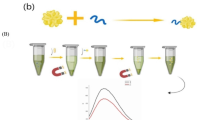
Abstract
The aim of this study was to develop a sensitive lateral flow immunoassay (LFIA) for the rapid detection of Escherichia coli (E. coli) O157:H7, a pathogen contributor to diseases and fatalities worldwide. Au nanoparticles with high stability, uniform size, and shape were synthesized and coated with heterobifunctional PEG polymer with carboxyl groups, and they were bioconjugated to be used as label in sandwich-LFIA. Then, a silver enhancement strategy was developed as an accessible, rapid, and cost-effective approach for signal amplification to reduce the limit of detection (LOD). The optimal results were achieved when a solution of silver nitrate and hydroquinone/citrate buffer was added to the strips for 4 min. This led to a decrease in the visual LOD from 2 × 106 (CFU mL−1) to 2 × 103 (CFU mL−1), resulting in a threefold improvement in sensitivity compared to the conventional LFIA system. The specificity of the system was evaluated by using non-target bacteria (E. coli BL21 and E. coli T515) and its reliability was determined by testing commercial food samples (milk, tap water, and orange juice), demonstrating its effectiveness for quickly detecting pathogenic bacteria in food products.
Graphical Abstract

Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
E. coli O157:H7 is a serotype of the bacterial species E. coli, and it belongs to a subset of Shiga toxin producers [1]. It causes severe health problems, usually through consuming contaminated or uncooked food, such as raw milk or ground beef that has not been adequately cooked. According to the World Health Organization (WHO), E. coli O157:H7 is a bacterial strain that can cause severe foodborne illness, including bloody diarrhea and kidney failure. Therefore, many countries have established regulations regarding the allowable concentration of E. coli O157:H7 in food to protect public health [2, 3]. In the USA, the Food and Drug Administration has set a zero tolerance policy for E. coli O157:H7 in certain foods, including ground beef. This means that any detectable amount of E. coli O157:H7 in these foods is illegal and can result in a product recall or enforcement action (https://www.usda.gov/topics/health-and-safety). The European Union has established regulatory limits for E. coli O157:H7 in various food categories, such as raw beef and beef products, sprouts, and milk, among others. For example, the maximum allowable concentration of E. coli O157:H7 in raw beef is 100 CFU g−1 (colony-forming units per gram), while the maximum allowable concentration in sprouts is 10 CFU g−1 [3]. Therefore, there is a strong need for quicker methods to detect pathogens to ensure public health and safety.
The available microbiological culture detection methods often require about 18–72 h to reach the suitable concentration for quantification. Immunological methods and techniques such as polymerase chain reaction and enzyme-linked immunosorbent assay require expensive equipment, laborious protocol, and technical expertise [4, 5]. It is therefore crucial to develop a fast, simple, straightforward, and sensitive technique for early detection of pathogens at low levels.
LFIAs as point-of-care diagnostic tools are cost-effective, robust, rapid, and user-friendly with diverse applications in various fields such as agriculture, food safety, environmental science, and medicine [6, 7]. However, LFIAs have certain drawbacks that can pose a challenge for researchers. These include limitations in terms of their sensitivity, non-specific binding, and inability to offer quantitative results [8]. Maintaining consistency and reliability in LFIAs is crucial, and this can be influenced by several factors, including the assay’s sensitivity and specificity, sample matrix, handling of assay components, and variability of instruments and operators [9].
