Abstract
Melon is one of the most important horticultural crops in Iran. There are a few studies on the genetic structure of Iranian melon. A set of 18 simple sequence repeat (SSR) primer pairs were used to assess the genetic diversity in a collection of 24 melon accessions representing different botanical groups of Iranian cultivated melons (vars. inodorus, cantalupensis and dudaim), along with 28 reference accessions from diverse geographic origin. All studied SSR loci were polymorphic that confirmed their usefulness for genetic analysis of melons. A total number of 141 alleles were detected, with an average of 7.8 alleles per locus for reference genotypes and 4.38 alleles per locus for Iranian accessions. The low variability within Iranian melon accessions is reflected by the low values of the observed heterozygosity (with an average of 0.119), indicating lack of intercrossing between accessions or a high rate of self-pollination. Values of observed homozygosity for “Suski-e-Sabz” and “Khatouni”, as the most cultivated melon in Iran, were 0.98 and 0.99, respectively. Cluster analysis divided Iranian accessions into two major groups. The highest level of polymorphism was detected among the dudaim group. The analysis of molecular variance indicated that the majority of variation (87 %) was due to the difference within accessions. The average pairwise genetic distance among Iranian accessions was 0.674. Our results showed a distinct separation of dudaim group from the rest of Iranian accessions, even separated two different groups of var. dudaim with different traits. There was a wide genetic distance between Honey Dew, as the most popular member of inodorus group worldwide and “Khatouni”, a major Iranian winter melon (GD = 0.809). This genetic distance shows the importance of Iranian accessions for conservation and use in breeding programs.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Melons, Cucumis melo L., are important horticultural crops in tropical and subtropical regions, which are also grown extensively in temperate climates. Worldwide, more than 25 million tons of melons were produced in 2010, where China, Turkey and Iran were the major producers (FAO 2010). In Iran, more than 50 % of total vegetable production is related to cucurbits. Among them, melon is the most important crop. In 2010, Iranian farmers grew almost 75,000 ha of melon with a total production of 1.31 million tons (FAO 2010).
Historical records indicate that melon was already cultivated in Persia (Iran) during the third millennia BC, and was imported to Europe from Iran and the Caucasus approximately 3,000 years ago (Walters 1989). Melons or muskmelon are native to Iran and adjacent countries toward the west and east. In fact, ‘Musk’ is a Persian word for a kind of perfume and ‘melon’ is derived from Greek words (Robinson and Decker-Walters 1997). The origin of diversity for melon was traditionally believed to be in Africa (Robinson and Decker-Walters 1997), although recent molecular systematic studies, suggested that it may be originated from Asia and then reached to Africa (Renner et al. 2007). Central Asia, Iran, Afghanistan, India, Transcaucasia, Turkmenistan, Tajikistan, and Uzbekistan, as well as Afghanistan and China (Robinson and Decker-Walters 1997) are considered primary diversity centre for melon (Tzitzikas et al. 2009).
Two formal infraspecific taxa within C. melo were recognized by Kirkbride (1993) subsp. melo and subsp. agrestis. Subsp. melo comprises the large-fruited and sweet dessert melons of commerce originating mostly in western Asia and Europe (Nesom 2011). Nesom proposed that no wild forms are represented within subsp. melo. Cultivated forms of C. melo have long been known to be similar to morphologically distinct wild and feral races generally identified as subsp. agrestis. The forms comprising subsp. melo are known strictly as cultivars and almost certainly have arisen from subsp. agrestis, thus their taxonomic treatment as conspecific subspecies is appropriate (Nesom 2011).
The high polymorphism of cultivated melons has led botanists to propose different infraspecific classifications (Pitrat et al. 2000) and several infra-specific classifications have been proposed for melon. Recently, classification focused mainly on central Asian diversity (Pitrat 2008). An overview of infra-specific nomenclature by Pitrat et al. (2000) proposed 16 botanical varieties: conomon, makuwa, chinensis, acidulus and momordica within the subsp. agrestis, and cantalupensis, reticulatus, adana, chandalak, ameri, inodorus, flexuosus, chate, tibish, dudaim and chito within subsp. melo. Nesom (2011) noted that molecular data have not supported the apportionment of the groups among the two subspecies in this classification. Recently, a simplified system is summarized by Nesom (2011) with four varieties [melo (including var. cantalupo], inodorus, reticulatus and flexuosus) within subsp. melo and seven varieties (agrestis, chito, conomon, texanus, dudaim, chate and momordica) within subsp. agrestis.
In Iran, various groups of melon cultigens are grown. The main commercial ones are the sweet type melon of vars. inodorus and cantalupensis. The Inodorus type Iranian accessions differ from other melons belonging to the var. inodorus in their netted skin surface and in occasional rugby ball-shaped fruits. Some researchers suggest that these melons should be considered in other group called iraniansis (Lotfi and Kashi 1999). Var. Cantalupensis are less important in Iran. There are two types of melons in this group: globular-shaped fruits with meridian stripes and soft, spongy flesh, and the other ones are larger, less sweet, always have orange flesh and do not have stripes. Var. dudaim is also cultivated in various areas of Iran. It is characterized by small reddish yellow fruits with ochre stripes, and a round or slightly oval shape with a velvety skin. It has a unique fragrant and musky aroma, and a whitish and insipid pulp that is barely edible. They are originated from Persia (Nesom 2011) and generally cultivated for ornamental or aromatic uses from Turkey and the Caucasus to Afghanistan (Aubert and Pitrat 2006). Some Iranian accessions of var. dudaim are edible, sweet, with typical aroma and big size.
