original sampling that included 29 taxa was also analysed using coalescent-based tree distance comparisons. The observed topological distances suggest that differences between gene phylogenies cannot be solely attributed to incomplete lineage sorting. Hybridisation is strongly suspected to have occurred in the history of many taxa in the genus, because of overlapping divergence times between suspected hybrids and each parental lineage, confirming earlier results based on only two genes. An attempt to reconcile the conflicting histories in a multispecies coalescent analysis, using multiple labels for taxa with hybrid histories, did not produce satisfactory results and may be fatally limited. We conclude that although the currently available data are not sufficient to clarify relationships in Medicago, many cases of hybridisation are probable. The phylogenetic history of the genus is therefore better understood as a network and not a single tree. This raises concerns over previous studies that have used single gene trees as summaries of the history of species relationships.








Similar content being viewed by others
References
Abbott RJ, Hegarty MJ, Hiscock SJ, Brennan AC (2010) Homoploid hybrid speciation in action. Taxon 59:1375–1386. doi:10.2307/20774035
Baquerizo-Audiot E, Desplanque B, Prosperi JM, Santoni S (2001) Characterization of microsatellite loci in the diploid legume Medicago truncatula (barrel medic). Molec Ecol Notes 1:1–3. doi:10.1046/j.1471-8278.2000.00001.x
Bena G (2001) Molecular phylogeny supports the morphologically based taxonomic transfer of the “Medicagoid” Trigonella species to the genus Medicago L. Pl Syst Evol 229:217–236. doi:10.1007/s006060170012
Bena G, Jubier MF, Olivieri I, Lejeune B (1998a) Ribosomal external and internal transcribed spacers: combined use in the phylogenetic analysis of Medicago (Leguminosae). J Molec Evol 46:299–306. doi:10.1007/PL00006306
Bena G, Lejeune B, Prosperi J-M, Olivieri I (1998b) Molecular phylogenetic approach for studying life-history evolution: the ambiguous example of the genus Medicago L. Proc R Soc London, Ser B, Biol Sci 265:1141–1151. doi:10.1098/rspb.1998.0410
Bena G, Prosperi JM, Lejeune B, Olivieri I (1998c) Evolution of annual species of the genus Medicago: a molecular phylogenetic approach. Molec Phylogen Evol 9:552–559. doi:10.1006/mpev.1998.0493
Bena G, Lyet A, Huguet T, Olivieri I (2005) Medicago–Sinorhizobium symbiotic specificity evolution and the geographic expansion of Medicago. J Evol Biol 18:1547–1558. doi:10.1111/j.1420-9101.2005.00952.x
Bertrand YJK, Scheen A-C, Marcussen T, Pfeil BE, Sousa F, Oxelman B (2015) Assignment of homoeologues to parental genomes in allopolyploids for species tree inference, with an example from Fumaria (Papaveraceae). Syst Biol 64:448–471. doi:10.1093/sysbio/syv004
Blanco-Pastor JL, Vargas P, Pfeil BE (2012) Coalescent simulations reveal hybridization and incomplete lineage sorting in Mediterranean Linaria. PLOS One 7:e39089. doi:10.1371/journal.pone.0039089
De Mita S, Santoni S, Ronfort J, Bataillon T (2007) Adaptive evolution of the symbiotic gene NORK is not correlated with shifts of rhizobial specificity in the genus Medicago. BMC Evol Biol 7:210. doi:10.1186/1471-2148-7-210
Delport W, Poon AF, Frost SDW, Pond SLK (2010) Datamonkey 2010: a suite of phylogenetic analysis tools for evolutionary biology. Bioinformatics 26:2455–2457. doi:10.1093/bioinformatics/btq429
Downie SR, Katz Downie DS, Rogers EJ, Zujewski HL, Small E (1998) Multiple independent losses of the plastid rpoC1 intron in Medicago (Fabaceae) as inferred from phylogenetic analyses of nuclear ribosomal DNA internal transcribed spacer sequences. Canad J Bot 76:791–803. doi:10.1139/b98-047
Drummond AJ, Suchard MA, Xie D, Rambaut A (2012) Bayesian phylogenetics with BEAUti and the BEAST 1.7. Molec Biol Evol 29:1969–1973. doi:10.1093/molbev/mss075