Numerous studies have aimed to enhance the reliability and sensitivity of LFIAs by utilizing various labeling methods, including colloidal metal nanoparticles [10], magnetic beads [11], and quantum dots [10], along with signal amplification techniques like electrochemistry [12], enzyme catalysis [13], and silver/gold enhancement [14, 15]. Among these techniques, silver/gold enhancement offers several advantages over other techniques for enhancing sensitivity of LFIAs including high sensitivity, simplicity, rapid results, cost-effectiveness, and portability [14]. Silver staining protocols for enhanced protein detection in electrophoresis gels or in electron microscopy are a source of inspiration for LFIA systems. Several methods have been developed for toxins and biomarkers [7], and they have been applied on commercial AuNPs. The silver/gold enhancement technique involves depositing a thin layer of silver on top of the AuNPs located on the test line of an LFIA strip [14]. This enhances the signal generated by the AuNPs, leading to a higher sensitivity of the assay and the ability to detect low levels of the target analyte in LFIAs. The silver deposition on the surface of AuNPs is carried out by mixing silver salts such as nitrate or acetate with a reducing agent including hydroquinone (1,4-dihydroxybenzene), catechol (1,2-dihydroxybenzene), and resorcinol (1,3-dihydroxybenzene) [16, 17] which causes a significant color change from red to black [18]. Our research group has compared several techniques to deposit silver on the AuNPs situated on the test line of the LFIA strip, including the immersion assay, sandwich immunochromatographic assay, and conjugated pad modified with silver [14]. We found that the highest signal was produced using the immersion method, which involved a 1:1 solution of silver nitrate and hydroquinone/citrate buffer. Using this approach, we were able to improve the sensitivity of the system one order of magnitude. In other work, Panferov et al. [19] developed a fast quality control method to lower the limit of detection (LOD) for R. solanacearum using silver-enhanced LFIA. They utilized fiberglass membranes pre-absorbed with silver lactate and hydroquinone, which were placed on the analytical zone, and added a drop of silver lactate for silver enhancement. The study found that this approach improved 10 times the sensitivity of the system.
Apart from the advantages of this technique, the use of silver enhancement in LFIA has some limitations and considerations, such as potential interference from sample matrix components, or the non-specific binding of silver to non-target molecules. These aspects should be considered for optimization of the assay.
Spherical AuNPs are the most frequently nanomaterials used as label in LFIAs. These materials are popular due to their favorable physical and chemical properties, such as their optical properties, ability to be easily functionalized and stabilized, strong affinity for biomolecules, non-toxicity, affordability, and quantum and plasmonic properties, which also have potential catalytic applications [20, 21]. However, the non-specific binding of the AuNPs on the test line is a frequent problem during LFIA development, and it is important to minimize false positive results. By reducing nonspecific binding, the specificity and sensitivity of the test can be improved, leading to more accurate results [22]. One common approach is to use biocompatible and hydrophilic coatings, such as polyethylene glycol (PEG), on the surface of the nanoparticles to prevent the adsorption of proteins and other molecules. Additionally, PEGylation of AuNPs improves their dispersion and stability, prevents aggregation and denaturation of the antibodies, and minimizes the non-specific binding which is a critical consideration when designing labels for LFIA systems [21, 23, 24].
In this study, a sandwich-type LFIA was developed for rapid and specific detection of E. coli O157:H7. AuNPs with an average size of 40 nm were synthesized and functionalized with heterobifunctional (SH-PEG-COOH, 5KDa) with carboxyl groups for bioconjugation with the antibody. A silver enhancement technique was developed for signal amplification. Quantitative detection was achieved by analysis of the signal intensity on the test line or the colorimetric signal after the silver enhancement through a portable strip reader. The specificity was investigated by using non-target bacteria (E. coli BL21 and E. coli T515) and to confirm the effectiveness of the developed assay, the strips were applied to detect E. coli O157:H7 in three different types of food samples: milk, drinking water, and fruit juice.
Experimental
Reagents and instruments for the immunoassay
Mouse monoclonal antibodies to Escherichia coli O157:H7 (MBS568290 and MBS568193) were purchased from Mybiosource (San Diego, CA, USA).
Goat anti-mouse IgG, N-hydroxysuccinimide (NHS), 1-ethyl-3-[3-dimethylaminopropyl]–carbodiimide hydrochloride (EDC), 2-(N-morpholino)ethanesulfonic acid (MES), bovine serum albumin (BSA), phosphate-buffered saline (PBS), silver nitrate (AgNO3), hydroquinone (C6H4-1,4-(OH)2), gold(III) chloride hydrate (HAuCl4), and sodium citrate tribasic dihydrate (C6H5Na3O7·2H2O) were obtained from Sigma-Aldrich (St. Louis, MO, USA). Sodium dodecyl sulfate (SDS) (C12H25NaO4S) was purchased from PanReac AppliChem (Barcelona, Spain).
HS-C2H4-CONH-PEG-C3H6-COOH, MW = 5000 g mol−1 (4900 Da), and CH3O-PEG-NH2, MW = 725–850 Da were purchased from RAPP POLYMER (Tuebingen, Germany). All the reagents were prepared using Milli-Q ultrapure water (resistivity 18.2 MΩ·cm at 25 °C), unless otherwise stated.
