Abstract
This paper examines macro and micro-level patterns of genome size evolution in the Brassicaceae. A phylogeny of 25 relatives of Arabidopsis thaliana was reconstructed using four molecular markers under both parsimony and Bayesian methods. Reconstruction of genome size (C value) evolution as a discrete character and as a continuous character was also performed. In addition, size dynamics in small chromosomal regions were assessed by comparing genomic clones generated for Arabidopsis lyrata and for Boechera stricta to the fully sequenced genome of A. thaliana. The results reveal a sevenfold variation in genome size among the taxa investigated and that the small genome size of A. thaliana is derived. Our results also indicate that the genome is free to increase or decrease in size across these evolutionary lineages without a directional bias. These changes are accomplished by insertions and deletions at both large and small-scales occurring mostly in intergenic regions, with repetitive sequences and transposable elements implicated in genome size increases. The focus upon taxa relatively closely related to the model organism A. thaliana, and the combination of complementary approaches, allows for unique insights into the processes driving genome size changes.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The fundamental riddle in genome size studies lies in the observations that across all of life there is large variation in genome size but low variation in the number of genes, and that genome size and phenotypic complexity seem to have little relationship with one another (Gregory 2005a; Nagl et al. 1983). This disconnect between genome size/complexity and phenotype was initially dubbed the “C value paradox” (Thomas 1971). Efforts to explain this conundrum have greatly increased our understanding of the genome but questions remain regarding the dominant mechanism for genome size change and the evolutionary forces that may be operating (Bennett and Leitch 2005b).
It has long been clear that dramatic increases in plant genome size can be accomplished via polyploidization (Bennetzen 2000; Bennetzen et al. 2005; Vitte and Panaud 2005; Wendel 2000). However, the lack of obvious mechanisms by which reductions in genome size could be accomplished led to a suggestion that plants have a “one-way ticket to genomic obesity” (Bennetzen and Kellogg 1997). Since then, potential mechanisms by which genome size can increase or decrease, especially via small and gradual changes, have been identified and demonstrated (Devos et al. 2002; Petrov 1997). Nevertheless, it remains unclear if these mechanisms are actually responsible for the changes observed across evolutionary time.
The small size of the A. thaliana genome (n = 5, C = 0.16 pg), among the smallest published (Bennett and Leitch 2005a), has made it a favored system in which to investigate questions of gene regulation and function, and genome organization. This has revealed a complicated history of genome and chromosome level duplication and rearrangement in A. thaliana as illustrated by Lysak et al. (2006) (for a review see Schranz et al. 2007). It is clear that simply adding together the micro-level processes will not necessarily provide us with a full understanding of genome evolution (Charlesworth et al. 2001; Gregory 2004, 2005b). If we are to have a realistic understanding of general processes in genome evolution then we must branch out from “model” species (Gregory 2005a; Koch and Kiefer 2005). Thus, greater clarity might be leveraged by analyzing the genomic data we now have for A. thaliana within a comparative framework that includes its relatives.
The family Brassicaceae has several attributes that make it attractive for the study of genome size evolution. The family is large, with ca. 340 genera and 3,700 species (Al-Shehbaz et al. 2006), has great variability in life history, habit, and ecology, and is known to have undergone recurrent polyploidization over the course of its evolution. The family displays a moderate diversity of genome sizes, (Johnston et al. 2005; Nagl et al. 1983) and a range of monoploid chromosome numbers (Warwick and Al-Shehbaz 2006). Finally, there is a considerable amount of genome level information already available from multiple genera and species within Brassicacae in addition to A. thaliana. These include emerging model species such as Arabidopsis lyrata, Boechera stricta, Brassica oleracea, and Capsella rubella.
Previous studies have begun to untangle the phylogenetic relationships within the Brassicaceae (Beilstein et al. 2006; Hall et al. 2002, 2004; Koch et al. 2001; O’Kane and Al-Shehbaz 2003; Warwick et al. 2002). A few studies have also approached the evolution of genome size within a phylogenetic context (Bennetzen and Kellogg 1997; Jakob et al. 2004; Soltis et al. 2003; Weiss-Schneeweiss et al. 2006). However, only one study has focused on genome size evolution among the taxa near to Arabidopsis thaliana (Johnston et al. 2005). But, it relied on only one marker for the reconstruction of the phylogeny and did not apply significance tests to the evolution of the character. Furthermore, no study has attempted to investigate the micro-level patterns that might reveal the actual mechanisms responsible for genome size changes among these evolutionary lineages. Thus, there is an opening for a study that combines a solid phylogeny of species related to A. thaliana with reconstruction and testing of genome size evolution upon that phylogeny and an investigation of possible mechanisms.
Here, we present the results from a comparative investigation into the evolutionary history of the small genome of A. thaliana that is centered around a robust and well-sampled phylogeny recovered using four genes and sampling of the genera thought closely related to Arabidopsis (Al-Shehbaz et al. 2006; Beilstein et al. 2006; Koch and Kiefer 2005). The character mapping of genome sizes onto this phylogeny thus gives a broad level view into the pattern of genome size evolution among A. thaliana and its close relatives. In addition, we delve into the patterns and mechanisms underlying the change in genome size. This is accomplished by comparing end-sequences of genomic shotgun clones from A. lyrata (O’Kane and Al-Shehbaz 1997) and B. stricta (Al-Shehbaz 2003) against the fully sequenced genome of A. thaliana. This combination of approaches allows for a robust analysis of genome size evolution in this area of Brassicaceae.
Materials and methods
Taxon sampling
We included 26 terminal taxa representing 23 genera of Brassicaceae (see Table 1), including as many genera as possible from the Cardaminoid, and Halimolobine genera near Arabidopsis (Koch et al. 2005). We used sequences from Aethionema as an outgroup based on the availability of sequences and the use of this taxon as an outgroup by other studies of this area of Brassicaceae (Bailey et al. 2002; Koch et al. 2001).
DNA isolation, amplification, and sequencing
Total genomic DNA was extracted from silica-gel dried material, herbarium specimens, or fresh leaf material using a modified 2× cetyltrimethylammoniumbromide (CTAB) method (Doyle and Doyle 1987) or, alternatively, using QIAGEN Genome DNA prep (Qiagen GmbH, Düsseldorf, Germany). The DNA was then used to amplify four markers by PCR under standard conditions: Chalcone Synthase (Chs), ITS, matK, and trnL-F. The Chs primers span the last exon and were the same as used by Koch et al. (1999). The primers used to amplify matK, trnK-710F and trnK-2R, originally derived from Johnson and Soltis (1995), were the same as used by Koch et al. (2001). The ITS primers amplified a region that included the 5.8S region and the flanking portions of the 18S and 26S regions (Baum et al. 1998; White et al. 1990). The trnL primers used corresponded to trnC and trnD from Taberlet et al. (1991). All of the primers used for amplification are summarized in Table S1 in Electronic supplementary material.
All of the PCR products were directly sequenced using the same primers as for PCR with BigDye chemistry (ABI Perkin-Elmer, Foster City, California, USA). Reactions that could not be directly sequenced were first cloned and 6–12 clones were then sequenced. Under this procedure PCR products were gel-cleaned and then cloned using the TOPO TA™ kit (Invitrogen Corp., Carlsbad, California, USA). Plasmid DNA was purified using Qiaquick Mini-Prep kits (Qiagen Corp.,) and then sequenced using the aforementioned amplification primers and the following internal primers for matK: matK-1010R, matK-1089R, and matK-1495R. These primer sequences are also included in Table S1.
