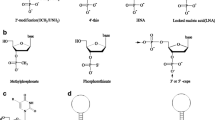
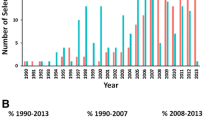
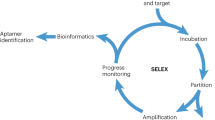
Abstract
Aptamers are functional single-stranded oligonucleotide fragments isolated from randomized libraries by Systematic Evolution of Ligands by Exponential Enrichment (SELEX), exhibiting excellent affinity and specificity toward targets. Compared with traditional antibody reagents, aptamers display many desirable properties, such as low variation and high flexibility, and they are suitable for artificial and large-scale synthesis. These advantages make aptamers have a broad application potential ranging from biosensors, bioimaging to therapeutics and other areas of application. However, the overall performance of aptamer pre-selected by SELEX screening is far from being satisfactory. To improve aptamer performance and applicability, various post-SELEX optimization methods have been developed in the last decade. In this review, we first discuss the key factors that influence the performance or properties of aptamers, and then we summarize the key strategies of post-SELEX optimization which have been successfully used to improve aptamer performance, such as truncation, extension, mutagenesis and modification, splitting, and multivalent integration. This review shall provide a comprehensive summary and discussion of post-SELEX optimization methods developed in recent years. Moreover, by discussing the mechanism of each approach, we highlight the importance of choosing the proper method to perform post-SELEX optimization.
Graphical Abstract





Similar content being viewed by others
References
Kim YS, Raston NHA, Gu MB (2016) Aptamer-based nanobiosensors. Biosens Bioelectron 76:2–19
Ni S, Zhuo Z, Pan Y, Yu Y, Li F, Liu J, Wang L, Wu X, Li D, Wan Y, Zhang L, Yang Z, Zhang B-T, Lu A, Zhang G (2021) Recent progress in aptamer discoveries and modifications for therapeutic applications. ACS Appl Mater Interfaces 13(8):9500–9519
Yao Y, Wang GX, Shi XJ, Li JS, Yang FZ, Cheng ST, Zhang H, Dong HW, Guo YM, Sun X, Wu YX (2020) Ultrasensitive aptamer-based biosensor for acetamiprid using tetrahedral DNA nanostructures. J Mater Sci 55(33):15975–15987
Jiao Z, Zhang H, Jiao S, Guo Z, Zhu D, Zhao X (2019) A turn-on biosensor-based aptamer-mediated carbon quantum dots nanoaggregate for acetamiprid detection in complex samples. Food Anal Meth 12(3):668–676
Sun H, Zhu X, Lu PY, Rosato RR, Tan W, Zu Y (2014) Oligonucleotide aptamers: new tools for targeted cancer therapy. Molecular Therapy-Nucleic Acids 3
Song K-M, Lee S, Ban C (2012) Aptamers and their biological applications. Sensors 12(1):612–631
Ng EWM, Shima DT, Calias P, Cunningham ET, Guyer DR, Adamis AP (2006) Pegaptanib, a targeted anti-VEGF aptamer for ocular vascular disease. Nat Rev Drug Discovery 5(2):123–132
Li T, Wang J, Zhu L, Li C, Chang Q, Xu W (2022) Advanced screening and tailoring strategies of pesticide aptamer for constructing biosensor. Crit Rev Food Sci Nutr
Chen X, Gao D, Sun F, Li Z, Wang Y, Qiu C, He K, Wang J (2022) Nanomaterial-based aptamer biosensors for ochratoxin A detection: a review. Anal Bioanal Chem 414(9):2953–2969
Yan J, Gao T, Lu Z, Yin J, Zhang Y, Pei R (2021) Aptamer-targeted photodynamic platforms for tumor therapy. ACS Appl Mater Interfaces 13(24):27749–27773
Devi S, Sharma N, Ahmed T, Huma ZI, Kour S, Sahoo B, Singh AK, Macesic N, Lee SJ, Gupta MK (2021) Aptamer-based diagnostic and therapeutic approaches in animals: current potential and challenges. Saudi J Biol Sci 28(9):5081–5093