A high level of molecular and morphological variability in leaf, plant, and fruit characteristics has been described in melon species (Akashi et al. 2002; Monforte et al. 2003; Stepansky et al. 1999). C. melo is, therefore, considered the most diverse species in Cucumis (Stepansky et al. 1999). Genetic diversity studies have used isozymes (Staub et al. 1997; Akashi et al. 2002), RFLPs (Zheng et al. 1999), RAPDs (Garcia et al. 1998; Stepansky et al. 1999; Mliki et al. 2001; López-Sesé et al. 2003; Staub et al. 2004; Sensoy et al. 2007; Tanaka et al. 2007), AFLPs (Garcia-Mas et al. 2000), ISSR and simple-sequence repeat (SSR) (Katzir et al. 1996; Staub et al. 2000; Daning-Poleg et al. 2001; López-Sesé et al. 2002; Monforte et al. 2003; Nakata et al. 2005; Tzitzikas et al. 2009) to analyze variability and relationships among melon groups. These studies have assisted in the elucidation of intraspecific relationships in melons from different origins.
In relation with Iranian melons, there are a few studies on the genetic structure which has been done by analysis of molecular markers. Feyzian et al. (2007) examined 38 melon accessions using RAPD analysis; however, they could not separate horticultural groups of melon. Soltani et al. (2010) studied melon accessions using RAPD markers, and observed a high genetic diversity in var. flexuosus and a genetic similarity with reference accessions of large-seed type (vars. inodorus and cantalupensis). This report showed a large variability in the Iranian melon germplasm, but it focused mainly on var. flexuosus. Less attention has been paid to inodorus and dudaim, with regard to their importance as genetic resources for melon improvement.
In the present research, we used a set of previously described SSR markers for defining the genetic relationships between 24 previously untested Iranian accessions, belonging to different melon groups (vars. inodorus, dudaim and cantalupensis), and comparing them with a set of 28 reference accessions to increase our understanding of the genetic variability of Iranian accessions and their relationship with melons from other areas.
Materials and methods
Plant materials and DNA extraction
Fifty-two different accessions were used: 24 Iranian accessions from geographically diverse areas of Iran, and 28 reference genotypes. The Iranian accessions were belonged to the vars. inodorus, cantalupensis and dudaim (Fig. 1), and the 28 reference genotypes were selected to include a broad spectrum of genetic variability. Details about the origin and classification of accessions into groups are given in Table 1.
Mature fruit of Iranian melon examined in the present study. Accession code is according to the number of accession that used in Table 1
Fifteen to 20 seeds of each Iranian accession were germinated at 30 °C in Petri dishes containing a wet filter paper for 48 h. Germinated seeds were planted in pots containing vermiculite under 20–24 °C, 300 μmols m2 s−1 light; 16/8 h (light/dark) photoperiod conditions, and transferred into a greenhouse at the Instituto de Hortofruticultura Subtropical y Mediterránea ‘La Mayora’ (IHSM, CSIC-UMA) in Málaga (Spain). When seedlings developed 2 or 3 true leaves, leaf tissues of 15 individual plants of each accession were used for genomic DNA extraction with Plant DNAzol Reagent (Invitrogen, Germany). The concentration and quality of extracted DNA was determined by reading at 230, 260 and 280 nm using Nanodrop spectrophotometer ND-100 (Nanodrop Technologies, Delaware, USA). DNA was diluted to get a working solution of 10 ng μl−1. DNA from the 15 individual plants per accession was analyzed individually with the SSRs. For the 28 reference genotypes, bulked DNA (5 plants per accession) available at IHSM ‘La Mayora’, was used.
Molecular evaluation
Eighteen SSR primer pairs described previously in the literature and distributed along the melon genome, according to the information of available genetic maps, were used. Their nomenclature assignment were carried out following the indications suggested by the different authors, being named as CM- and CS- (Daning-Poleg et al. 2001), CMBR (Ritschel et al. 2004), CM—N and TJ (Gonzalo et al. 2005), and CMN (Fukino et al. 2007). Reverse primers of each SSR primer pairs were fluorescently labeled with WellRED fluorescent dyes D2, D3 or D4 (Proligo, Paris, France), and PCR amplifications were carried out using a thermal cycler (cycler, Bio-Rad Laboratories, Hercules, CA, USA). Amplification reactions of SSR loci were carried out, with slight variations depending on the primer, as follows an initial cycle at 94 °C for 2–3 min followed by 36 cycles of 15–30 s of denaturing at 94 °C, 51–60 °C for annealing for 15–30 s, and then extension at 72 °C for 5–30 s. Amplicon sizes for these SSR markers were analyzed by capillary electrophoresis using a CEQ 8000/GenomeLab GeXP capillary DNA analysis system (Beckman Coulter, Fullerton, CA, USA).