Phylogenetic analyses
Sequences were assembled and edited using SeqMan™ (DNASTAR Inc., Madison, Wisconsin, USA), and manually aligned using MacClade™ (Sinauer Associates Inc., Sunderland, Massachusetts, USA). There was little ambiguity in alignment. However, when there were alternatives that seemed equally plausible, we chose the alignment that minimized the number of potentially informative characters to avoid biasing the results. Sequences generated for this study were submitted to GenBank with accession numbers as given in Table 1. Voucher specimens were deposited at M and DUKE.
Parsimony analyses were performed using PAUP* version 4.0b10 (Swofford 2002). All searches for most parsimonious trees were conducted with tree bisection-reconnection (TBR) heuristic searches and 100 random taxon addition replicates. Bootstrap analyses were done with 100 bootstrap replicates (Felsenstein 1985), as implemented in PAUP* version 4.0b10, with the search conditions the same as described previously. In order to test congruence between our nuclear and chloroplast markers, partition homogeneity tests were conducted in PAUP* 4.0. We used the heuristic search strategy previously described with 999 random replicates to generate the distribution resulting in the P values.
Bayesian phylogenetic analysis on the combined dataset was performed using Metropolis-coupled Markov-chain Monte Carlo (MC3) analysis, as implemented in the program MrBayes 3.1 (Huelsenbeck and Ronquist 2001). Given that Bayesian analysis requires a model of molecular evolution to be specified, we used the program ModelTest 2.2 (Posada and Crandall 1998) to select an appropriate model of DNA substitution. For this dataset, ModelTest 2.2 selected a general time-reversible model (GTR) with a discrete approximation to the gamma distribution for rate variation among sites (Γ) and a proportion of invariant sites (I) (Yang 1994a, b).
Tree searching using MrBayes was performed by running five coupled chains initiated from random trees (sequential heat = 0.2) for 1,000,000 generations with trees sampled every 100th generation (Huelsenbeck and Ronquist 2001). At the end of the run, convergence was evaluated by visual inspection of a graph of likelihood as a function of generation. A conservative burn-in period was identified, and only post-burn-in trees were saved. This analysis was repeated four times, and the majority-rule consensus trees from each run were compared to evaluate mixing. The four sets of post-burn-in trees were then pooled to form a majority rule consensus tree, and this pool was taken as the best representation of the posterior distribution of tree topology and model parameters (Huelsenbeck et al. 2002; Miller et al. 2002).
The phylogenetic placements of taxa that were uncertain were further examined using the Templeton test (Templeton 1983) and the Shimodaira-Hasegawa test (Kishino and Hasegawa 1989; Shimodaira and Hasegawa 1999) as implemented in PAUP* 4.0b10 (Swofford 2002) on the combined dataset. In each case, the most optimal tree was compared to a tree recovered in searches that constrained the taxon of interest to its alternative placement. The Shimodaira-Hasegawa tests assumed the GTR + Γ model parameters (Yang 1994a, b). If the P values were less than 0.05, the two trees were considered significantly different.
Reconstruction of genome size evolution
Genome size measurements for each taxon were obtained from fresh material of diploid individuals (i.e. we excluded polyploid individuals) using a Partec™ CCA-II flow-cytometer and CyStain UV precise P nuclei extract and staining kit (Partec GmbH, Münster, Germany). All measurements used B. oleracea, B. rapa, Matthiola incana, or some mixture of those three species as standards and were recorded as a ratio of the size of the plant being measured to B. oleracea. These ratios were converted into 2C values using a published value of 0.710 pg for B. oleracea (Johnston et al. 2005). One C value (Cardamine amara) was obtained from Johnston et al. (2005). The values we obtained agreed with those available in the C values Database (http://www.kew.org/uk/cval/homepage.html). These values and the phylogeny were used to examine the phylogenetic constraints on genome size evolution as well as its directionality, mode, and tempo.
The genome sizes (2C) were converted into a discrete character by grouping them into nine size ranges of 0.1-pg intervals and each interval designated as a character state. This character of genome size was mapped onto the phylogeny using MacClade™ under ACCTRAN parsimony optimization under both an ordered and unordered treatment of the character. In order to test for phylogenetic constraint of genome size evolution, a permutation test was conducted in MacClade™ that resembled a test of phylogenetic autocorrelation for discrete characters. To conduct this test, the character of genome size was replicated 999 times (for a total of 1,000 characters) and then the states were shuffled among the taxa. This resulted in a distribution of steps across the 1,000 characters that approximated the normal distribution with a slight skew toward longer trees. If the number of steps necessary to explain the original character was within the lower or upper 5% of the distribution, then the distribution of genome-size on the phylogeny was considered to be significantly non-random.
Analysis of genome size (2C) as a continuous character was done using Continuous (Pagel 1997, 1999). This program is able to test whether there is directionality to genome size changes by comparing likelihood ratio statistics recovered from a random walk model and a directional random walk model. The program also allows for the estimation of parameters (Lambda, Kappa, and Delta), which, respectively, capture the degree to which the evolution of the character is explained by the phylogeny, whether the changes occur gradually, and whether the pattern of changes is consistent with an adaptive scenario. In order to conduct a more conservative test, the values for these parameters were first estimated under Maximum Likelihood and the random walk model and then used to conduct a Maximum Likelihood ratio test between the directional and non-directional random walk models.
The evolution of chromosome numbers (2n values) across the phylogeny was also analyzed using MacClade™ and Continuous. The values were obtained from published sources (Al-Shehbaz et al. 2006; Bailey 2006; Lysak et al. 2003; Warwick and Al-Shehbaz 2006). For the MacClade™ analyses, the chromosome counts (2n) for each taxon were converted into a discrete character state (a range of 14 possible states) and treated as unordered or ordered as described earlier. We also performed the modified permutation test as described for genome size measurements on the character of chromosome number. The Continuous analyses used the same methods as used for genome size. We also used Continuous to test whether or not the evolution of genome size (2C, pg) and chromosome number (2n) are correlated on the phylogeny. In this case, the values of Kappa, Delta and Lamda were first optimized using both characters. Then all of the possible combinations of directional and non-directional models were compared while forcing the covariance of the two characters in the null model to be zero (i.e. complete independence).
Genomic library construction
Size-selected genomic shotgun libraries for both B. stricta and A. lyrata were constructed in the pZERO-II plasmid vector (Invitrogen, Karlsruhe, Germany). Approximately 5 μg of plant genomic DNA were partially digested with Sau3AI (New England Biolabs, Frankfurt am Main, Germany). The DNA was then dephosphorylated by incubation with 0.1 U of CIAP (Amersham, Freiburg, Germany), extracted once with phenol/chloroform, ethanol-precipitated, dissolved in TE, and size-fractionated by means of gel electrophoresis on a 0.7% agarose gel. DNA fragments in the size-interval from 4.5 to 5.5 kb were excised in-block and purified using MiniElute columns (Qiagen). Approximately 40 ng of insert DNA were ligated to 10 ng of BamHI-digested pZERO-II vector with t4-ligase (Invitrogen). Into DHB10 ElectroMax Escherichia coli cells (Invitrogen), 0.5-μL aliquots were electroporated and they were then screened with kanamycin. The primary B. stricta library contained 12,000 colony-forming units (cfu) with an average insert size of approximately 5.0 kb and the primary A. lyrata library contained 4,000 cfu with an average insert size of approximately 5.5 kb.
End sequencing of genomic clones and comparison to A. thaliana genome
Nine hundred and sixty (960) genomic shotgun plasmid clones from both A. lyrata and B. stricta were end-sequenced, resulting in two sequences per clone. Sequencing was performed using BigDye chemistry and bases were called with the phred algorithm in the Phred/cross_match/Phrap program (Ewing and Green 1998; Ewing et al. 1998). Vector sequences and low quality regions were then removed with the algorithm cross_match of the Phred/cross_match/Phrap program. A base-call was considered to be of high quality if it had a phred quality score of at least 30, and the five left and right neighboring base-calls had a phred score of at least 20. We also removed all sequences having similarity to plastid (i.e. chloroplast or mitochondrial) sequence.