Oliveira R, Pinho E, Sousa AL, DeStefano JJ, Azevedo NF, Almeida C (2022) Improving aptamer performance with nucleic acid mimics: de novo and post-SELEX approaches. Trends Biotechnol 40(5):549–563
Tuerk C, Gold L (1990) Systematic evolution of ligands by exponential enrichment - RNA ligands to bacteriophage-T4 DNA-polymerase. Science 249(4968):505–510
Yu H, Pan C, Zhu J, Shen G, Deng Y, Xie X, Geng X, Wang L (2022) Selection and identification of a DNA aptamer for fluorescent detection of netilmicin. Talanta 250
Lyu C, Khan IM, Wang Z (2021) Capture-SELEX for aptamer selection: a short review. Talanta 229
Yang Y, Tang Y, Wang C, Liu B, Wu Y (2021) Selection and identification of a DNA aptamer for ultrasensitive and selective detection of lambda-cyhalothrin residue in food. Anal Chim Acta 1179
Kong Q, Yue F, Liu M, Huang J, Yang F, Liu J, Li J, Li F, Sun X, Guo Y, Zhu Y (2022) Non-immobilized GO-SELEX of aptamers for label-free detection of thiamethoxam in vegetables. Anal Chim Acta 1202
Nguyen V-T, Kwon YS, Kim JH, Gu MB (2014) Multiple GO-SELEX for efficient screening of flexible aptamers. Chem Commun 50(72):10513–10516
Ouellet E, Foley JH, Conway EM, Haynes C (2015) Hi-Fi SELEX: a high-fidelity digital-PCR based therapeutic aptamer discovery platform. Biotechnol Bioeng 112(8):1506–1522
Chen M, Yu Y, Jiang F, Zhou J, Li Y, Liang C, Dang L, Lu A, Zhang G (2016) Development of Cell-SELEX technology and its application in cancer diagnosis and therapy. Int J Mol Sci 17(12)
Guo K-T, Paul A, Schichor C, Ziemer G, Wendel HP (2008) Cell-SELEX: novel perspectives of aptamer-based therapeutics. Int J Mol Sci 9(4):668–678
Darmostuk M, Rimpelova S, Gbelcova H, Ruml T (2015) Current approaches in SELEX: an update to aptamer selection technology. Biotechnol Adv 33(6):1141–1161
Wang T, Chen C, Larcher LM, Barrero RA, Veedu RN (2019) Three decades of nucleic acid aptamer technologies: lessons learned, progress and opportunities on aptamer development. Biotechnol Adv 37(1):28–50
Green LS, Jellinek D, Bell C, Beebe LA, Feistner BD, Gill SC, Jucker FM, Janjic N (1995) Nuclease-resistant nucleic acid ligands to vascular permeability factor/vascular endothelial growth factor. Chem Biol 2(10):683–695
Gao S, Zheng X, Jiao B, Wang L (2016) Post-SELEX optimization of aptamers. Anal Bioanal Chem 408(17):4567–4573
Qi S, Duan N, Khan IM, Dong X, Zhang Y, Wu S, Wang Z (2022) Strategies to manipulate the performance of aptamers in SELEX, post-SELEX and microenvironment. Biotechnol Adv 55:107902–107902
Nie J, Yuan L, Jin K, Han X, Tian Y, Zhou N (2018) Electrochemical detection of tobramycin based on enzymes-assisted dual signal amplification by using a novel truncated aptamer with high affinity. Biosens Bioelectron 122:254–262
Cong Quang V, Rotkrua P, Tantirungrotechai Y, Soontornworajit B (2017) Oligonucleotide hybridization combined with competitive antibody binding for the truncation of a high-affinity aptamer. Acs Comb Sci 19(10):609–617
Cowperthwaite MC, Ellington AD (2008) Bioinformatic analysis of the contribution of primer sequences to aptamer structures. J Mol Evol 67(1):95–102
Zhou J, Soontornworajit B, Snipes MP, Wang Y (2011) Structural prediction and binding analysis of hybridized aptamers. J Mol Recognit 24(1):119–126
Li Q, Wang Y-D, Shen G-L, Tang H, Yu R-Q, Jiang J-H (2015) Split aptamer mediated endonuclease amplification for small-molecule detection. Chem Commun 51(20):4196–4199
Jo E-J, Byun J-Y, Mun H, Kim M-G (2017) Luminescence resonance energy transfer (LRET) aptasensor for ochratoxin A detection using upconversion nanoparticles. International Conference on Nano-Bio Sensing, Imaging, and Spectroscopy, Jeju, South Korea