Statistical analysis
SSR markers were scored as codominant, so homozygous and heterozygous genotypes could be distinguished in individual plants. The analysis of the pooled DNA samples was carried out based on the assumption that the observation of two or more SSR alleles in a single genotype could have resulted from the presence of several heterozygous plants, homozygous plants for the alternative alleles, or a combination of both. All monomorphic loci were discarded for analysis. Statistics of genetic variation (number of observed and effective alleles, Nei’s gene diversity, Shannon’s information index, heterozygosity and polymorphic) were calculated using allelic frequency estimates obtained from genotypic frequencies of SSR loci using the computer program POPGENE (Yeh et al. 1997). In addition, Chi-square test (1:2:1) for Hardy–Weinberg equilibrium for each population was obtained for SSR alleles using this program. Comparison of P values allowed for an assessment of the level of fixation among accessions. The microsatellite data matrix were used to calculate Nei’s distance (Nei 1978), and to generate the corresponding matrix of genetic distance estimates among accessions using POPGENE (Yeh et al. 1997). Cluster analyses were performed on the genetic distance matrix by using UPGMA method to determine the relationships among accessions (dendrograms) based on estimated similarity by Dice and Jaccard coefficient using the NTSYS-pc program version 2.2 (Rohlf 2000). Jaccard’s similarity coefficient with cophenetic correlation coefficient of 0.94 showed greater value than the other method. Therefore, for cluster analysis was used obtained similarity matrix based on the jaccard coefficient, which has high ability for analysis of codominant markers.
The polymorphism information content (PIC) of the SSR used or gene diversity value was calculated as PIC = 1−∑f 2 ij ; where f ij is the frequency of the ith allele for the jth SSR locus (Anderson et al. 1993). PIC values provided an estimate of the discriminatory power of any locus by considering the number of alleles per locus and the relative frequencies of those alleles in the population.
The average number of alleles per locus, within each horticultural group, was corrected for sampling size of each group by dividing the average number of alleles per locus by the number of accessions in each group. The analysis of molecular variance was done using GenAlEx 6.1 software to describe the population structure.
Results
With the 18 SSRs loci used, a total of 141 alleles were detected among all melon genotypes examined herein, with average of 7.8 allele per locus, ranging from three for ‘CMBR34’ and ‘CMN01-54’ to 14 for ‘CMBR98’ and ‘CMCTN86’ loci (data not shown). Variation of allele sizes ranged from 107 to 268 bp. All SSR loci were polymorphic, confirming their usefulness for genetic analysis.
Intra-accession variation and genetic diversity of Iranian melons
All SSRs were polymorphic among Iranian accessions and informative for describing their genotypic variation (i.e., PIC values different from zero). PIC values for SSRs ranged from 0.19 to 0.85 (Table 2), with a mean PIC of 0.49. Four of these SSRs were very informative (PIC > 0.7), with the highest PIC value recorded for CMBR98 (0.85) and followed by CMCT134b, CMCTN86 and TJ24. The mean number of allele and effective alleles for SSR loci were 4.11 and 2.38, respectively (data not shown). Few heterozygous individuals were observed in general for all SSRs, with an average of 0.12, ranging between 0.04 and 0.21 (Table 2). The values of expected heterozygosity (He) for each SSR locus, considering all studied accessions, were always higher than the observed heterozygosity (average He = 0.49), indicating an excess of homozygosity. The Hardy–Weinberg equilibrium was not significant for any of the SSR loci in the Iranian accessions examined.
For the 24 Iranian melon accessions, a total of 79 alleles were detected using the 18 loci, ranging from 18 for accessions “Khatouni”, “Mirpanji”, “Zanjan”, “Dastanbou-Ksa1”, “Dastanbou-Brj1” and “Dastanbou-Ksa2” to 47 for “Dastanbou-Ara2” (Table 3). The average values for observed and effective alleles per accession were 1.46 and 1.23, respectively (Table 3).
The number of heterozygotes observed ranged from 1 to 15 per accession (6.6–100 %) for any locus (data not shown). In all within-accession polymorphic loci cases only two alleles were detected within an accession. The analysis of allelic pattern showed that the number of within-accession polymorphic alleles ranged between 0 and 4 and the number of plant having polymorphic alleles per accession ranged between 0 (“Dastanbou-Ksa1”, “Dastanbou-Ksa2”) and 15 (“Dastanbou-Kurd”, “Dastanbou-Ksa3”) (data not shown). The number of polymorphic loci within accessions ranged from zero in “Mirpanji”, “Dastanbou-Ksa1” and “Dastanbou-Ksa2” to all loci in “Dastanbou-Ara2” (data not shown).