In order to examine the influence of repetitive elements in our sequences, two bioinformatic filtering regimes were implemented. These filtering regimes differed principally in the inclusion versus exclusion in the resulting datasets of end-sequences having similarity to repetitive sequences. Thus, two datasets were derived from the full set of end-sequences, one for each filtering regime. Comparison of these two datasets allows us to infer the role of repetitive DNA in genome size changes (e.g. transposable elements, tandem repeats, etc.).
To compile the first dataset (hereafter “unfiltered”), BLASTn with default parameters was used to compare each set of end-sequences to itself in order to produce a non-redundant collection of inserts with paired end-sequences (Altschul et al. 1997). BLASTn (gap-opening penalty = 5, gap-extension penalty = 2, all other parameters set to default) was used to identify similarity between every non-redundant end-sequence and the A. thaliana nuclear genome (chromosome pseudomolecules NC_003070, NC_003071, NC_003074, NC_003075 and NC_003076; http://ftp.ncbi.nlm.nih.gov/genomes/Arabidopsis_thaliana). The resultant BLAST output was parsed, and end-sequences with at least one match to the A. thaliana nuclear genome with an E value greater than 10e−20 were scored as a “hit”; end-sequences that failed to meet this criterion were deemed as not having similarity to the A. thaliana nuclear genome (no hit).
To compile the second dataset (hereafter “filtered”), a number of additional bioinformatic filtering steps were applied. First, in order to avoid over-sampling regions of the genome resulting from the non-random cutting of the restriction enzyme, all sequence reads were clustered separately for each species with CAP3 (Huang and Madan 1999) and the end-sequences from clones that formed clusters were discarded from further analysis. Second, the remaining end-sequences were compared against the A. thaliana genome with BLASTn (arabi all dna v170902) and scored as a “hit” if they had at least one match with an E value greater than 10e−20. End-sequences that did not meet this criterion and end-sequences for which the logarithm of the difference of the E value of the best and the second best hit was less than 10 (to exclude repetitive sequences) were scored as “no hit”. The third and final step removed from further analysis end-sequences from clones for which the two end-sequences either matched to different chromosomes or spanned a region of more than 25,000 bp on the same chromosome of the A. thaliana genome.
Both bioinformatic filtering regimes resulted in three classes of clones: those from which both end-sequences passed all filtering steps (two-hit clones), those from which only one end-sequence passed all filtering steps (one-hit clones), and those from which neither end-sequence passed all filtering steps (zero-hit clones). For both datasets, we tested whether the two ends of the genomic clones were independent from one another using a chi-square test on the number of clones in each category. The chi-square test examined if the observed distribution of clones among these three categories was significantly different from the distribution expected under an assumption of independence between the two ends in their “hit” versus “no-hit” status.
Gel-based estimation of clone insert sizes
In order to conduct a conservative comparative analysis, only two-hit clones from the filtered dataset were used for pair-wise comparisons of clone insert sizes to their A. thaliana homolog. To measure the size of the insert, clones were digested to completion with Mph1103I (MBI Fermenta) and size-fractionated on a 0.8% agarose gel, together with size standards. Sizing of fragments was performed on an UV visualizer. To get the size of the homologous region in A. thaliana, the end-sequences of these clones were pair-wise aligned with their best matching A. thaliana BAC sequence using cross_match, which employs the Smith–Waterman algorithm for local pairwise alignments (Ewing et al. 1998; Smith and Waterman 1981). Using the alignment, the start and end positions of the query sequence and of the corresponding BAC clone were determined. These positions were then mapped onto the A. thaliana pseudochromosomes using the MIPS annotation (v170902).
Fully sequencing a s-set of 15 clones of A. lyrata
Inserts of 15 of the two-hit clones from the filtered dataset were selected for complete sequencing using the GeneJumper Primer Insertion Kit for Sequencing (Invitrogen). The two-hit clones were first assigned to one of three categories based on their size relative to their homologous A. thaliana region (larger, smaller, or the same size). Within each category, clones were randomly chosen. After electroporation into GeneHogs electrocompetent E. coli cells (Invitrogen), colonies were picked, grown, and the plasmid isolated using standard procedures. Sequencing, base-calling, and low-quality sequence trimming were performed as described earlier. Sequences were assembled with CAP3 and manually edited.
Sequences were compared with the A. thaliana genome using BLASTn. Where significant matches existed, sequences were pair-wise aligned with A. thaliana BACs using DOTTER (Sonnhammer and Durbin 1995) and VISTA (Bray et al. 2003; Couronne et al. 2003). Sequence annotation was performed manually by comparison of the sequences with homologous A. thaliana regions, as well as with the help of the GENSCAN gene prediction algorithm (Burset and Guigo 1996), NetGene2 splice-site prediction on-line server (Brunak et al. 1991; Hebsgaard et al. 1996), and RepBase transposon database (Jurka et al. 2005).
Results
Phylogenetic analysis
The aligned matrix of all four markers was 3,376 bp in length, which comprised 491 (14.5%) parsimony informative characters. For the nuclear markers, the aligned matrix of Chs sequences was 706 bp long, of which 173 (24.5%) were parsimony informative, whereas the aligned matrix of ITS sequences was 651 bp in length, of which 165 (25.0%) were parsimony informative. Among our chloroplast markers, the aligned matrix of matK sequences was 1,517 bp long, of which 117 (7.7%) were parsimony informative, whereas the aligned matrix of trnL sequences was 502 bp long, of which 36 (7.2%) were parsimony informative. The genus Aethionema was used as an outgroup.
Parsimony analysis of the combined dataset recovered a single most-parsimonious tree with a length of 2,165 (CI = 0.624, RI = 0.533). In all four replicate Bayesian analyses of the combined dataset, the same topology was recovered with a log-likelihood score of approximately −16,260. The parsimony and Bayesian trees conflicted in the placement of two lineages, one leading to Lepidium and the other a clade consisting of the genera Fourraea, Sisymbrium, and Eutrema. The positions of these two lineages are reversed between the two analyses. The Bayesian topology, however, is only one parsimony step longer.
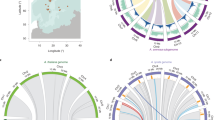
The topology recovered via Bayesian analyses was used in all subsequent analyses and is the tree shown in Fig. 1. Bayesian posterior probabilities are placed above branches leading to clades and parsimony bootstrap values below (Fig. 1). This tree, rather than the parsimony tree, was used because: (1) among the individual markers, only Chs supports a basal position for Lepidium; (2) topological tests could not reject the more nested placement of Lepidium (indeed, there is only a one-step difference under parsimony); and, (3) another study using a different mix of markers supports a distant relationship between Sisymbrium, Eutrema (Thellungiella), and the “Halimolobine” Brassicaceae (Hall et al. 2002).
Genome size measurements
The genome size (2C) values for the taxa included in this study and their chromosome counts (2n) are listed in Table 1 and also shown in Fig. 1. Conversion of the continuous measurements of genome size (2C) into discrete size categories resulted in nine possible character states (see Fig. 1). The small size of the A. thaliana genome is clearly revealed to be a derived state under a parsimony reconstruction of character evolution. Similarly, B. stricta also has a genome size that is smaller than its immediate relatives.