Yang C, Du C, Su R, Wang J, Li Y, Ma X, Li Z, Sun C (2022) A signal-on fluorescent aptasensor by sensitized Tb3+ luminescence for detection of melamine in milk. Talanta 236
Kimoto M, Nakamura M, Hirao I (2016) Post-ExSELEX stabilization of an unnatural-base DNA aptamer targeting VEGF(165) toward pharmaceutical applications. Nucleic Acids Res 44(15):7487–7494
Tian Y, Wang Y, Sheng Z, Li T, Li X (2016) A colorimetric detection method of pesticide acetamiprid by fine-tuning aptamer length. Anal Biochem 513:87–92
Mok W, Li Y (2008) Recent progress in nucleic acid aptamer-based biosensors and bioassays. Sensors 8(11):7050–7084
Stoltenburg R, Reinemann C, Strehlitz B (2007) SELEX-A (r)evolutionary method to generate high-affinity nucleic acid ligands. Biomol Eng 24(4):381–403
Chun HJ, Kim S, Han YD, Kim DW, Kim KR, Kim H-S, Kim J-H, Yoon HC (2018) Water-soluble mercury ion sensing based on the thymine-Hg2+-thymine base pair using retroreflective Janus particle as an optical signaling probe. Biosens Bioelectron 104:138–144
Xu A, Chao L, Xiao H, Sui Y, Liu J, Xie Q, Yao S (2018) Ultrasensitive electrochemical sensing of Hg2+ based on thymine-Hg2+-thymine interaction and signal amplification of alkaline phosphatase catalyzed silver deposition. Biosens Bioelectron 104:95–101
Yang D, Okamoto K (2010) Structural insights into G-quadruplexes: towards new anticancer drugs. Future Med Chem 2(4):619–646
Cai S, Yan J, Xiong H, Liu Y, Peng D, Liu Z (2018) Investigations on the interface of nucleic acid aptamers and binding targets. Analyst 143(22):5317–5338
Shinomiya R, Katahira Y, Araki H, Shibata T, Momotake A, Yanagisawa S, Ogura T, Suzuk A, Neya S, Yamamoto Y (2018) Characterization of catalytic activities and heme coordination structures of heme-DNA complexes composed of some chemically modified hemes and an all parallel-stranded tetrameric G-quadruplex DNA formed from d(TTAGGG). Biochemistry 57(41):5930–5937
Shibata T, Nakayama Y, Katahira Y, Tai H, Moritaka Y, Nakano Y, Yamamoto Y (2017) Characterization of the interaction between heme and a parallel G-quadruplex DNA formed from d(TTGAGG). BBA-Gen Subjects 1861(5):1264–1270
Zhao L, Qi X, Yan X, Huang Y, Liang X, Zhang L, Wang S, Tan W (2019) Engineering aptamer with enhanced affinity by triple helix-based terminal fixation. J Am Chem Soc 141(44):17493–17497
Dragan AI, Pavlovic R, McGivney JB, Casas-Finet JR, Bishop ES, Strouse RJ, Schenerman MA, Geddes CD (2012) SYBR green I: fluorescence properties and interaction with DNA. J Fluoresc 22(4):1189–1199
Shangguan D, Tang Z, Mallikaratchy P, Xiao Z, Tan W (2007) Optimization and modifications of aptamers selected from live cancer cell lines. ChemBioChem 8(6):603–606
Bing T, Yang X, Mei H, Cao Z, Shangguan D (2010) Conservative secondary structure motif of streptavidin-binding aptamers generated by different laboratories. Bioorg Med Chem 18(5):1798–1805
Yang X, Bing T, Mei H, Fang C, Cao Z, Shangguan D (2011) Characterization and application of a DNA aptamer binding to L-tryptophan. Analyst 136(3):577–585
Zuker M (2003) Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res 31(13):3406–3415
Kilgour M, Liu T, Walker BD, Ren P, Simine L (2021) E2EDNA: simulation protocol for DNA aptamers with ligands. J Chem Inf Model 61(9):4139–4144
Chen Y, Wang Z, Liu S, Zhao G (2021) A highly sensitive and group-targeting aptasensor for total phthalate determination in the environment. J Hazard Mater 412
Esposito V, Galeone A, Mayol L, Randazzo A, Virgilio A, Virno A (2007) A mini-library of tba analogues containing 3’-3’ and 5’-5’ inversion of polarity sites. Nucleos Nucleot Nucl 26(8–9):1145–1149