An IsoFlow reagent dispensing system (ImageneTechnology, USA) was used to dispense the detection lines (dispense rate 0.100μL mm−1) and the strips were cut with a guillotine Fellowes Gamma (Spain).
A portable strip reader ESE Quant LR3 lateral flow system (Qiagen Inc., Germany) was used to quantify the intensity of the test line by reflectance measurements.
Bacterial strains
E. coli O157:H7, E. coli BL21, and E. coli CEC T515 were cultured in 50 ml of Tryptic Soy Broth medium at 37°C and agitation speed of 240 rpm, overnight. The culture media was centrifuged and resuspended in 5ml of PBS. In order to inactivate the E. coli O157:H7 for safe handling, the cells were heated at 100°C for 15min. Samples were frozen until further use.
Synthesis of citrate-AuNPs and PEGylation
The colloidal AuNPs coated with PEG polymer were synthesized through a two-step process that involved: (i) synthesizing the AuNPs using a seeded growth method and (ii) subsequent coating with the SH-PEG-COOH 5KDa polymer. To achieve an average nanoparticle size of 40 nm, the ratio of HAuCl4 to sodium citrate (1:0.4; 1:2; 1:4) was optimized. In the first step, a seed solution was made by preparing a solution of HAuCl4 (77, 51, or 25.4 mg) in 293 ml of Milli-Q water and sodium citrate (33, 99, or 99 mg) in 996.3 μl of Milli-Q water separately. The pH of the HAuCl4 solution was set to 5.4±0.5 using a pH meter, and it was then added to a three-neck round-bottomed flask and heated to 100°C with vigorous stirring (900 rpm) and reflux. Upon reaching 100°C, the sodium citrate was quickly injected, and the stirring was continued at that temperature for 90 min. After the reaction was stopped, the mixture was cooled to room temperature and protected from light. The size of the resulting nanoparticles was measured using UV-Vis spectroscopy after cooling.
In the following step, the suspension was stabilized using a heterobifunctional HS-PEG, 5kD via the Au-SH functional groups of the PEG polymer. The synthesized nanoparticles were first concentrated to 10 nM through centrifugation at 10000 rpm for 10 min. Then, 125 μl of SDS 10% and 120 μl of HS-PEG-COOH (458 Da) at 10 mg mL−1 were added to the solution and mixed to reach a final volume of 50 ml after the addition of Milli-Q water. Finally, 675 μL of 2 M NaOH was added to the solution, which was covered with aluminum foil and incubated overnight at room temperature with gentle mixing. After incubation, the nanoparticles were centrifuged three times at 10000 rpm for 10 min to purify them, removing any extra PEG polymer and unreacted starting materials. The size and concentration of the nanoparticles were determined by measuring their absorbance.
Characterization of AuNPs
UV-Vis spectroscopy
The spectral analysis was performed in the 200–800 nm range at room temperature with a UV-Vis spectrophotometer (PG Instrument, LTD). The concentration of the AuNPs was calculated with the following equation:
where A is the absorbance at 450 nm, ε is the molar absorption coefficient of AuNPs 1.76 × 108 cm M−1 or 11.1 ml cm−1 mg−1 at their localized surface plasmon resonance (LSPR) 450 nm [25, 26], C is the molar concentration, and l is path length of light traveling in the cuvette.
The size of nanoparticles was determined based on the absorbance at the maximum absorption peak by UV-Vis spectroscopy, following a method reported by Heiss et al. at the literature [25].
Transmission electron microscopy
The size and shape of AuNPs were characterized by TEM (JEOL-2000 Ex II TEM (Japan)).
Dynamic light scattering
The surface properties and particles size were determined by the ζ-potential value and dynamic light scattering respectively (Zetasizer Nano ZS (Malvern Instruments Ltd., UK).
Conjugation of AuNPs with E. coli O157:H7 antibody
The AuNPs coated with PEG-COOH were then functionalized with an anti-E. coli monoclonal antibody to create the immunoassay. Carbodiimide chemistry was used to activate the carboxyl groups: 1.5 ml of EDC (3 mM in MES 10 mM, pH 6) and 1.5 ml of NHS (7 mM in MES 10 mM, pH 6) were mixed with AuNPs with the concentration of 0.5 mg mL−1 for 30 min. To remove the excess of EDC and NHS, the nanoparticles were washed twice with MES buffer 10 mM by centrifugation at 10,000 rpm for 10 min. After centrifugation, E. coli detection antibody with different final concentration of 2, 4, 6, and 8 μg mL−1 in MES buffer 10 mM, pH 5.5 mixed with nanoparticles for 1.5 h. Then, 500 μl of CH3O-PEG-NH2 as a blocking agent was added to the solution and incubated at the same operational condition for 2h (concentration of 2.5% of 500 μl of MES buffer 10 mM, pH 5.5). The nanoparticle solution was centrifuged at 10,000 rpm for 10 min and washed once with MES buffer then with HEPES buffer 10 mM, pH 7.4. After removing the supernatant, the mixture was resuspended in 200 μl of stabilizer solution including HEPES buffer 10 mM, pH 7.4, Tween 20 0.1%, and BSA 0.1% and stored at 4°C.