Reconstruction of the evolution of genome size (2C) in MacClade required 13 steps when the states were considered to be unordered and 17 when considered as ordered. These comprise six unambiguous transitions to a larger genome and seven unambiguous transitions to a smaller genome (see Fig. 1). Under our modified permutation test, this total number of transitions is not statistically different from what is expected if the character states were randomly distributed across the tree when the character is unordered. However, the 17 steps when the character was treated as ordered fall into the smallest 1.7% of a random distribution of possible steps in the modified permutation test, suggesting phylogenetic constraint. Analysis of the evolution chromosome numbers (2n) in MacClade revealed that the observed number of character state changes is lower than the average value under a random distribution, suggesting phylogenetic constraint. Under the permutation method in MacClade described earlier, this result was significant when the character was treated as unordered but not significant when the character was treated as ordered.
Our analyses of genome size (2C) and chromosome number (2n) as continuous characters in the program Continuous (Pagel 1997, 1999) were unable to reject the null-hypothesis that genome size and chromosome number each evolved under a constant-variance random walk model (Lamba not significantly different from 1; Model A not rejected). Furthermore, we failed to reject the null hypothesis that genome size (2C) and chromosome number (2n) evolved independently (i.e. not correlated; ρ = 0.205). However, whereas there was significant support for genome size having evolved under a model of punctuated equilibrium in which most change happened at the tips (Kappa = 0, Delta = 2.048), the evolution of chromosome number was found to be significantly dependent on branch length (Kappa = 3).
End sequencing of Arabidopsis lyrata and Boechera stricta genomic clones
The 960 clones we end-sequenced from each of the A. lyrata and B. stricta libraries resulted in a total of 3,840 sequence reads with a mean read length of 390 bp. The unfiltered dataset contained 619 clones from the A. lyrata library and 686 from the B. stricta library. The filtered dataset comprised 477 clones for A. lyrata and 568 for B. stricta. The BLASTn comparisons to A. thaliana (cutoff E value of 1−20) revealed that a higher proportion of A. lyrata clones than B. stricta clones had a significant match (46 vs. 31%). The full complement of end-sequences (over 10 bp) was submitted to GenBank with accession numbers DX922572–DX924258 for A. lyrata and DX924259–DX926011 for B. stricta.
The additional quality trimming and filtering steps used to created the filtered dataset reduced the numbers of clones used for the analysis of segment lengths (i.e. two-hit clones) to 172 for A. lyrata and 150 for B. stricta. Almost all of the clones have the expected insert size range of 5–6 kb. Pairwise comparisons of the estimated sizes of the inserts from these clones to the lengths of homologous regions in A. thaliana are shown in Fig. 2.
Graphs comparing sizes of A. lyrata and B. stricta inserts from two-hit clones in the filtered dataset to their homologous region in the A. thaliana genome. The top panels show the direct comparison of the sizes of the A. lyrata and B. stricta inserts from the genomic shotgun libraries to the sizes of their A. thaliana homologs. The bottom panels show the distribution of the size differences between the A. lyrata and B. stricta inserts to their A. thaliana homologs
Both A. lyrata and B. stricta homologous regions are slightly but significantly larger compared with A. thaliana in a pairwise Wilcoxon Signed Rank test (Table 2). The mean and median for the size differences between A. lyrata clones and their homologous A. thaliana regions were, respectively, 396.9 and 205.5 bp larger (SD = 1,795.2). Meanwhile, the mean and median for the size differences between B. stricta clones and their homologous A. thaliana regions were, respectively, 82.7 and 277.5 bp larger (SD = 2,664.3). The distribution of comparative sizes in both species is unimodel but with a tail of smaller clone sizes (see Fig. 2). These measurements were not made for the set of two-hit clones in the unfiltered dataset because the ambiguity of homology to the highly similar regions in A. thaliana could make those values unreliable.
The distributions of the clones among the categories of two-hit (both end-sequences of a clone passed all filtering steps), one-hit (only one end-sequence passed all filtering steps), and zero-hit (neither end-sequence passed all filtering steps) were different between the two filtering regimes for both A. lyrata and B. stricta (Table 3). The numbers observed for the unfiltered dataset were significantly different from the expectations for all three categories under the chi-square test (A. lyrata: χ 2 = 124.13, P < 0.001; B. stricta: χ 2 = 88.85, P < 0.001) due to a deficit of one-hit clones. For the filtered dataset, the observed numbers were not significantly different from the expected according to the chi-square test (A. lyrata: χ 2 = 2.28, P = 0.320; B. stricta: χ 2 = 0.06, P = 0.971). Given that these two datasets differ mainly in the rigorousness with which they exclude repetitive sequences, comparing the results from these two bioinformatic regimes suggests that repetitive DNA is responsible for excesses in the zero-hit and two-hit categories for the unfiltered dataset. This pattern is predicted under a coarse indel model of genome size changes, where insertions and deletions >5 kb result in the hit-status of paired clone ends being correlated.
Fully sequenced clones
Complete sequencing of 15 of the two-hit clones from A. lyrata revealed that the average value by which the gel based estimation erred from the sequenced length was 59 bp (±57 bp). In the clones sequenced, much of the size difference between the homologous A. lyrata and A. thaliana regions was found to be due to the presence of transposon-like and repetitive sequence in the larger homologous region (see supplementary data). The transposon-like sequences had similarity to a variety of Class I and Class II elements. For those sequences with similarity to retrotransposons (Class I), this included Ta1, subfamily Tf (LINE), other LINEs, and non-LTRs. For those sequences with similarity to transposons (Class II), this included En/Spm, ARNOLDY, and MITE. Those clones that were similarly sized to their A. thaliana homolog exhibited a high sequence similarity between the two species. These results are consistent with those observed by Windsor et al. (2006).
The length of exons and introns within genes of the sequenced A. lyrata clones were compared to their homologs in A. thaliana. This comparison included 13 complete genes and 22 incomplete genes for a total of 119 exons and 97 introns, excluding two introns (79 and 93 bp) found in A. lyrata but not in A. thaliana (Table 4). No significant difference in the length of either exons or introns was detected under a Wilcoxon Signed Ranks test. This suggests that most of the addition or deletion of genetic material occurred in intergenic regions. The complete annotated sequences of these clones were submitted to GenBank and have the accession numbers EU379001–EU379015.
Discussion
The use of four molecular markers (two nuclear and two chloroplast) and sampling among the genera thought closely related to A. thaliana provides a robust hypothesis of the phylogenetic relationships among these taxa. In addition, the mapping of genome sizes onto the phylogeny, and the subsequent examination of the mode, tempo, and directionality of genome size changes, provides a more critical assessment of the evolution of genome size than heretofore available. Finally, comparisons of genomic clones from A. lyrata and B. stricta to A. thaliana provide an in-depth look at the underlying patterns of genome size evolution. We are thus able to not only describe the history of genome size evolution in this area of the Brassicaceae but also illustrate where patterns in our observed data are consistent with theoretical mechanisms generating diversity in genome size. This should provide a guide for further research on this topic.
Genome size changes across the phylogeny
The phylogeny recovered in this study (Fig. 1) is strongly supported and is similar to previous studies that have incorporated some of the taxa included here (Bailey et al. 2002; Hall et al. 2002; Johnston et al. 2005; Koch et al. 2001, 2003; O’Kane and Al-Shehbaz 2003). The one ambiguity is the placement of two basal lineages, which are swapped between the Bayesian and parsimony analyses. However, this does not affect our reconstruction of genome sizes since Lepidium and the lineage leading to Fourraea, Sisymbrium, and Eutrema have the same character state for genome size (Fig. 1).