Odeh F, Nsairat H, Alshaer W, Ismail MA, Esawi E, Qaqish B, Al Bawab A, Ismail SI (2020) aptamers chemistry: chemical modifications and conjugation strategies. Molecules 25(1)
Chinnappan R, AlZabn R, Mir TA, Bader M, Zourob M (2019) Fluorometric determination of okadaic acid using a truncated aptamer. Microchim Acta 186(7)
He X, Guo L, He J, Xu H, Xie J (2017) Stepping library-based post-SELEX strategy approaching to the minimized aptamer in SPR. Anal Chem 89(12):6559–6566
Chinnappan R, AlAmer S, Eissa S, Rahamn AA, Abu Salah KM, Zourob M (2018) Fluorometric graphene oxide-based detection of Salmonella enteritis using a truncated DNA aptamer. Microchim. Acta 185(1)
Ma P, Guo H, Duan N, Ma X, Yue L, Gu Q, Wang Z (2021) Label free structure-switching fluorescence polarization detection of chloramphenicol with truncated aptamer. Talanta 230
Ye H, Duan N, Gu H, Wang H, Wang Z (2019) Fluorometric determination of lipopolysaccharides via changes of the graphene oxide-enhanced fluorescence polarization caused by truncated aptamers. Microchim Acta 186(3)
Wei H, Cai R, Yue H, Tian Y, Zhou N (2020) Screening and application of a truncated aptamer for high-sensitive fluorescent detection of metronidazole. Anal Chim Acta 1128:203–210
Jia M, Sha J, Li Z, Wang W, Zhang H (2020) High affinity truncated aptamers for ultra-sensitive colorimetric detection of bisphenol A with label-free aptasensor. Food Chem 317
Guo H, Sun Y, Ma P, Khan IM, Duan N, Wang Z (2022) Sensitive detection of patulin based on DNase I-assisted fluorescent aptasensor by using AuNCs-modified truncated aptamer. Food Control 131
Lu Q, He W, Junlin H, Shuming Y, Ailiang C (2021) Truncated affinity-improved aptamers for 17beta-estradiol determination by AuNPs-based colorimetric aptasensor. Food Chem 340:128181–128181
Matulakul P, Vongpramate D, Kulchat S, Chompoosor A, Thanan R, Sithithaworn P, Sakonsinsiri C, Puangmali T (2020) Development of low-cost AuNP-based aptasensors with truncated aptamer for highly sensitive detection of 8-Oxo-dG in urine. ACS Omega 5(28):17423–17430
Zhilei Z, He W, Wenlei Z, Xiaoyuan F, Xia F, Ailiang C, Meng W (2020) A lateral flow strip based on a truncated aptamer-complementary strand for detection of type-B aflatoxins in nuts and dried figs. Toxins 12(2):136–136
Wang C, Zhu K, Shi P, Ding X, Zhang S (2022) Rapid and label-free detection of aflatoxin B1 using a rationally truncated aptamer and via circular dichroism measurement. Chem Commun 58(30):4779–4782
Zhou S-S, Zhang L, Cai Q-Y, Dong Z-Z, Geng X, Ge J, Li Z-H (2016) A facile label-free aptasensor for detecting ATP based on fluorescence enhancement of poly(thymine)-templated copper nanoparticles. Anal Bioanal Chem 408(24):6711–6717
Heydari Shayesteh O, Ghafouri Khosroshahi A (2020) A polyA aptamer-based label-free colorimetric biosensor for the detection of kanamycin in human serum. Anal Methods 12(14):1858–1867
Reza M, Heydari Shayesteh O, Katayoun D, Akram R, Fereshteh M, Ali H (2022) A novel label-free colorimetric polyA aptasensing approach based on cationic polymer and silver nanoparticles for detection of tobramycin in milk. Food Chem 382:132580–132580
Jiang H, Ling K, Tao X, Zhang Q (2015) Theophylline detection in serum using a self-assembling RNA aptamer-based gold nanoparticle sensor. Biosens Bioelectron 70:299–303
Zheng X, Hu B, Gao SX, Liu DJ, Sun MJ, Jiao BH, Wang LH (2015) A saxitoxin-binding aptamer with higher affinity and inhibitory activity optimized by rational site-directed mutagenesis and truncation. Toxicon 101:41–47