Ellison NW, Liston A, Steiner JJ, Williams WM, Taylor NL (2006) Molecular phylogenetics of the clover genus (Trifolium—Leguminosae). Molec Phylogen Evol 39:688–705. doi:10.1016/j.ympev.2006.01.004
Heled J, Drummond AJ (2010) Bayesian inference of species trees from multilocus data. Molec Biol Evol 27:570–580. doi:10.1093/molbev/msp274
Ho SYW, Lanfear R (2010) Improved characterisation of among-lineage rate variation in cetacean mitogenomes using codon-partitioned relaxed clocks. Mitochondrial DNA 21:138–146. doi:10.3109/19401736.2010.494727
Holland BR, Benthin S, Lockhart PJ, Moulton V, Huber KT (2008) Using supernetworks to distinguish hybridization from lineage-sorting. BMC Evol Biol 8:202. doi:10.1186/1471-2148-8-202
Huson DH, Bryant D (2006) Application of phylogenetic networks in evolutionary studies. Molec Biol Evol 23:254–267. doi:10.1093/molbev/msj030
Kay KM, Whittall JB, Hodges SA (2006) A survey of nuclear ribosomal internal transcribed spacer substitution rates across angiosperms: an approximate molecular clock with life history effects. BMC Evol Biol 6:36. doi:10.1186/1471-2148-6-36
Kelchner SA, Thomas MA (2007) Model use in phylogenetics: nine key questions. Trends Ecol Evol 22:87–94. doi:10.1016/j.tree.2006.10.004
Lavin M, Herendeen PS, Wojciechowski MF (2005) Evolutionary rates analysis of Leguminosae implicates a rapid diversification of lineages during the Tertiary. Syst Biol 54:575–594. doi:10.1080/10635150590947131
Lesins KA, Lesins I (1979) Genus Medicago (Leguminosae). A taxogenetic study. Dr. W Junk, The Hague. doi:10.1007/978-94-009-9634-2
Maddison WP, Maddison DR (2006) Mesquite: a modular system for evolutionary analysis. Available at: http://mesquiteproject.org
Martin DP, Lemey P, Lott M, Moulton V, Posada D, Lefeuvre P (2010) RDP3: a flexible and fast computer program for analyzing recombination. Bioinformatics 26:2462–2463. doi:10.1093/bioinformatics/btq467
Maureira-Butler IJ, Pfeil BE, Muangprom A, Osborn TC, Doyle JJ (2008) The reticulate history of Medicago (Fabaceae). Syst Biol 57:466–482. doi:10.1080/10635150802172168
Maynard Smith J (1992) Analyzing the mosaic structure of genes. J Molec Evol 34:126–129. doi:10.1007/BF00182389
Ramadugu C, Pfeil BE, Manjunath KL, Lee RF, Maureira-Butler IJ, Roose ML (2013) Coalescence simulation testing of hybridization versus lineage sorting in Citrus (Rutaceae) using six nuclear genes. PLOS One 8:e68410. doi:10.1371/journal.pone.0068410
Rambaut A (2012) Figtree v 1.4.0. Available at: http://tree.bio.ed.ac.uk/software/figtree/
Robinson D, Foulds L (1981) Comparison of phylogenetic trees. Math Biosci 53:131–147. doi:10.1016/0025-5564(81)90043-2
Şakiroğlu M, Doyle JJ, Brummer EC (2010) Inferring population structure and genetic diversity of broad range of wild diploid alfalfa (Medicago sativa L.) accessions using SSR markers. Theor Appl Genet 121:403–415. doi:10.1007/s00122-010-1319-4
Simmons MP, Ochoterena H (2000) Gaps as characters in sequenced-based phylogenetic analyses. Syst Biol 49:369–381. doi:10.1093/sysbio/49.2.369
Small E (2011) Alfalfa and relatives: evolution and classification of Medicago. NRC Research Press, Ottawa
Small E, Brooks B (1985) Medicago lesinsii, a new Mediterranean species. Canad J Bot 63:728–734
Small E, Jomphe M (1989) A synopsis of the genus Medicago (Leguminosae). Canad J Bot 67:3260–3294. doi:10.1139/b89-405
Small E, Warwick SI, Brookes BS (1999) Allozyme variation in relation to morphology in Medicago Sect. Spirocarpos subsect. Intertextae (Fabaceace). Pl Syst Evol 214:29–47. doi:10.1007/BF00985730
Sousa F, Bertrand YJK, Nylinder S, Oxelman B, Eriksson JS, Pfeil BE (2014) Phylogenetic properties of 50 nuclear loci in Medicago (Leguminosae) generated using multiplexed sequence capture and next-generation sequencing. PLOS One 9:e109704. doi:10.1371/journal.pone.0109704