The most parsimonious explanation for the small genome size of A. thaliana is a reduction in that lineage, not an increase in the sister clade (Fig. 1). This agrees with the conclusions of Johnston et al. (2005), although they found Arabidopsis to have a sister relationship with a clade containing Capsella, Olimarabidopsis, and Crucihimalaya. The differences in our topologies are likely due to our increased sampling of genera, denser taxonomic sampling near Arabidopsis, and the addition in our dataset of three other molecular markers. The genome size of 0.176 pg that we measured for A. thaliana, while slightly higher than the commonly reported value of 0.16 pg, is not much different from those reported by Johnston et al. (2005). Our analysis also indicated a genome size reduction in B. stricta relative to its sister taxa.
The results from our character mapping in MacClade™ and Continuous find significant support for species-specific changes in genome size (2C). This is consistent with findings in other groups of organisms in which genome size changes occur along terminal branches (Grover et al. 2004; Neafsey and Palumbi 2003). In our MacClade™ analyses, there is a greater tendency for genome size to increase (nine transitions) than for genome size to decrease (three transitions) (Fig. 1), but this is likely due to the fact that there are more taxa with small (0.0–0.5 pg) genomes than with large (0.5–0.9 pg) genomes (22 vs. 5, respectively). Indeed, the failure to reject the null-hypothesis that genome sizes are distributed randomly across the tree under an unordered treatment of the character is consistent with no phylogenetic constraint. Although the result was significant when the character was treated as ordered, this was not as conservative a test. Our analyses using the program Continuous failed to support a model of directional evolution for either genome size (2C) or chromosome number (2n), but did find significant support for phylogenetic constraint in the evolution of both characters. The analyses in Continuous also found that the evolution of genome size (2C) and chromosome number (2n) is not correlated, and that these two characters appear to have different modes and tempos of evolution. The final picture suggests that genome size, at the scale we investigated, is free to increase and decrease over evolutionary time.
Mechanisms of change in genome size
An important distinction must be drawn in the evolution of genome size between the most common mechanistic event and the mechanism by which the majority of DNA addition or removal occurs. That is, a few events resulting in dramatic change of the total amount of DNA may overshadow the fact that most events have a very small effect on total genome size. In addition, mechanisms shown to work in the laboratory are not necessarily responsible for the variation in genome size that arises over the course of evolution. Nevertheless, our current understanding of genome size evolution does point toward a few generally important mechanisms. Furthermore, our data shed light on distinguishing between the most common mechanistic events and the events that likely account for most of the observed change in genome size among these lineages.
Two important mechanisms for genome size increases are polyploidization and the insertion of transposable elements (Bennetzen 2000; Kellogg and Bennetzen 2004). We do not address polyploidization in this paper but it is a relatively well understood phenomenon and evidence for multiple rounds of polyploidization in the evolutionary past of A. thaliana has already been documented (Simillion et al. 2002; Wendel 2000). Furthermore, polyploidy is known to occur at the tips of the Brassicaceae tree, for example in the genera Arabidopsis and Boechera (Clauss and Koch 2006; Schranz et al. 2005). Meanwhile, the importance of transposable elements in genome evolution is also well documented (Bartolome et al. 2002; Bennetzen 2000; Biemont and Vieira 2005; Kumar and Bennetzen 1999; Lenoir et al. 2005). Thus, it is not surprising that our sequenced clones revealed a correlation between the presence of transposable elements and increases in the size of homologous regions (see online supplementary data associated with this article).
Mechanisms that might account for reductions in genome size across evolutionary time are not as obvious (Rabinowicz 2000). One proposed mechanism is the loss of whole chromosomes or genes following polyploidization events (Leitch and Bennett 2004). But our data reveal no correlation between chromosome number and genome size from an evolutionary perspective. Petrov (2002) and Petrov and Hartl (1997) have also suggested that biased mutation could lead to genome size reduction. And, evidence from Devos et al. (2002) indicates that “illegitimate recombination” is another means by which plants can avoid genomic obesity. In this scenario, breaks in double stranded DNA are repaired via non-homologous end joining, which leads to small-scale deletions (Devos et al. 2002; Gregory 2005a; Puchta 2005; Vitte and Panaud 2005). In the laboratory, DNA loss has been observed in A. thaliana during Double Strand Break repair (Kirik et al. 2000). Other conceivable mechanisms include unequal homologous recombination (Bennetzen et al. 2005), increases in the rate of deletions relative to insertions, or increases in the size of deletions relative to insertions.
Patterns from our genomic clones can help to identify which of the theoretical mechanisms were actually involved in genome size changes among the taxa studied here. For example, the majority of changes to genome size are small but there are also a considerable number of large increases and decreases (Fig. 2). This suggests that at least two processes are at work. Meanwhile, the similar characteristics of the A. lyrata and B. stricta datasets, suggests that the processes underlying the flux of genome sizes in these taxa are similar (Table 3). Although our data point toward certain explanations for the change in genome size as more probable than others, it is clear that the modest but significant size difference seen between our clones and the homologous regions from A. thaliana does not completely account for the change in overall genome size.
The contrasting results from the chi-square analyses of our genomic clone classes from the unfiltered and filtered datasets highlight the significant role of insertions and deletions larger than 5 kb containing repetitive sequence in genome size changes. Nevertheless, the results from the filtered dataset also suggest that a portion of the changes to genome size is made of up insertion or deletion events that are smaller than 5 kb. Thus, the analyses indicate that both coarse- (changes larger than 5 kb) and fine-grained models (changes smaller than 5 kb) are at work in the evolution of genome size. Consequently, our data paint a picture of genome size change in which much of the reduction or increase of genome size results from large insertions/deletions involving repetitive sequences or transposable elements, but where most of the insertion/deletion events involve mechanisms by which relatively small amounts of DNA are inserted or deleted.
Further support for a fine-grained model comes from the many small insertion and deletion events observed in the fully sequenced two-hit clones of A. lyrata. This is congruent with the predictions of the “illegitimate recombination” model for small deletion sizes in LTR-regions (Bennetzen et al. 2005; Devos et al. 2002). In addition, the lack of significant size difference in exons or introns of the clones sequenced from A. lyrata relative to A. thaliana reveals that insertion and deletion events are generally intergenic (Table 4). This has also been observed in a previous study comparing A. thaliana and A. lyrata (Kusaba et al. 2001), Drosophila (Bartolome et al. 2002), and a study comparing the genomes of A. thaliana and C. rubella (Boivin et al. 2004).
Evidence in support of a coarse grained model, whereby relatively few changes have a large effect, can be found in the results from our chi-square analysis and our analysis using Continuous (Kappa = 0). This scenario agrees with suggestions that transposon activity may occur in bursts within specific lineages, thus leading to relatively abrupt increases in genome size (Vitte and Panaud 2005). This pattern is also consistent with general observations of dramatic genomic rearrangements and more rapid evolution following speciation events (Navarro and Barton 2003). However, the fact that our character mapping finds no support for genome size increases in the A. lyrata and B. stricta lineages and that the transposon-like sequences in our fully sequenced clones corresponded to a variety of transposon types, suggests that transposon infection and expansion, such as seen in maize (SanMiguel et al. 1998), is not responsible for the differences in genome size we observe.
Genome size reduction in the A. thaliana lineage
The possible causes, as opposed to the mechanisms, for a small genome size remain unclear (Charlesworth and Barton 2004; Petrov 2001). In the case of A. thaliana, genome size reduction is correlated with changes in other characteristics such as a shift to an annual lifestyle, unique within the genus. This lifestyle is characterized by more rapid development, which might have selected for a smaller genome size (Bennett et al. 1998; Charlesworth and Barton 2004). The shift to self-fertilization, also apomorphic along this lineage, would have led to a small effective population size that could have acted as a barrier to the establishment of transposable elements, thwarting a major mechanism of genome size increase (Lynch and Conery 2003). And, once a small genome size became established, it could itself have exerted a selective pressure for removal of non-coding DNA, creating a positive feedback loop (Gregory 2005b).