The mean Shannon’s information index (I) in Iranian accessions was 0.19, and ranged from 0 to 0.70 (Table 3). The average observed and expected homozygosity values in Iranian accessions were 0.88 and 0.86, respectively, and this parameter was 1.00 in 4 accessions (“Mirpanji”, “Zanjan”, “Dastanbou-Ksa1” and “Dastanbou-Ksa2”), which indicated that all their individuals were homozygous for all SSR loci examined (no heterozygotes were observed). The average observed and expected heterozygosity values per accession were 0.118 and 0.13, respectively (Table 3), and the average heterozygosity for all the Iranian accessions was 0.126 (data not shown). The average Nei’s expected heterozygosity (gene diversity across alleles) values per accession was 0.125, and ranged from 0 to 0.447. The lowest values for this parameter were detected in “Mirpanji”, “Dastanbou-Ksa1” and “Dastanbou-Ksa2”, as expected given their homozygosity. The highest values were identified in “Dastanbou-Ara2” (Table 3).
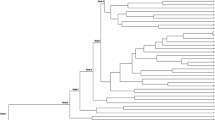
The dendrogram obtained for the Iranian accessions (Fig. 2a) showed two main branches (I and II). Within these groups, three subclusters, two (SC1 and SC2) within cluster I and one (SC3) within cluster II were defined. The SC1 includes thirteen accessions, all from the vars. inodorus and cantalupensis. Within this subcluster, certain associations with geographic origin could be detected. Partitioning of variation at SC1 subcluster resulted in three branches containing accessions from different geographical areas: accessions from north-eastern province (Khorasan), south-eastern province (Kerman) and western and central Iran (Esfahan, Semnan, Hamedan and Azarbayjan) grouped separately from each other. Accessions “Garmak” and “Golpayegan” (var. cantalupensis) and inodorus accessions grouped in a single cluster. Dudaim accession are located in two different branches, there are 2 big groups in dudaim, according to the 18 SSR loci examined. The SC2 subcluster included six accessions, all from the var. dudaim. Most of these accessions (five accessions), have edible sweet fruits, which are bigger in size than the other dudaims. Subcluster grouping SC3 consisted of five accessions, also belonged to the var. dudaim. Most cases (four accessions) in this subcluster had small and insipid fruits that were not edible.
Cluster analysis of 24 Iranian melon accessions (a) and 52 accessions (Iranian and references melon accessions) (b) by UPGMA grouped using genetic distances as estimated by 18 SSR loci (Nei’s distance). In the dendrogram b, accession code is according to the number of accession that used in Table 1 followed by origin (i.e., Iran, Spn, USA, Ger, Gre, Zim, Tur, Lib, Chi, Ind, Jap, Ned and Zam) and, either cultivar group designation as inodorus (In), cantalupensis (Ca), dudaim (Du), conomon (Co), agrestis (Ag) and flexuosus (Fl)
Relationships among Iranian accessions were also visualized by MSD graph including the first and second principal components (Fig. 3). It showed a relatively distinct grouping among accessions by type. The value of first 2 Eigen vectors was 3.15 and 1.20, respectively, and the percent variance accounted with a cumulative of 44.02 (31.89 and 12.13, respectively by each Eigenvalue).
Associations among 24 Iranian melon accessions after principal coordinate analysis (accessions codes according Table 1)
The average genetic distance between any two pairs of Iranian accessions was relatively high (0.674). Distances ranged between 0.0581 (most related accessions; “Khatouni” vs. “Ghasri”) and 6.286 (distantly related; “Zanjan” vs. “Dastanbou-Brj1”). The accession highest distant from all other Iranian accessions was “Dastanbou-Brj1” belonged to var. dudaim, with an average GD of 2.011 (data not shown).
Various population parameters of Iranian accessions were assessed to identify the degree of intra accession variation among horticultural grouping (Table 4). The var. dudaim has the highest values of observed and expected heterozygosity (0.203 and 0.53, respectively), proportion of polymorphic loci (1.00) and specific alleles (17 alleles) among Iranian accessions (Table 4).
The analysis of molecular variance of Iranian accessions (AMOVA) (Table 5) showed that the inter-accession variation was 13 %, and the majority of variation was due to the difference within accessions (87 %). The majority of variation detected could not be cited to differences between horticultural groups (3 %), but to the differences within groups.
Genetic relationship among Iranian and reference accessions
The total numbers of observed and effective alleles for SSR loci among the 52 accessions examined (Iranian and reference accessions) were 4.11 and 2.38, respectively. As expected, reference accessions had a larger genetic variability, with an average of 6.55 alleles per locus, than the Iranian accessions, with 4.05 alleles per locus. Moreover, allelic frequencies were more balanced among reference accessions, which had an average major allele frequency of 0.51 (data not shown) when compared with 0.62, which was the average major allele frequency of Iranian accessions (Table 2). SSRs tested were more informative in references genotypes with a mean PIC of 0.63 (data not shown) compare with 0.49 in Iranian accessions (Table 2).