Lateral flow strip manufacturing and assembly
A sandwich LFIA was carried out for detection of E. coli O157 bacteria. The nitrocellulose membrane was incorporated onto a backing card. Then, capture (anti-E. coli) and control (goat anti-mouse IgG) antibodies (1 mg mL−1) were dispensed on the membrane at a rate of 0.100 μL mm−1 for the test and control lines, respectively. The membrane was then dried for 1 h at 37°C. To assemble the strips, the sample pad and the absorbent pad were stuck onto the backing card, with an overlap between them of 2 mm. The complete strip was cut into individual 5-mm-wide dipsticks. For the optical density quantification, a portable reader (ESEQuant, Qiagen) was used.
Silver enhancement of LFIA test strips
After optimization of the immunoassay, the strips were washed with Milli-Q water. The immersion protocol for silver enhancement was carried out according to a protocol developed in our research group with some modifications [14]. A solution of silver nitrate (0.3%w/v in water) and hydroquinone (3% w/v in 0.5 M citrate buffer pH 4.0) as a reducing agent was prepared and stored at room temperature protected from light. Just before use, the enhancing solution was freshly prepared by 1:1 mixing the two solutions and the volume of 40 μl was dropped onto the test zone in a dark place and left there until the signal appeared. Then, the strips were washed with Mili-Q water to stop the reaction, and they were kept protected from light. All experiments were carried out by triplicate.
Food sample analysis
To evaluate the applicability of the proposed assay, three real food samples were used: orange juice, liquid milk, and tap water. Orange juice and milk were purchased from a local market. The samples (orange juice and liquid milk) firstly were diluted with PBS buffer (1:10). Then, the diluted samples were spiked with different concentrations of E. coli O157 bacteria (2×105, 2×103, 2×102 CFU mL−1). Control samples (non-spiked) were also used to investigate the effect of the complex matrix on the proposed sandwich assay and silver enhancement strategy.
Results and discussion
Optimization and characterization of synthesis of AuNPs
The optimal size for spherical AuNPs in LFIAs is typically within the range of 20–40 nm. This size range is considered optimal because nanoparticles produce a bright red color after accumulation in the test-line, as a result of the LSPR effect. This way, the line can be detected with naked-eyes or recorded by optical readers for qualitative and quantitative measurement, respectively [27]. AuNPs that are too large may result in aggregation, while smaller AuNPs are less favorable at the LSPR and reduce sensitivity. Hence, it is crucial to have the appropriate size of AuNPs to attain the best results and precision in LFIA [27, 28].
The size of AuNPs is mainly dependent on the concentration of sodium citrate during the synthesis [29, 30]. However, other factors such as the concentration of gold salt, the optimal pH, and temperature during preparation also play a role in determining the final particle size of AuNPs [29]. These factors can all impact the nucleation, growth, and stability of the AuNPs and, therefore, must be carefully considered and controlled to achieve the desired particle size. In this work, spherical AuNPs with an average size of 40 nm were synthesized by the reduction of HAuCl4 using citrate as a capping and stabilizing agent. Briefly, the synthesis of spherical AuNPs consists of the reduction of gold by sodium citrate and the stabilization of the colloidal AuNPs in an aqueous suspension [31]. During the synthesis, the citrate forms a larger layer around the colloids and partially blocks their growth. Thus, the final size of the nanoparticles is strongly related to the ratio between HAuCl4 and sodium citrate. The pH of the solution also has a dramatic effect on the size, morphology, and stability of AuNPs [20, 32]. The initial pH of HAuCl4 solution before adding sodium citrate was set at 5.4±0.5 in all the experiments.