Steele KP, Ickert-Bond SM, Zarre S, Wojciechowski MF (2010) Phylogeny and character evolution in Medicago (Leguminosae): evidence from analyses of plastid trnK/matK and nuclear GA3ox1 sequences. Amer J Bot 97:1142–1155. doi:10.3732/ajb.1000009
Turini FG, Braeuchler C, Heubl G (2010) Phylogenetic relationships and evolution of morphological characters in Ononis L. (Fabaceae). Taxon 59:1077–1090. doi:10.2307/20773978
Wojciechowski MF, Lavin M, Sanderson MJ (2004) A phylogeny of legumes (Leguminosae) based on analysis of the plastid matK gene resolves many well-supported subclades within the family. Amer J Bot 91:1846–1862. doi:10.3732/ajb.91.11.1846
Wood TE, Takebayashi N, Barker MS, Mayrose I, Greenspoon PB, Rieseberg LH (2009) The frequency of polyploid speciation in vascular plants. Proc Natl Acad Sci USA 106:13875–13879. doi:10.1073/pnas.0811575106
Yoder JB, Briskine R, Mudge J, Farmer A, Paape T, Steele K, Weiblen GD, Bharti AK, Zhou P, May GD, Young ND, Tiffin P (2013) Phylogenetic signal variation in the genomes of Medicago (Fabaceae). Syst Biol 62:424–438. doi:10.1093/sysbio/syt009
Acknowledgments
This work was supported by grants from the Swedish Research Council, the Royal Swedish Academy of Sciences (Grant 2009-5206), Lars Hiertas Minne fund, The Royal Physiographic Society in Lund, Helge Ax:son Johnsons fund, and the Lundgrenska fund to B.E.P; from the P. A. Larssons fund and Lars Hiertas Minne fund to F.S. The authors would like to thank two anonymous reviewers for their input, which helped to improve this manuscript.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflicts of interest in the research.
Additional information
Handling editor: Hervé Sauquet.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplementary material 1
Online Resource 1. Accession Table - GenBank accession numbers of sequences used in the study. Multiple accession numbers listed under ETS-ITS are either partial 18S-ITS1-5.8S-ITS2 sequences (numbers prefixed by AY, DQ, or GQ) or ETS, ITS1, ITS2 sequences (prefixed by AJ, AF or Z). Sequences come from the following sources: Bcop, CNGC5 and rps14-cob (Maureira-Butler et al. 2008), GA3ox1 and matK (Steele et al. 2010), ETS-ITS (Bena 2001), ITS only (Campbell, T.A., unpublished GenBank submission; Ellison et al. 2006; Turini et al. 2010). (XML 93 kb)
Supplementary material 2
Online Resource 2. SplitsTree species network obtained from the concatenated dataset of six loci and 29 taxa. (NEX 8 kb)
Supplementary material 3
Online Resource 3-20. Nexus files with sequence and gap data and xml files for Bcop, CNGC5, rps14-cob, GA3ox1, matK and ITS. (NEX 61 kb)
Information on Electronic Supplementary Material
Information on Electronic Supplementary Material
Online Resource 1. Accession Table - GenBank accession numbers of sequences used in the study. Multiple accession numbers listed under ETS-ITS are either partial 18S-ITS1-5.8S-ITS2 sequences (numbers prefixed by AY, DQ, or GQ) or ETS, ITS1, ITS2 sequences (prefixed by AJ, AF or Z). Sequences come from the following sources: Bcop, CNGC5 and rps14-cob (Maureira-Butler et al. 2008), GA3ox1 and matK (Steele et al. 2010), ETS-ITS (Bena 2001), ITS only (Campbell, T.A., unpublished GenBank submission; Ellison et al. 2006; Turini et al. 2010).
Online Resource 2. SplitsTree species network obtained from the concatenated dataset of six loci and 29 taxa.
Online Resource 3-20. Nexus files with sequence and gap data and xml files for Bcop, CNGC5, rps14-cob, GA3ox1, matK and ITS.
Rights and permissions
About this article
Cite this article
de Sousa, F., Bertrand, Y.J.K. & Pfeil, B.E. Patterns of phylogenetic incongruence in Medicago found among six loci. Plant Syst Evol 302, 493–513 (2016). https://doi.org/10.1007/s00606-016-1278-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00606-016-1278-6