The interplay of several of these mechanisms could account for the dramatic reduction in genome size in A. thaliana. The 3.1–9 million years age range for the lineage (Koch et al. 2000) encompasses the 6 million years half-life reported for LTR-retrotransposons in rice (Ma et al. 2004), meaning that transposable elements present at the time of divergence would have lost much of their ability to increase genome size over the time that has since elapsed. Meanwhile, the concomitant shift to a weedy, annual, self-fertilizing lifestyle in A. thaliana could have significantly impaired mechanisms that would otherwise have increased genome size, such as the spread of newly introduced transposons into the genome. Mechanisms that act to decrease genome size, however, would have remained unaffected. In this scenario, evolutionary pressure does not directly act to reduce genome size but merely hinders genome size increase.
The scenario of a reduced rate of transposon insertion but unhindered deletion has been found in other organisms. Neafsey and Palumbi (2003) find the probable explanation for the reduced genome size in the smooth pufferfish lineage to be a decline in the rate of large-scale insertions. The situation in A. thaliana may be, however, a little more complicated. Although a small effective population size could thwart the establishment of new transposable elements, it would also be expected to lower the efficacy of selection for the removal of transposable elements. However, this does not seem to have been the case in A. thaliana, relative to A. lyrata, which might be due to a population expansion following the evolution of selfing (Wright et al. 2002). In addition, A. thaliana seems to have a more complicated population structure than would be expected for a self-fertilizing annual, suggesting that its effective population size might not be as small as previously thought (Nordborg et al. 2005; Schmid et al. 2006).
Further genome–genome comparisons
We have a robust phylogeny that includes multiple species of Arabidopsis and closely related genera. This allows us to polarize the overall direction of genome size changes with confidence and claim that most of the processes leading to genome size reduction in A. thaliana occurred on the branch leading to that taxon. We also have genomic data from a close relative of A. thaliana (A. lyrata) and a more distant relative (B. stricta). This allows us to compare the micro-level changes in genome composition among the three taxa and claim that multiple mechanisms are driving the evolution of genome size. There are, however, two caveats. The first is that since only the genome of A. thaliana has been fully sequenced, we do not have a true three-way comparison between A. thaliana, A. lyrata and B. stricta and thus cannot polarize the individual indel events identified in our fully sequenced genomic clones. The second is that we do not have sufficient population or species level sampling to consider inter- versus intraspecific variation in genome size. Thus, confirmation of our claims will require greater population level measurements of genome size for the numerous Brassicaceae taxa in our phylogeny and direct comparisons of the complete genome sequences of the three taxa.
The sequencing of the A. lyrata genome will help to address the problem of polarizing the indel events by making direct comparisons between B. stricta and A. lyrata possible. It would also provide the potential to identify the transposons responsible for genome size changes. However, as was discovered with Drosophila, the whole-genome shotgun sequencing strategy may not be able to accurately place repetitive elements or indels, and thus it will be difficult to use the data generated to infer processes of genome size evolution (Benos et al. 2001). This means that even after the completion of this project, and the effort to sequence the genome of Capsella, the role of repetitive sequences in the evolution of the A. thaliana genome may remain unclear.
Conclusion
The summary picture that emerges from this study is that the small size of the A. thaliana genome is derived relative to its sister taxa, and that this is likely due to both large, coarse-grained insertion/deletion events and smaller, fine-grained insertion/deletion events. Furthermore, our data implicate transposable elements in the insertion events leading to increases in genome size or at least the maintenance of a larger genome size. We also found evidence that insertion/deletion events occurring in a fine-grained way happen in intergenic regions scattered across the genome. This study illustrates how we can extract much more from the data generated by genomics techniques when we integrate them into a comparative study that includes closely related non-model species.
References
Al-Shehbaz IA (2003) Transfer of most North American species of Arabis to Boechera (Brassicaceae). Novon 13:381–391
Al-Shehbaz IA, Beilstein MA, Kellogg EA (2006) Systematics and phylogeny of the Brassicaceae (Cruciferae): an overview. Pl Syst Evol 259:89–120
Altschul SF, Madden TL, Schaeffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25:3389–3402
Bailey CD (2006) Toward a global phylogeny of the Brassicaceae. Molec Biol Evol 23:2142–2160
Bailey CD, Price RA, Doyle JJ (2002) Systematics of the halimolobine Brassicaceae: evidence from three loci and morphology. Syst Bot 27:318–332
Bartolome C, Maside X, Charlesworth B (2002) On the abundance and distribution of transposable elements in the genome of Drosophila melanogaster. Molec Biol Evol 19:926–937
Baum DA, Small RL, Wendel JF (1998) Biogeography and floral evolution of baobabs (Adansonia, Bombacaceae) as inferred from multiple data sets. Syst Biol 47:181–207
Beilstein MA, Al-Shehbaz IA, Kellogg EA (2006) Brassicaceae phylogeny and trichome evolution. Amer J Bot 93:607–619
Bennett MD, Leitch IJ (2005a) Nuclear DNA amounts in angiosperms: progress, problems and prospects. Ann Bot 95:45–90
Bennett MD, Leitch IJ (2005b) Plant genome size research: a field in focus. Ann Bot 95:1–6
Bennett MD, Leitch IJ, Hanson L (1998) DNA amounts in two samples of angiosperm weeds. Ann Bot 82:121–134
Bennetzen JL (2000) Transposable element contributions to plant gene and genome evolution. Pl Molec Biol 42:251–269
Bennetzen JL, Kellogg EA (1997) Do plants have a one-way ticket to genomic obesity? Pl Cell 9:1509–1514
Bennetzen JL, Ma JX, Devos K (2005) Mechanisms of recent genome size variation in flowering plants. Ann Bot 95:127–132
Benos PV, Gatt MK, Murphy L, Harris D, Barrell B, Ferraz C, Vidal S, Brun C, Demaille J, Cadieu E, Dreano S, Gloux S, Lelaure V, Mottier S, Galibert F, Borkova D, Minana B, Kafatos FC, Bolshakov S, Siden-Kiamos I, Papagiannakis G, Spanos L, Louis C, Madueno E, de Pablos B, Modolell J, Peter A, Schottler P, Werner M, Mourkioti F, Beinert N, Dowe G, Schafer U, Jackle H, Bucheton A, Callister D, Campbell L, Henderson NS, McMillan PJ, Salles C, Tait E, Valenti P, Saunders RDC, Billaud A, Pachter L, Glover DM, Ashburner M (2001) From first base: the sequence of the tip of the X chromosome of Drosophila melanogaster, a comparison of two sequencing strategies. Genome Res 11:710–730