Cluster analysis of Iranian and references melon accessions using Nei genetic distances is shown in Fig. 2b. “Africanus” was the most distant genotype, as expected since it is an out-group accession belonging to a species different from melo. Var. conomon (“Ginsenmakuwa” and “PI-161375”), var. momordica (“PI-414723” and “PI-124112 –B”), subsp. agrestis (“TGR-1937” and “TGR-1551”) and genotypes “Africanus” (C. africanus) and “Kiwano” (C. metuliferus) had very distinctive positions. There was a relatively definite clustering among Iranian vars. inodorus and cantalupensis with the other genotypes. Four related references genotypes (e.g., “Sudbalkan-4”, “Kirkagac”, “Enfurter netzmelone.1”, and “China-2”), were grouped at node 14 with Iranian accessions of vars. inodorus and cantalupensis. Nine accessions of Iranian inodorus and cantalupensis related closely with each other and were grouped in the same subcluster, even though they were collected from different areas. Inodorus type of Iranian accessions have some differences with ‘Honey Dew’ as the most popular member of inodorus group worldwide, such as netted skin surface, crisp flesh and various fruit shape. In addition, all of Iranian dudaim accessions were grouped in one subcluster of node 8, but “Dastanbou-Brj1” which is closely related with ‘Honey Dew’ at node 2. Relative position of Iranian dudaim accessions in both dendrograms (Fig. 2) are similar, except for “Dastanbou-Brj1”. Also Iranian accessions of vars. inodorus and cantalupensis clustered together and were located in similar positions, with the exception that two reference accessions were placed between “Golpayegan” and “Khatouni” and another two between “Tashkandi” and “Zanjan”.
There was a wide genetic distance between ‘Honey Dew’ and “Khatouni”, a major Iranian winter melon (GD = 0.809). Of all evaluated genotypes (Iranian and references), the most similar genotypes were ‘Honey Dew’ with “Dastanbou-Brj1” (GD = 0.0); the most dissimilar ones were “Ginsenmakuwa” and “Africanus” (GD = 3.526) (data not shown).
Among the references genotypes, a total of 118 alleles were detected, compared with 79 for Iranian accessions. The total number of alleles for all the accessions (Iranian and reference accessions) was 141, thus 62 more alleles were added when the references genotypes were used (data not shown). Seventeen specific alleles were observed in the Iranian accessions of var. dudaim (Table 4).
Discussion
In previous studies, Soltani et al. (2010) used RAPD markers to characterize a number of Iranian melon germplasm focused on var. flexuosus. In this study, we assessed the genetic diversity of Iranian germplasm, including other cultivar groups (inodorus, dudaim, cantalupensis) and some commercial melon cultigens in Iran. All Iranian accessions used in this study were assessed for the first time. The 18 SSRs used herein were sufficient to distinguish all the tested accessions, indicating the usefulness of the chosen marker set to study the genetic variability among the analyzed accessions.
The mean number of allele and effective alleles for SSR loci for the 24 Iranian accessions were 4.11 and 2.38, respectively. Daning-Poleg et al. (2001) found 3.5 alleles using 30 SSR primers on 13 genotypes, while López-Sesé et al. (2002) found 2.4 alleles on 15 Spanish melons, and Tzitzikas et al. (2009) 2.47 alleles on 14 Greek and Cypriot melons. Monforte et al. (2003) detected 6.3 alleles on 27 wild and cultivated melons, which was similar to the mean number of allele for reference genotypes in our study (6.55). This high value was due to various subspecies of melons which they examined. The percentage of gene loci polymorphism that Katzir et al. (1996) obtained using seven SSR primers on eight genotypes was 71 %, while Monforte et al. (2003) found 100 % polymorphism, which was similar to our results. The difference between number of alleles in each locus and number of effective loci obtained herein shows the existence of rare alleles (alleles which have low frequency), so we can use these alleles to identify the Iranian melon genotypes by combination of some genetic loci.
Heterozygosity can be considered a measure of genetic variability. This parameter refers to how much of that variation exists in the population and how that variation is distributed across the alleles of an analyzed locus. Low heterozygosity means little genetic variability. The observed heterozygosity (Ho) is the proportion of heterozygous individuals in population samples, expected heterozygosity is the probability of an individual being heterozygous in any locus. In this study, the highest observed heterozygosity values (Table 2) were achieved with locus CMCTN86 (Ho = 0.21). The observed heterozygosity showed low values in relation at expected heterozygosity values in all loci, indicating an excess of homozygosity. Mean observed heterozygosity per locus in the Iranian accessions examined herein was higher than those in López-Sesé et al. (2002) and Tzitzikas et al. (2009), probably due to the greater diversity of genotypes used in the present study. The tested accessions have been primarily developed and maintained by local farmers and, therefore cross-pollination with other accessions would be expected, resulting in high levels of heterozygosity. Our results showed, however, low observed heterozygosity values among Iranian accessions, indicating lack of intercrossing between them or with other accessions, or a high rate of self-pollination. Another possibility is that the accessions originated from small populations or high levels of inbreeding. The low variability within accession is reflected by the observed heterozygosity between gene loci with an average of 0.119, being lower than expected for all markers (Table 2). There were four accessions with 100 % observed homozygosity. Values of this parameter for “Suski-e-Sabz” and “Khatouni”, as the most cultivated melon in Iran, were 0.98 and 0.99, respectively. This unexpected result is similar to the results of López-Sesé et al. (2002) and Tzitzikas et al. (2009) for other accessions, which were attributed to the use of similar seed among related growers or selection by farmers (Staub et al. 2004). Tzitzikas et al. (2009) assumed that, if out crossing occurred, farmers have made efforts to maintain the genetic originality of the accessions, probably to keep original fruit traits because of regional consumer preferences. It is likely that selection for flavor, good growth, disease resistance, etc. has been practiced by farmers, and this may partially explain the relatively high degree of homozygosity (within accessions) observed in this study. Also the Hardy–Weinberg equilibrium was not significant for any of the SSR loci in Iranian studied accessions, which might be due to the artificial selection which benefits one allele and causes the loss of other alleles through human intervention that prevents the movement of genotypes toward reaching equilibrium.