Figure 1 displays the UV-Vis absorption spectrum of AuNPs that were prepared by using different ratios of HAuCl4 to sodium citrate. The results show that the maximum LSPR peak of AuNPs with the ratio of 1:0.4, 1:2, and 1:4 was at 520 nm, 524 nm, and 530 nm, respectively, corresponding to particle sizes of 15 nm, 20 nm, and 40 nm. The data suggests that a higher concentration of sodium citrate leads to larger particle sizes of AuNPs after 90 min of synthesis. In the other work, Ji et al. [33] reported that, by increasing the Na3Ct/HAuCl4 ratio from 0.7:1 to 3.5:1, the size of synthesized AuNPs decreased. However, a further increasing trend of average size was observed from 3.5: 1 to 28:1, instead of a continuous reduction of the average size. The presence of unreacted citrates could be one possible explanation for the observed increase in the size of AuNPs at high Na3Ct/HAuCl4 ratio. Citrates are known to have a high affinity for the surface of Au nanoparticles, and excess citrate can form a stable layer on the surface of the nanoparticles, preventing further growth [33, 34].
AuNPs with the maximum LSPR peak at 530 nm were considered in this work. They were subsequently coated with heterobifunctional PEG of a length of 5 kDa (SH-PEG-COOH). The concentration of AuNPs was determined using the formula provided in the “UV-Vis spectroscopy” section and the method from reference [25], and subsequently, a fixed value of 10 nM for AuNP PEGylation was set for this work. The PEGylation of AuNPs is essential to avoid aggregation and non-specific adsorption of the nanoparticles [23]. It also provides a suitable surface modification by providing the most adequate chemical groups for covalent binding with several biomolecules. One of the most common approaches to coat plasmonic nanoparticles is by using thiol-terminated derivatives (SH-PEG-COOH) that graft onto the surfaces of NPs covalently. In the case of the synthesis of AuNPs with citrate, by adding thiol-terminated derivatives (SH-PEG-COOH), the citrate ligands progressively exchange the SH-PEG-COOH on the surface of NPs increasing the stability of colloidal solution at high ionic strength solutions [35, 36]. On the other hand, it has been reported in the literature that the addition of SDS helps to improve the dispersion of the nanoparticles and prevent aggregation, while the use of NaOH creates an alkaline environment for coating and functionalizing the nanoparticles [37]. Fig. S1 shows the reaction of synthesized nanoparticles on the strip before and after PEGylation. As it can be seen, the PEGylation of AuNPs effectively eliminates the non-specific binding between the AuNPs and the capture antibody on the test line. Figure 2 displays the characterization of the UV-Vis spectra of the AuNPs both before and after PEGylation, as well as the distribution and size of the nanoparticles, as seen in a TEM micrograph. After the PEGylation of AuNPs, the maximum LSPR peak has only slightly increased (from 530 to 531 nm). Further characterization by means of dynamic light scattering and Z-potential measurements after PEGylation (Fig. S2 also) show a size of 42.9 ± 0.2 nm and a Z-potential of −31 ± 2 mV, respectively. These results confirm the successful PEGylation of the nanoparticles.
Development of a sandwich lateral flow immunoassay method
The first step of development of the immunoassay was the optimization of the amount of E. coli O157 detection antibody for conjugation with AuNPs. Different total concentrations of (2, 4, 6, and 8 μg mL−1 of antibody) were mixed with 0.5 mg mL−1 of AuNPs. The minimum concentration was set by analyzing the signal intensity generated on the testing line using a portable strip reader (Fig. S3). The running buffer in all the experiments was optimized with BSA 2%, Tween 20 2%, and PBS 10 mM, at pH 7.4. The concentration E. coli O157 bacteria used at this stage to compare the intensity in the different concentration of E. coli detection antibody was 2×107 (CFU mL−1). By applying sandwich LFIA, the result in Fig. S3 indicated there is no difference in signal intensity at concentrations higher than 6 μg mL−1 of E. coli detection antibody on the test-line, and this value was selected.