Nonaka Y, Yoshida W, Abe K, Ferri S, Schulze H, Bachmann TT, Ikebukuro K (2013) Affinity improvement of a VEGF aptamer by in silico maturation for a sensitive VEGF-detection system. Anal Chem 85(2):1132–1137
Sun Y, Duan N, Ma P, Liang Y, Zhu X, Wang Z (2019) Colorimetric aptasensor based on truncated aptamer and trivalent DNAzyme for Vibrio parahemolyticus Determination. J Agric Food Chem 67(8):2313–2320
Ptacek J, Zhang D, Qiu L, Kruspe S, Motlova L, Kolenko P, Novakova Z, Shubham S, Havlinova B, Baranova P, Chen S-J, Zou X, Giangrande P, Barinka C (2020) Structural basis of prostate-specific membrane antigen recognition by the A9g RNA aptamer. Nucleic Acids Res 48(19):11130–11145
He H, Sun D-W, Pu H, Huang L (2020) Bridging Fe3O4@Au nanoflowers and Au@Ag nanospheres with aptamer for ultrasensitive SERS detection of aflatoxin B1. Food Chem. 324
Zhang Y, Li L, Zhang H, Shang J, Li C, Naqvi SMZA, Birech Z, Hu J (2022) Ultrasensitive detection of plant hormone abscisic acid-based surface-enhanced Raman spectroscopy aptamer sensor. Anal Bioanal Chem 414(8):2757–2766
Wang N, Liu Z, Wen L, Zhang B, Tao C-A, Wang J (2022) Aptamer-binding zirconium-based metal-organic framework composites prepared by two conjunction approaches with enhanced bio-sensing for detecting isocarbophos. Talanta 236
Klose AM, Miller BL (2020) A Stable biotin-streptavidin surface enables multiplex, label-free protein detection by aptamer and aptamer-protein arrays using arrayed imaging reflectometry. Sensors 20(20)
Uppala JK, Ghosh C, Sabat G, Dey M (2022) Pull-down of biotinylated RNA and associated proteins. Bio-Protocol 12(4)
Heemstra JM (2014) Small molecule-dependent split aptamer ligation. University of Utah Research Foundation
Zuo X, Xiao Y, Plaxco KW (2009) High specificity, electrochemical sandwich assays based on single aptamer sequences and suitable for the direct detection of small-molecule targets in blood and other complex matrices. J Am Chem Soc 131(20):6944
Sun Y, Yuan B, Deng M, Wang Q, Huang J, Guo Q, Liu J, Yang X, Wang K (2018) A light-up fluorescence assay for tumor cell detection based on bifunctional split aptamers. Analyst 143(15):3579–3585
Yuan B, Sun Y, Guo Q, Huang J, Yang X, Chen Y, Wen X, Meng X, Liu J, Wang K (2017) High signal-to-background ratio detection of cancer cells with activatable strategy based on target-induced self-assembly of split aptamers. Anal Chem 89(17):9347–9353
Zimmermann GR, Jenison RD, Wick CL, Simorre JP, Pardi A (1997) Interlocking structural motifs mediate molecular discrimination by a theophylline-binding RNA. Nat Struct Biol 4(8):644–649
Anderson PC, Mecozzi S (2005) Unusually short RNA sequences: design of a 13-mer RNA that selectively binds and recognizes theophylline. J Am Chem Soc 127(15):5290–5291
Kent AD, Spiropulos NG, Heemstra JM (2013) General approach for engineering small-molecule-binding DNA split aptamers. Anal Chem 85(20):9916–9923
Yu H, Canoura J, Guntupalli B, Lou X, Xiao Y (2017) A cooperative-binding split aptamer assay for rapid, specific and ultra-sensitive fluorescence detection of cocaine in saliva. Chem Sci 8(1):131–141
Yu H, Canoura J, Guntupalli B, Alkhamis O, Xiao Y (2018) Sensitive detection of small-molecule targets using cooperative binding split aptamers and enzyme-assisted target recycling. Anal Chem 90(3):1748–1758
Abnous K, Danesh NM, Ramezani M, Taghdisi SM, Emrani AS (2018) A novel amplified double-quenching aptasensor for cocaine detection based on split aptamer and silica nanoparticles. Anal Methods 10(26):3232–3236
Luo Y, Yu H, Alkhamis O, Liu Y, Lou X, Yu B, Xiao Y (2019) Label-free, visual detection of small molecules using highly target-responsive multimodule split aptamer constructs. Anal Chem 91(11):7199–7207