Genetic variability parameters of reference accessions were higher than the Iranian accessions (i.e., PIC value 0.49 for Iranian accessions compare with 0.63 for references genotypes). With regard to differences among reference accessions, which are from various cultivar groups and species of Cucumis, the genetic variability observed among Iranian accessions is significantly comparable with reference accessions. These results demonstrate that the Iranian melon is diversified and supports the idea of their origin in Asia (Renner et al. 2007). In addition, it indicates the importance of Iranian inodorus and dudaim accessions for the study of origin and diversification of vars. inodorus and dudaim, because of retaining an important level of genetic variability. Unlike foreign commercial cultivars, Iranian analyzed accessions are not obtained from controlled crosses between cultivars and are often result of open pollination, which are grown in various climates of Iran. This could be the reason for their greater genetic diversity. In previous studies when RAPD markers were used (Soltani et al. 2010), dudaim accessions of Iranian germplasm did not clearly differ from inodorus accessions. This difference might be the result of different germplasm used or the higher power of discrimination of SSR markers when compared with RAPDs. Our results showed a distinct separation of var. dudaim from the rest of Iranian accessions, even separated two different groups of var. dudaim with different traits.
Some of the Iranian genotypes belonging to the same geographical area were grouped near each other (i.e., ‘Khorasan’ accessions grouped separately from ‘Esfahan’ and ‘Semnan’ accessions). The most populated among the three observed subclusters was SC1, which includes accessions from inodorus and cantalupensis groups. The present results showed that accessions from the same botanical groups (i.e. inodorus, cantalupensis) clustered together. This result is in accordance with the work of López-Sesé et al. (2003) on Spanish melon, Tzitzikas et al. (2009) on Greek and Cypriot melon, and Monforte et al. (2003) on a broad range of wild and cultivated melon. These observations in various population support mixing between vars. inodorus and cantalupensis and lead us to the idea that former inodorus accessions would have been probably related with cantalupensis as a result of intercrossing and further selection by farmers. Thus, molecular resolution between vars. cantalupensis and inodorus is slight (Staub et al. 1997; Silberstein et al. 1999; Stepansky et al. 1999), despite significant differences between them in morphology and physiology (Nesom 2011). At the present research vars. flexuosus and cantalupensis were grouped together. Var. flexuosus is variable in hypanthium vestiture but molecular data in previous studies also place it within subsp. melo (Silberstein et al. 1999; Stepansky et al. 1999; López-Sesé et al. 2003; Soltani et al. 2010).
All accessions in SC2 subcluster included var. dudaim and most of them have edible sweet fruits and it separated from dudaim accessions in SC3 which have small and insipid fruits. The accessions of SC2 and SC1 which includes inodorus and cantalupensis types clustered together and accessions in both subclusters have edible fruits. This indicated the possibility of outcrossing between vars. dudaim and inodorus or cantalupensis and later selection of the edible types by farmers. Edible types of dudaim group are distributed in various provinces of Iran. There is a possibility that sweet Iranian melon genotypes belonging to vars. inodorus and cantalupensis have been crossed with other non-sweet types such as var. dudaim. Beside researcher believe that the occurrence of sweet-fruited genotypes at least in vars. agrestis and conomon of subsp. agrestis indicating multiple domestications have occurred in parallel with domestication in subsp. melo (Stepansky et al. 1999; Pitrat et al. 2000; Sebastian et al. 2010). Our results according to the 18 SSRs loci examined, showed that the Iranian dudaim group was not clearly grouped with the other varieties in subsp. agrestis base on Nesom (2011) proposed classification, although all of dudaim accessions were relatively located near to wild and feral races of subsp. agrestis in the dendrogram (Fig. 2b).