Then, a serial dilution of E. coli O157 bacteria starting at the initial concentration of 2×109(CFU mL−1) was prepared in 10 mM PBS pH 7.4. The sandwich LFIA was carried out in dipstick format. For the first step, 10 μl of complex of AuNPs-E. coli O157 Ab (0.2 mg mL−1) and 25 μl of different concentrations of standard solution of E. coli O157 bacteria were transferred into a microtube. Then, running buffer was added with final volume of 100 μL. The components of running buffer included BSA (2%), Tween 20 (2%), and PBS (10 mM), at pH 7.4. After mixing, the solution was incubated for 15 min and then, the strips were immersed in it and the immunochromatography was carried out in dipstick format for 15 min. After this time, the strips were washed with Mili-Q water to remove extra nanoparticles and avoiding a high background at silver enhancement. Figure 3 A and B show the schematic of sandwich LFIA and the strips in the presence of different concentration of standard solution of E. coli O157:H7 bacteria. The first line from the bottom is the test line, where capture E. coli O157:H7 antibody was immobilized by physical adsorption. The immunochromatography was performed in all the experiments for 15 min and then the strips were washed with Mili-Q water. The minimum concentration that could be detected with the naked eye was 2×106 (CFU mL−1). Fig. S4 shows the results of repeatability of the assay with different batches of AuNPs conjugated with E. coli O157: H7 detection antibody in response to different concentrations of E. coli O157:H7 at lower concentration of 2×106 (CFU mL−1) before applying the silver enhancement procedure. At concentration lower than 2×106 (CFU mL−1), it was not possible to detect any line visually. A silver enhancement protocol was developed for that concentration range and combined with LFIA.
Silver enhancement for E. coli O157 bacteria detection
The silver enhancement strategy is based on the catalytic reduction of silver ions on the surface of the AuNPs, which changes the color of the line from red to black (Fig. 4A). Our protocol was based on the immersion method, as explain in the previous section. The reaction time of silver deposition was optimized over by preparing a series of strips from 2×106 (CFU mL−1) to 2×103 (CFU mL−1). After silver deposition, the signal of false positive on the blank strips began to appear after 5 min, as shown in Fig. S5 A. Therefore, the ideal reaction time was determined to be 4 min, but longer times resulted in increased color intensity at that line.
A Schematics of the silver enhancement method and its effect on LFIA strips in the presence of various concentrations of E. coli O157:H7; B The minimum detectable concentration with AuNPs was 2 × 106 (CFU mL−1) and it dropped to 2 × 103 (CFU mL−1) after silver enhancement; C Signal intensities obtained for different concentrations of E. coli O157:H7 using AuNPs (red bars) an after silver enhancement (gray bars). The graph shows the mean ± SD of three independent measurements. D The calibration plot using the developed LFIA in the presence of different concentration of E. coli O157:H7 after silver deposition for 4 min
A series of strips with different concentration of bacteria between 2×102 (CFU mL−1) to 2×106 (CFU mL−1) was prepared and the silver enhancement was performed following a 4-min silver deposition process (Fig. 4B). After silver enhancement, the visual LOD for E. coli bacteria decreased to 2×103 CFU mL−1. This led to a threefold improvement in sensitivity compared to the conventional LFIA system. Figure 4 C shows a comparison of the results obtained for the E. coli O157:H7 detection before and after silver enhancement. The findings of the experiments conducted using different batches were evaluated for their repeatability, and the results are illustrated in Fig. S5 (B and C). A portable strip reader was used to quantify the color intensity of the test line in different concentration by reflectance measurements after silver enhancement. Based on the generated signals, the linear relationship was established between 2 × 103 and 2 × 106 (CFU mL−1), and the linear regression equation was obtained for the intensity (I) and the bacteria concentration (CFU mL−1): Log I (m.V) = 0.2948 Log [E. coli] (CFU mL−1) + 0.5008, R2=0.9925 (Fig. 4D). The LOD with the instrumental reader was 11 (CFU mL−1). It was calculated by using the (3 ×σ) / S) criterium, where σ is the standard deviation of the y-intercept and S is the slope of the calibration curve (S).
The coefficient of variation of the method (CV%) was between the range of 2.2 to 9.8% for different concentration of bacteria, which demonstrates acceptable precision for a rapid method [38].