Zhang S, Wang K, Li J, Li Z, Sun T (2015) Highly efficient colorimetric detection of ATP utilizing a split aptamer target binding strategy and superior catalytic activity of graphene oxide-platinum/gold nanoparticles. RSC Adv 5(92):75746–75752
Lu N, Shao C, Deng Z (2008) Rational design of an optical adenosine sensor by conjugating a DNA aptamer with split DNAzyme halves. Chem Commun 46:6161–6163
Gu Y, Li J, Qian K, Zhang Z, Wang S, Wang J (2020) Integrated dual-signal aptasensor based on magnet-driven operations and miniaturized analytical device for on-site analysis. Sens Actuator B-Chem. 310
Ma X, Guo Z, Mao Z, Tang Y, Miao P (2018) Colorimetric theophylline aggregation assay using an RNA aptamer and non-crosslinking gold nanoparticles. Microchim Acta 185(1)
Wang J, Cheng W, Meng F, Yang M, Pan Y, Miao P (2018) Hand-in-hand RNA nanowire-based aptasensor for the detection of theophylline. Biosens Bioelectron 101:153–158
Liang Y, Guo T, Zhou L, Offenhaeusser A, Mayer D (2020) Label-free split aptamer sensor for femtomolar detection of dopamine by means of flexible organic electrochemical transistors. Materials 13(11)
Guo T, Wu C, Offenhaeusser A, Mayer D (2020) A novel ratiometric electrochemical biosensor based on a split aptamer for the detection of dopamine with logic gate operations. Physica Status Solidi a-App Mater Sci 217(13)
Li X, Tang X, Chen X, Qu B, Lu L (2018) Label-free and enzyme-free fluorescent isocarbophos aptasensor based on MWCNTs and G-quadruplex. Talanta 188:232–237
Wang R, Zhang Q, Zhang Y, Shi H, Kim Truc N, Zhou X (2019) Unconventional split aptamers cleaved at functionally essential sites preserve biorecognition capability. Anal Chem 91(24):15811–15817
Du Y, Zhou Y, Wen Y, Bian X, Xie Y, Zhang W, Liu G, Yan J (2019) Multiplexed aptasensing of food contaminants by using terminal deoxynucleotidyl transferase-produced primer-triggered rolling circle amplification: application to the colorimetric determination of enrofloxacin, lead (II), Escherichia coli O157:H7 and tropomyosin. Microchim Acta 186(12)
Neves MAD, Slavkovic S, Reinstein O, Shoara AA, Johnson PE (2019) A proof of concept application of aptachain: ligand-induced self-assembly of a DNA aptamer. RSC Adv 9(3):1690–1695
Sheng W, Chen T, Tan W, Fan ZH (2013) Multivalent DNA nanospheres for enhanced capture of cancer cells in microfluidic devices. ACS Nano 7(8):7067–7076
Mahlknecht G, Maron R, Schechter B, Yarden Y, Sela M (2015) Multimerization of ERBB2/HER2 specific aptamer leads to improved receptor binding. Biochem Biophys Res Commun 465(2):218–224
Song Y, Shi Y, Huang M, Wang W, Wang Y, Cheng J, Lei Z, Zhu Z, Yang C (2019) Bioinspired engineering of a multivalent aptamer-functionalized nanointerface to enhance the capture and release of circulating tumor cells. Angewandte Chemie-Int Ed 58(8):2236–2240
Chen Y, Tyagi D, Lyu M, Carrier AJ, Nganou C, Youden B, Wang W, Cui S, Servos M, Oakes K, He S, Zhang X (2019) Regenerative NanoOctopus based on multivalent-aptamer-functionalized magnetic microparticles for effective cell capture in whole blood. Anal Chem 91(6):4017–4022
Kwon PS, Ren S, Kwon S-J, Kizer ME, Kuo L, Xie M, Zhu D, Zhou F, Zhang F, Kim D, Fraser K, Kramer LD, Seeman NC, Dordick JS, Linhardt RJ, Chao J, Wang X (2020) Designer DNA architecture offers precise and multivalent spatial pattern-recognition for viral sensing and inhibition. Nat Chem 12(1):26–35
Gao M-L, Yin B-C, Ye B-C (2019) Construction of a DNA-AuNP-based satellite network for exosome analysis. Analyst 144(20):5996–6003