Biemont C, Vieira C (2005) What transposable elements tell us about genome organization and evolution: the case of Drosophila. Cytogenet Genome Res 110:25–34
Boivin K, Acarkan A, Mbulu RS, Clarenz O, Schmidt R (2004) The Arabidopsis genome sequence as a tool for genome analysis in Brassicaceae. A comparison of the Arabidopsis and Capsella rubella genomes. Pl Physiol 135:735–744
Bray N, Dubchak I, Pachter L (2003) AVID: a global alignment program. Genome Res 13:97
Brunak S, Engelbrecht J, Knudsen S (1991) Prediction of human messenger-RNA donor and acceptor sites from the DNA-sequence. J Molec Biol 220:49–65
Burset M, Guigo R (1996) Evaluation of gene structure prediction programs. Genomics 34:353–367
Charlesworth B, Barton N (2004) Genome size: does bigger mean worse? Curr Biol 14:R233–R235
Charlesworth D, Charlesworth B, McVean GAT (2001) Genome sequences and evolutionary biology, a two-way interaction. Trends Ecol Evol 16:235–242
Clauss MJ, Koch MA (2006) Poorly known relatives of Arabidopsis thaliana. Trends Plant Sci 11:449–459
Couronne O, Poliakov A, Bray N, Ishkhanov T, Ryaboy D, Rubin E, Pachter L, Dubchak I (2003) Strategies and tools for whole-genome alignments. Genome Res 13:7
Devos KM, Brown JKM, Bennetzen JL (2002) Genome size reduction through illegitimate recombination counteracts genome expansion in Arabidopsis. Genome Res 12:1075–1079
Doyle JJ, Doyle JL (1987) A rapid DNA isolation procedure for small quantities of fresh leaf material. Phytochem Bull 19:11–15
Ewing B, Green P (1998) Base-calling of automated sequencer traces using phred. II. Error probabilities. Genome Res 8:186–194
Ewing B, Hillier L, Wendl MC, Green P (1998) Base-calling of automated sequencer traces using phred. I. Accuracy assessment. Genome Res 8:175–185
Felsenstein J (1985) Confidence-limits on phylogenies—an approach using the bootstrap. Evolution 39:783–791
Gregory TR (2004) Macroevolution, hierarchy theory, and the C value enigma. Paleobiology 30:179–202
Gregory TR (2005a) The C value enigma in plants and animals: a review of parallels and an appeal for partnership. Ann Bot 95:133–146
Gregory TR (2005b) Synergy between sequence and size in large-scale genomics. Nat Rev Genet 6:699–708
Grover CE, Kim HR, Wing RA, Paterson AH, Wendel JF (2004) Incongruent patterns of local and global genome size evolution in cotton. Genome Res 14:1474–1482
Hall JC, Iltis HH, Sytsma KJ (2004) Molecular phylogenetics of core brassicales, placement of orphan genera Emblingia, Forchhammeria, Tirania, and character evolution. Syst Bot 29:654–669
Hall JC, Sytsma KJ, Iltis HH (2002) Phylogeny of Capparaceae and Brassicaceae based on chloroplast sequence data. Amer J Bot 89:1826–1842
Hebsgaard SM, Korning PG, Tolstrup N, Engelbrecht J, Rouze P, Brunak S (1996) Splice site prediction in Arabidopsis thaliana pre-mRNA by combining local and global sequence information. Nucleic Acids Res 24:3439–3452
Huang XQ, Madan A (1999) CAP3: a DNA sequence assembly program. Genome Res 9:868–877
Huelsenbeck JP, Larget B, Miller RE, Ronquist F (2002) Potential applications and pitfalls of Bayesian inference of phylogeny. Syst Biol 51:673–688
Huelsenbeck JP, Ronquist F (2001) MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics 17:754–755
Jakob SS, Meister A, Blattner FR (2004) Considerable genome size variation of Hordeum species (Poaceae) is linked to phylogeny, life form, ecology, and speciation rates. Molec Biol Evol 21:860–869
Johnson LA, Soltis DE (1995) Phylogenetic inference in Saxifragaceae sensu-stricto and Gilia (Polemoniaceae) using matK sequences. Ann Missouri Bot Gard 82:149–175
Johnston JS, Pepper AE, Hall AE, Chen ZJ, Hodnett G, Drabek J, Lopez R, Price HJ (2005) Evolution of genome size in Brassicaceae. Ann Bot 95:229–235
Jurka J, Kapitonov VV, Pavlicek A, Klonowski P, Kohany O, Walichiewicz J (2005) Repbase update, a database of eukaryotic repetitive elements. Cytogenet Genome Res 110:462–467
Kellogg EA, Bennetzen JL (2004) The evolution of nuclear genome structure in seed plants. Amer J Bot 91:1709–1725
Kirik A, Salomon S, Puchta H (2000) Species-specific double-strand break repair and genome evolution in plants. Embo J 19:5562–5566
Kishino H, Hasegawa M (1989) Evaluation of the maximum-likelihood estimate of the evolutionary tree topologies from DNA-sequence data, and the branching order in Hominoidea. J Molec Evol 29:170–179
Koch M, Al-Shehbaz IA, Mummenhoff K (2003) Molecular systematics, evolution, and population biology in the mustard family (Brassicaceae). Ann Missouri Bot Gard 90:151–171
Koch M, Bishop J, Mitchell-Olds T (1999) Molecular systematics and evolution of Arabidopsis and Arabis. Plant Biol 1:529–537
Koch M, Haubold B, Mitchell-Olds T (2001) Molecular systematics of the Brassicaceae: evidence from coding plastidic matK and nuclear Chs sequences. Amer J Bot 88:534–544
Koch MA, Dobes C, Matschinger M, Bleeker W, Vogel J, Kiefer M, Mitchell-Olds T (2005) Evolution of the trnF(GAA) gene in Arabidopsis relatives and the Brassicaceae family: monophyletic origin and subsequent diversification of a plastidic pseudogene. Molec Biol Evol 22:1032–1043
Koch MA, Haubold B, Mitchell-Olds T (2000) Comparative evolutionary analysis of chalcone synthase and alcohol dehydrogenase loci in Arabidopsis, Arabis, and related genera (Brassicaceae). Molec Biol Evol 17:1483–1498
Koch MA, Kiefer M (2005) Genome evolution among cruciferous plants: a lecture from the comparison of the genetic maps of three diploid species—Capsella rubella, Arabidopsis lyrata subsp Petraea, and A. thaliana. Amer J Bot 92:761–767
Kumar A, Bennetzen JL (1999) Plant retrotransposons. Annu Rev Genet 33:479–532
Kusaba M, Dwyer K, Hendershot J, Vrebalov J, Nasrallah JB, Nasrallah ME (2001) Self-incompatibility in the genus Arabidopsis: characterization of the S locus in the outcrossing A. lyrata and its autogamous relative A. thaliana. Pl Cell 13:627–643
Leitch IJ, Bennett MD (2004) Genome downsizing in polyploid plants. Biol J Linn Soc 82:651–663
Lenoir A, Pelissier T, Bousquet-Antonelli C, Deragon JM (2005) Comparative evolution history of SINEs in Arabidopsis thaliana and Brassica oleracea: evidence for a high rate of SINE loss. Cytogenet Genome Res 110:441–447
Lynch M, Conery JS (2003) The origins of genome complexity. Science 302:1401–1404
Lysak MA, Berr A, Pecinka A, Schmidt R, McBreen K, Schubert I (2006) Mechanisms of chromosome number reduction in Arabidopsis thaliana and related Brassicaceae species. Proc Natl Acad Sc USA 103:5224–5229
Lysak MA, Pecinka A, Schubert I (2003) Recent progress in chromosome painting of Arabidopsis and related species. Chromosome Res 11:195–204
Ma JX, Devos KM, Bennetzen JL (2004) Analyses of LTR-retrotransposon structures reveal recent and rapid genomic DNA loss in rice. Genome Res 14:860–869
Miller RE, Buckley TR, Manos PS (2002) An examination of the monophyly of morning glory taxa using Bayesian phylogenetic inference. Syst Biol 51:740–753