Compared to previously published studies regarding the improvement of the sensitivity of the conventional LFIA (Table 1), AuNPs coated with the heterobifunctional PEG polymer with carboxyl groups followed by the silver enhancement has several advantages. Firstly, the use of functionalized AuNPs allows for greater control over the orientation and density of the antibodies on the surface of the nanoparticles. This can improve the specificity and sensitivity of the LFIA, as well as reduce non-specific binding of other proteins or biomolecules in the sample. Secondly, the functionalized AuNPs can also improve the stability and shelf-life of the LFIA, as the surface coating can prevent aggregation or denaturation of the antibodies and maintain their activity for longer periods of time [39]. Thirdly, the carboxyl groups on the surface of the functionalized AuNPs can also be used for conjugation with other molecules, such as enzymes or fluorescent dyes, which can further enhance the detection and visualization of the target analyte [40, 41]. Compared to magnetic nanoparticles, heterobifunctional AuNPs are more consistent in size and shape, more stable, and more adaptable for creating assays for diverse purposes [42]. Moreover, compared to other techniques like fluorescence microscopy that require specialized equipment and costly fluorescent dyes, silver enhancement offers a more accessible and cost-effective approach for signal amplification and detection. Silver enhancement does not require specialized imaging equipment to detect the signal, and the silver-enhanced lines or dots can be visualized with the naked eye or using a simple benchtop scanner. This makes silver enhancement a more accessible option for point-of-care testing or field use, especially in resource-limited settings where access to specialized equipment and expensive reagents may be limited [43].
In terms of the selection technique for silver deposition, the immersion protocol is a more simple, cost-effective, and adaptable method. It involves immersing the test strip in a microtube containing a mixture of silver nitrate and hydroquinone/citrate buffer. In contrast, the other techniques require the preparation of glass fibers separately which can increase the risk of false positives in the results due to the sensitivity of silver nitrate to light [14].
Furthermore, the specificity and selectivity of the newly developed assay was tested against non-target bacteria E. coli BL21 and E. coli CEC T515 (at a concentration of 1010 CFU mL−1). The results in Fig. 5 reveal that the current intensity towards E. coli O157:H7 is significantly higher compared to the intensity obtained for the analysis of the other bacteria, indicating that the system exhibits good selectivity for this particular serotype.
Therefore, this method has advantages in comparison with the other conventional techniques for the detection of E. coli O157:H7, with sensitivity, and reliability for on-site applications. Moreover, it is very simple and low cost, and can be completed within 30 min without complex handling procedures, expensive instruments, and specialized staff.
Food sample analysis
An assessment was conducted to evaluate the accuracy of the immunoassay in detecting bacteria in food samples. To determine the capability of the proposed assay to quickly detection of E. coli O157:H7 within 30 min, three samples (tap water, orange juice, and liquid milk) were selected for testing and spiked with the concentration of bacteria that were not visible to the naked eye (2 × 102, 2 × 103, and 2 × 105 CFU mL−1). The samples (liquid milk and orange juice) were diluted as explained in the section “Food sample analysis” and spiked with E. coli O157:H7.
Figure 6 demonstrates that this newly developed assay could detect E. coli O157:H7 at concentrations as low as 2 × 102 (CFU mL−1) in milk and 2 × 103 (CFU mL−1) in tap water and orange juice. To check the reproducibility of the results, three replicates of the tests were carried out, as shown in Fig. S6. The accuracy of the developed assay was assessed by using the recovery rate of spiked samples with varying concentrations of E. coli O157:H7. Table 2 shows the recovery rates for each concentration. The results show a possible systematic excess error. This could be related to data following a log-normal distribution, where the most probable value is lower than the mean in a normal distribution. In this case, we had to assume a normal distribution, since we work with small series of data. Anyway, the extent of the deviation seems to be highly dependent on the matrix of the sample, being lower for tap water than for orange juice and milk, at the same level of concentration. It seems therefore that there could be an effect related to the viscosity of the sample. This could cause accumulation of errors during the dilution step, since the volumes were taken by conventional micropipettes which are designed for non-viscuous aqueous solutions. Nevertheless, the excess error in this method would protect the consumer in all cases.
On the other hand, the reproducibility of the method was good, with coefficients of variation (CV%) lower than 2% for orange juice, lower than 5% for tap water, and lower than 9% for the milk sample. These values are lower than those of other LFIAs reported in the literature [46]. The CV was also calculated assuming a normal distribution (the median and the mean of the small series of data were very close). The values at Table 2 show that the CV is also dependent on the type of sample, and higher for the lower concentration detected, as expected when results are close to the LOD. The observed difference at the ability of the developed LFIA assay to detect low concentrations of E. coli O157:H7 in milk, down to 2 × 102 CFU mL−1 could be attributed to the different composition of each matrix. Milk is a complex biological fluid with high protein and fat content, whereas orange juice and tap water are less complex matrices with lower protein and fat content. In another work from our research group using biological samples, better detectability was also found when moving from standard solutions to serum samples, due to a similar effect of the higher chemical complexity of the biofluid matrix [50].