Yang J, Li X, Jiang B, Yuan R, Xiang Y (2020) In situ-generated multivalent aptamer network for efficient capture and sensitive electrochemical detection of circulating tumor cells in whole blood. Anal Chem 92(11):7893–7899
Chang EK, Eckert MA, Ali MM, Riazifar H, Pone EJ, Liu L, Zhao W (2015) Facile supermolecular aptamer inhibitors of L-selectin. PLoS One 10(3)
Zhang P, Ye J, Liu E, Sun L, Zhang J, Lee S-J, Gong J, He H, Yang VC (2017) Aptamer-coded DNA nanoparticles for targeted doxorubicin delivery using pH-sensitive spacer. Front Chem Sci Eng 11(4):529–536
Jiang Y, Qiu Z, Le T, Zou S, Cao X (2020) Developing a dual-RCA microfluidic platform for sensitive E. coli O157:H7 whole-cell detections. Anal Chim Acta 1127:79–88
Kim YS, Chung J, Song MY, Jurng J, Kim BC (2014) Aptamer cocktails: enhancement of sensing signals compared to single use of aptamers for detection of bacteria. Biosens Bioelectron 54:195–198
Qin W, Chen L, Wang Z, Li Q, Fan C, Wu M, Zhang Y (2020) Bioinspired DNA nanointerface with anisotropic aptamers for accurate capture of circulating tumor cells. Adv Sci 7(19)
Zhao L, Tang C, Xu L, Zhang Z, Li X, Hu H, Cheng S, Zhou W, Huang M, Fong A, Liu B, Tseng H-R, Gao H, Liu Y, Fang X (2016) Enhanced and differential capture of circulating tumor cells from lung cancer patients by microfluidic assays using aptamer cocktail. Small 12(8):1072–1081
Lin Y, Jiang L, Huang Y, Yang Y, He Y, Lu C, Yang H (2019) DNA-mediated reversible capture and release of circulating tumor cells with a multivalent dual-specific aptamer coating network. Chem Commun 55(37):5387–5390
Dou B, Xu L, Jiang B, Yuan R, Xiang Y (2019) Aptamer-functionalized and gold nanoparticle array-decorated magnetic graphene nanosheets enable multiplexed and sensitive electrochemical detection of rare circulating tumor cells in whole blood. Anal Chem 91(16):10792–10799
Duan Y, Zhang C, Wang Y, Chen G (2022) Research progress of whole-cell-SELEX selection and the application of cell-targeting aptamer. Mol Biol Rep
Shoaib M, Shehzad A, Mukama O, Raza H, Niazi S, Khan IM, Ali B, Akhtar W, Wang Z (2020) Selection of potential aptamers for specific growth stage detection ofYersinia enterocolitica. RSC Adv 10(41):24743–24752
Ma P, Duan N, Ye H, Xia Y, Ding Z, Wang Z (2022) Selection, truncation and fluorescence polarization based aptasensor for Weissella viridescens detection. Talanta 246:123499–123499
Dai Z, Gao Q, Cheung MC, Leung HM, Lau TCK, Sleiman HF, Lai KWC, Lo PK (2016) A highly versatile platform based on geometrically well-defined 3D DNA nanostructures for selective recognition and positioning of multiplex targets. Nanoscale 8(43):18291–18295
Li J, Chen K, Zhu L, Li X, Li C, Chang Q, Xu W (2022) Multiple recognition-based sensor for pesticide residues. Front Chem 10
Chen H, Bian F, Guo J, Zhao Y (2022) Aptamer-functionalized barcodes in herringbone microfluidics for multiple detection of exosomes. Small Methods
Qi L, Liu S, Jiang Y, Lin J-M, Yu L, Hu Q (2020) Simultaneous detection of multiple tumor markers in blood by functional liquid crystal sensors assisted with target-induced dissociation of aptamer. Anal Chem 92(5):3867–3873
Funding
This work was supported by a grant from the Major Science & Technology Programs of Yunnan Province (No. 202102AE090015) and the National Key Research and Development Program (2017YFD0800704).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Yu, H., Zhu, J., Shen, G. et al. Improving aptamer performance: key factors and strategies. Microchim Acta 190, 255 (2023). https://doi.org/10.1007/s00604-023-05836-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00604-023-05836-6