Nagl W, Jeanjour M, Kling H, Kuhner S, Michels I, Muller T, Stein B (1983) Genome and chromatic organization in higher plants. Biol Zentralbl 102:129–148
Navarro A, Barton NH (2003) Chromosomal speciation and molecular divergence—accelerated evolution in rearranged chromosomes. Science 300:321–324
Neafsey DE, Palumbi SR (2003) Genome size evolution in pufferfish: a comparative analysis of diodontid and tetraodontid pufferfish genomes. Genome Res 13:821–830
Nordborg M, Hu TT, Ishino Y, Jhaveri J, Toomajian C, Zheng HG, Bakker E, Calabrese P, Gladstone J, Goyal R, Jakobsson M, Kim S, Morozov Y, Padhukasahasram B, Plagnol V, Rosenberg NA, Shah C, Wall JD, Wang J, Zhao KY, Kalbfleisch T, Schulz V, Kreitman M, Bergelson J (2005) The pattern of polymorphism in Arabidopsis thaliana. Plos Biol 3:1289–1299
O’Kane SL Jr, Al-Shehbaz IA (1997) A synopsis of Arabidopsis (Brassicaceae). Novon 7:326
O’Kane SL Jr, Al-Shehbaz IA (2003) Phylogenetic position and generic limits of Arabidopsis (Brassicaceae) based on sequences of nuclear ribosomal DNA. Ann Missouri Bot Gard 90:603–612
Pagel M (1997) Inferring evolutionary processes from phylogenies. Zool Scr 26:331–348
Pagel M (1999) Inferring the historical patterns of biological evolution. Nature 401:877–884
Petrov D (1997) Slow but steady: reduction of genome size through biased mutation. Pl Cell 9:1900–1901
Petrov DA (2001) Evolution of genome size: new approaches to an old problem. Trends Genet 17:23–28
Petrov DA (2002) DNA loss and evolution of genome size in Drosophila. Genetica 115:81–91
Petrov DA, Hartl DL (1997) Trash DNA is what gets thrown away: high rate of DNA loss in Drosophila. Gene 205:279–289
Posada D, Crandall KA (1998) MODELTEST: testing the model of DNA substitution. Bioinformatics 14:817–818
Puchta H (2005) The repair of double-strand breaks in plants: mechanisms and consequences for genome evolution. J Exp Bot 56:1–14
Rabinowicz PD (2000) Are obese plant genomes on a diet? Genome Res 10:893–894
SanMiguel P, Gaut BS, Tikhonov A, Nakajima Y, Bennetzen JL (1998) The paleontology of intergene retrotransposons of maize. Nature Genet 20:43–45
Schmid K, Torjek O, Meyer R, Schmuths H, Hoffmann MH, Altmann T (2006) Evidence for a large-scale population structure of Arabidopsis thaliana from genome-wide single nucleotide polymorphism markers. Theor Appl Genet 112:1104–1114
Schranz ME, Dobes C, Koch MA, Mitchell-Olds T (2005) Sexual reproduction, hybridization, apomixis, and polyploidization in the genus Boechera (Brassicaceae). Amer J Bot 92:1797–1810
Schranz ME, Song B-H, Windsor AJ, Mitchell-Olds T (2007) Comparative genomics in the Brassicaceae: a family-wide perspective. Curr Opin Pl Biol 10:168–175
Shimodaira H, Hasegawa M (1999) Multiple comparisons of log-likelihoods with applications to phylogenetic inference. Molec Biol Evol 16:1114–1116
Simillion C, Vandepoele K, Van Montagu MCE, Zabeau M, Van de Peer Y (2002) The hidden duplication past of Arabidopsis thaliana. Proc Natl Acad Sci USA 99:13627–13632
Smith TF, Waterman MS (1981) Identification of common molecular subsequences. J Molec Biol 147:195–197
Soltis DE, Soltis PS, Bennett MD, Leitch IJ (2003) Evolution of genome size in the angiosperms. Amer J Bot 90:1596–1603
Sonnhammer ELL, Durbin R (1995) A dot-matrix program with dynamic threshold control suited for genomic DNA and protein sequence analysis. Gene 167:GC1–GC10
Swofford DL (2002) PAUP*: phylogenetic analysis using parsimony. Sinauer Press, Sunderland
Taberlet P, Gielly L, Pautou G, Bouvet J (1991) Universal primers for amplification of three noncoding regions of chloroplast DNA. Plant Molec Biol 17:1105–1109
Templeton AR (1983) Phylogenetic inference from restriction endonuclease cleavage site maps with particular reference to the evolution of humans and the apes. Evolution 37:221–244
Thomas CA (1971) Genetic organization of chromosomes. Annual Rev Genet 5:237–256
Vitte C, Panaud O (2005) LTR retrotransposons and flowering plant genome size: emergence of the increase/decrease model. Cytogenet Genome Res 110:91–107
Warwick SI, Al-Shehbaz IA (2006) Brassicaceae: chromosome number index and database on CD-Rom. Pl Syst Evol 259:237–248
Warwick SI, Al-Shehbaz IA, Price RA, Sauder C (2002) Phylogeny of Sisymbrium (Brassicaceae) based on ITS sequences of nuclear ribosomal DNA. Canad J Bot 80:1002–1017
Weiss-Schneeweiss H, Greilhuber J, Schneeweiss GM (2006) Genome size evolution in holoparasitic Orobanche (Orobanchaceae) and related genera. Amer J Bot 93:148–156
Wendel JF (2000) Genome evolution in polyploids. Plant Molec Biol 42:225–249
White TJ, Bruns T, Lee S, Taylor J (1990) Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis MA, Gelfand D, Sninsky J, White T (eds) PCR protocols: a guide to methods and applications. Academic Press, San Diego, pp 315–322
Windsor AJ, Schranz ME, Formanova N, Gebauer-Jung S, Bishop JG, Schnabelrauch D, Kroymann J, Mitchell-Olds T (2006) Partial shotgun sequencing of the Boechera stricta genome reveals extensive microsynteny and promoter conservation with Arabidopsis. Pl Physiol 140:1169–1182
Wright SI, Lauga B, Charlesworth D (2002) Rates and patterns of molecular evolution in inbred and outbred Arabidopsis. Molec Biol Evol 19:1407–1420
Yang ZB (1994a) Estimating the pattern of nucleotide substitution. J Molec Evol 39:105–111
Yang ZH (1994b) Maximum-likelihood phylogenetic estimation from DNA-sequences with variable rates over sites—approximate methods. J Molec Evol 39:306–314
Acknowledgments
The authors thank I. Al-Shehbaz for taxonomic identifications and seeds, E. Schranz for unpublished data, and S. Renner for comments on an earlier draft. This project was financed by the Max Planck Society.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
606_2008_17_MOESM1_ESM.tif
Supplemental Figure 1: DOTTER plots of fully sequenced inserts from the genomic shotgun clones of A. lyrata vs. the homologous region in the A. thaliana genome, where the inserts were smaller than the homologous region in A. thaliana. (TIF 2040 kb)
606_2008_17_MOESM2_ESM.tif
Supplemental Figure 2: DOTTER plots of fully sequenced inserts from the genomic shotgun clones of A. lyrata vs. the homologous region in the A. thaliana genome, where the inserts were similarly sized to the homologous region in A. thaliana. (TIF 1782 kb)
606_2008_17_MOESM3_ESM.tif
Supplemental Figure 3: DOTTER plots of fully sequenced inserts from the genomic shotgun clones of A. lyrata vs. the homologous region in the A. thaliana genome, where the inserts were larger than the homologous region in A. thaliana. (TIF 1560 kb)
Rights and permissions
Open Access This is an open access article distributed under the terms of the Creative Commons Attribution Noncommercial License (https://creativecommons.org/licenses/by-nc/2.0), which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
About this article
Cite this article
Oyama, R.K., Clauss, M.J., Formanová, N. et al. The shrunken genome of Arabidopsis thaliana . Plant Syst Evol 273, 257–271 (2008). https://doi.org/10.1007/s00606-008-0017-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00606-008-0017-z