According to our results, there was a wide genetic distance between ‘Honey Dew’, as the most popular member of inodorus group worldwide and “Khatouni” as one of the most cultivated melon in Iran (GD = 0.809). Other genotypes from foreign sources had wide genetic distance with Iranian accessions. The average genetic distance that López-Sesé et al. (2002) obtained on Spanish genotypes was 0.285 and the maximum genetic distance between genotypes was 0.491, while at the present study the average on Iranian accessions was 0.674. This genetic distance shows the important of Iranian accessions for conservation and use in breeding programs. In Iran, there are five known melon groups (in Persian word) consisting ‘Kharboze’, ‘Talebi’, ‘Garmak’, ‘Dastanbou’ and ‘Khiarchanbar’. Based on the classification proposed by Pitrat et al. (2000) characters of ‘Talebi’, ‘Garmak’, ‘Dastanbou’ and ‘Khiarchanbar’ is adapted to vars. cantalupensis, reticulatus, dudaim and flexuosus, respectively. However, features of ‘Kharboze’ (inodorus type of Iranian accessions) which is the most cultivated groups of melon in Iran, although almost near to vars. inodorus and ameri, but several unique characters, such as netted skin surface, crisp flesh and various fruit shape were observed in inodorus type of Iranian accessions, which are different from features mention for ‘Honey Dew’ as the most popular member of inodorus group worldwide. One has a rugby ball-shaped with completely netted skin while the other is spindle-shaped with longitudinal stripes with netting between these stripes. “Khatouni” placement in var. ameri (Pitrat 2008), also doesn’t quite match with features of this accession. Because climacteric fruit mentioned as a feature for var. ameri, but “Khatouni” is no climacteric with long shelf life and is quite opposite with ‘Ananas’ which mentioned another instance of var. ameri (Pitrat et al. 2000), because of its quite firm and creamy white flesh, compelling scent and rather short shelf life. Since features of inodorus type of Iranian accessions are not mentioned in other melon groups, it is better to call these the iraniansis group as previously suggested (Lotfi and Kashi 1999).
References
Akashi Y, Fukuda N, Wako T, Masuda M, Kato K (2002) Genetic variation and phylogenetic relationships in East and South Asian melons, Cucumis melo L., based on the analysis of five isozymes. Euphytica 125:385–396
Anderson JA, Churchill GA, Autrique JE, Tanksley SD, Sorrels ME (1993) Optimizing parental selection for genetic linkage maps. Genome 36:181–186
Aubert C, Pitrat M (2006) Volatile compounds in the skin and pulp of Queen Anne’s pocket melon. J Agric Food Chem 54:8177–8182
Daning-Poleg Y, Reis N, Baudracco-Arnas S, Pitrat M, Staub JE, Oliver M, Arús P, de Vicente CM, Katzir N (2001) Development and characterization of microsatellite markers in Cucumis. Theor Appl Genet 102:61–72
FAO (2010) FAOSTAT agricultural database. http://apps.fao.org
Feyzian E, Javaran MJ, Dehghani H, Zamyad H (2007) Analysis of the genetic diversity among some of Iranian melon (Cucumis melo L.) landraces using morphological and RAPD molecular markers. J Sci Technol Agric Natur Resour 11:151–162
Fukino N, Sakata Y, Kunihisa M, Matsumoto S (2007) Characterization of novel simple sequence repeat (SSR) markers for melon (Cucumis melo L.) and their use for genotype identification. J Hort Sci Biotechnol 82:330–334
Garcia E, Jamilena M, Alvarez JI, Arnedo T, Oliver JL, Lozano R (1998) Genetic relationships among melon breeding lines revealed by DNA markers and agronomic traits. Theor Appl Genet 96:878–885
Garcia-Mas J, Oliver M, Gomez-Paniagua H, De Vicente MC (2000) Comparing AFLP, RAPD and RFLP markers for measuring genetic diversity in melon. Theor Appl Genet 101:860–864
Gonzalo MJ, Oliver M, García-Mas J, Monfort A, Dolcet-Sanjuan R, Katzir N, Arús P, Monforte AJ (2005) Simple-sequence repeat markers used in merging linkage maps of melon (Cucumis melo L.). Theor Appl Genet 110:802–811
Katzir N, Danin-Poleg Y, Tzuri G, Karchi Z, Lavi U, Cregan PB (1996) Length polymorphisms and homologies of microsatellites in several Cucurbitaceae species. Theor Appl Genet 93:1282–1290
Kirkbride JH (1993) Biosystematic monograph of the genus Cucumis (Cucurbitaceae). Parkway Publishers, Boone
López-Sesé AI, Staub JE, Katzir N, Gómez-Guillamón ML (2002) Estimation of between and within accession variation in selected Spanish melon germplasm using RAPD and SSR markers to assess strategies for large collection evaluation. Euphytica 127:41–51
López-Sesé AI, Staub JE, Gómez-Guillamón ML (2003) Genetic analysis of Spanish melon (Cucumis melo L.) germplasm using a standardized molecular marker array and reference accessions. Theor Appl Genet 108:41–52
Lotfi M, Kashi A (1999) The Iranian melon as a new cultivar group. In: Andrews S, Leslie AG, Alexander C (eds) Taxonomy of cultivated plants: third inter symposium. Royal Botanic Gardens, Kew, pp 447–449
Mliki A, Staub JE, Zhangyong S, Ghorbel A (2001) Genetic diversity in melon (Cucumis melo L.): an evaluation of African germplasm. Genet Res Crop Evol 48:587–597
Monforte AJ, Garcia-Mas J, Arus P (2003) Genetic variability in melon based on microsatellite variation. Plant Breed 122:153–157
Nakata E, Staub JE, López-Sesé AI, Katzir N (2005) Genetic diversity of Japanese melon cultivars as assessed by random amplified polymorphic DNA and simple sequence repeat markers. Genet Res Crop Evol 52:405–419
Nei M (1973) Analysis of gene diversity in subdivided populations. Proc Natl Acad Sci USA 70:3321–3323
Nei M (1978) Estimation of average heterozygosity and genetic distance from a small number of individuals. Genetics 89:583–590
Nesom GL (2011) Toward consistency of taxonomic rank in wild/domesticated Cucurbitaceae. Phytoneuron 13:1–33
Pitrat M (2008) Melon (Cucumis melo L.). In: Prohens J, Nuez F (eds) Handbook of plant breeding—Vegetables, vol 1. Springer, New York, pp 283–315
Pitrat M, Hanelt P, Hammer K (2000) Some comments on infraspecific classification of cultivars of melon. Proc Cucurbitaceae Acta Hortic 510:29–36
Renner SS, Schaefer H, Kocyan A (2007) Phylogenetics of Cucumis (Cucurbitaceae): Cucumber (C. sativus) belongs in an Asian/Australian clade far from melon (C. melo). BMC Evol Biol 7:58–69
Ritschel PS, Lins TCL, Tristan RL, Buso GSC, Buso JA, Ferreira ME (2004) Development of microsatellite markers from an enriched genomic library for genetic analysis of melon (Cucumis melo L.). BMC Plant Biol 4:9
Robinson RW, Decker-Walters DS (1997) Cucurbits. University Press, New York
Rohlf FJ (2000) NTSYS-pc numerical taxonomy and multivariate analysis system version 22. Exeter Publications, Setauket
Sebastian P, Schaefer H, Telford IR, Renner SS (2010) Cucumber (Cucumis sativus) and melon (C. melo) have numerous wild relatives in Asia and Australia, and the sister species of melon is from Australia. Proc Natl Acad Sci USA 107:14269–14273
Sensoy S, Buyukalaca S, Abak K (2007) Evaluation of genetic diversity in Turkish melons based on phenotypic characters and RAPD markers. Genet Res Crop Evol 54:1351–1365
Silberstein L, Kovalski I, Huang R, Anagnostou K, Jahn MMK, Perl-Treves R (1999) Molecular variation in melon (Cucumis melo L.) as revealed by RFLP and RAPD markers. Scientia Hortic 79:101–111
Soltani F, Akashi Y, Kashi A, Zamani Z, Mostofi Y, Kato K (2010) Characterization of Iranian melon landraces of Cucumis melo L. Groups Flexuosus and Dudaim by analysis of morphological characters and random amplified polymorphic DNA. Breed Sci 60:34–45
Staub JE, Box J, Meglic V, Horejsi TF, Mccreight JD (1997) Comparison of isozyme and random amplified polymorphic DNA data for determining intraspecific variation in Cucumis. Genet Res Crop Evol 44:257–269
Staub JE, Danin-Poleg Y, Fazio G, Horejsi T, Reis N, Katzir N (2000) Comparative analysis of cultivated melon groups (Cucumis melo L.) using random amplified polymorphic DNA and simple sequence repeat markers. Euphytica 115:225–241
Staub JE, López-Sesé AI, Fanourakis N (2004) Diversity among melon landraces (Cucumis melo L.) from Greece and their genetic relationship with other melon germplasm of diverse origin. Euphytica 136:151–166
Stepansky A, Kovalski I, Perl-Treves R (1999) Intraspecific classification of melons (Cucumis melo L.) in view of their phenotypic and molecular variation. Plant Syst Evol 217:313–332
Tanaka K, Akashi Y, Nishitani A, Sakata Y, Nishida H, Yoshino H, Kato K (2007) Molecular characterization of South and East Asian melon Cucumis melo L., and the origin of Group Conomon var. makuwa and var. conomon revealed by RAPD analysis. Euphytica 153:233–247
Tzitzikas EN, Monforte AJ, Fatihi A, Kypriotakis Z, Iacovides TA, Ioannides IM, Kalaitzis P (2009) Genetic diversity and population structure of traditional Greek and Cypriot melon cultigens (Cucumis melo L.) based on simple sequence repeat variability. Hortscience 44(7):1820–1824
Walters TW (1989) Historical overview on domesticated plants in China with special emphasis on the Cucurbitaceae. Econ Bot 43:297–313
Yeh FC, Yang RC, Boiley T, Ye ZH, Mao JX (1997) POPGENE, the user-friendly shareware for population genetic analysis. Molecular Biology and Biotechnology Center, University of Alberta, Canada
Zheng XY, Wolff DW, Baudracco-Arnas S, Pitrat M (1999) Development and utility of cleaved amplified polymorphic sequences (CAPS) and restriction fragment length polymorphism (RFLPs) linked to the Fom-2 Fusarium wilt resistance gene in melon (Cucumis melo). Theor Appl Genet 99:453–463
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution License which permits any use, distribution, and reproduction in any medium, provided the original author(s) and the source are credited.
About this article
Cite this article
Raghami, M., López-Sesé, A.I., Hasandokht, M.R. et al. Genetic diversity among melon accessions from Iran and their relationships with melon germplasm of diverse origins using microsatellite markers. Plant Syst Evol 300, 139–151 (2014). https://doi.org/10.1007/s00606-013-0866-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00606-013-0866-y