The results demonstrate the effectiveness of the developed LFIA in detecting E. coli O157:H7 in various food samples.
Conclusion
In conclusion, this study has introduced a highly sensitive LFIA combined with a silver enhancement strategy that allows for rapid detection of E. coli O157:H7. This approach provides several benefits over other LFIA methods that have been previously reported in the literature for this system. The assay is rapid and can provide results in 30 minutes. The use of heterobifunctional PEG polymer AuNPs with carboxyl groups makes the system more adaptable for detecting different target analytes, improving its stability and shelf-life, specific, and minimize the non-specific binding, which ensures reliable results. Silver enhancement technique was able to assess a linear relation ranging between 2 × 103 (CFU mL−1) and 2 × 106 (CFU mL−1), and it dropped the LOD to 2 × 103 (CFU mL−1). The assay’s specificity, selectivity, and accuracy were tested against non-target bacteria and actual food samples, with promising outcomes. Recovery analysis showed that the method can efficiently detect at least 2 × 102 (CFU mL−1) of E. coli O157:H7 in milk samples and 2 × 103 (CFU mL−1) in orange juice and tap water, with good recoveries and CV (< 10%) in all cases tested. Therefore, compared to other LFIAs reported in the literature, this method is highly specific, rapid, low-cost, and reliable to protect the consumer. These characteristics could make this method a good candidate for the rapid detection of E. coli O157:H7 bacteria and other pathogens in industry and medical fields.
References
Gally DL, Stevens MP (2017) Microbe profile: Escherichia coli O157: H7 – notorious relative of the microbiologist’s workhorse. Microbiology (United Kingdom) 163:1–3. https://doi.org/10.1099/mic.0.000387
World Health Organization Food and Agriculture Organization of the United Nations. In: Enterohaemorrhagic Escherichia coli in raw beef and beef products: approaches for the provision of scientific advice: meeting report. Microbiological Risk Assessment Series No. 18. Geneva: FAO/WHO. 126 pp.
Griffin PM, Ostroff SM, Tauxe RV et al (1998) Illnesses associated with Escherichia coli 0157:H7 infections a broad clinical spectrum. Ann Intern Med 109(9):705–712. https://doi.org/10.7326/0003-4819-109-9-705
Nnachi RC, Sui N, Ke B Luo Z, Bhalla N, He D, Yang Z (2022) Biosensors for rapid detection of bacterial pathogens in water, food and environment. Environ Int 166:107357. https://doi.org/10.1016/j.envint.2022.107357
Mohammad Lukman Y, Nor Dyana Z, Rahmah N, Khairunisak AR (2018) Development and evaluation of colloidal gold lateral flow immunoassays for detection of Escherichia coli O157 and Salmonella typhi. J Phys Conf Ser 1082(1):012049
Sena-Torralba A, Álvarez-Diduk R, Parolo C et al (2022) Toward next generation lateral flow assays: integration of nanomaterials. Chem Rev. https://doi.org/10.1021/acs.chemrev.1c01012
Bishop JD, Hsieh HV, Gasperino DJ, Weigl BH (2019) Sensitivity enhancement in lateral flow assays: a systems perspective. Lab Chip 19:2486–2499
Hristov DR, Rodriguez-Quijada C, Gomez-Marquez J, Hamad-Schifferli K (2019) Designing paper-based immunoassays for biomedical applications. Sensors (Switzerland) 19
Waugh C, Clark G (2021) Factors affecting test reproducibility among laboratories. OIE. Revue Scientifique et Technique 40:131–143. https://doi.org/10.20506/rst.40.1.3213
Chen X, Ding L, Huang X, Xiong Y (2022) Tailoring noble metal nanoparticle designs to enable sensitive lateral flow immunoassay. Theranostics 12:574–602
Moyano A, Serrano-Pertierra E, Salvador M, Martínez-García JC, Rivas M, Blanco-López MC (2020) Magnetic lateral flow immunoassays. Diagnostics 10(5):288. https://doi.org/10.3390/diagnostics10050288
Cheng J, Yang G, Guo J et al (2022) Integrated electrochemical lateral flow immunoassays (eLFIAs): Recent advances. Analyst 147:554–570. https://doi.org/10.1039/D1AN01478A
Calabria D, Calabretta MM, Zangheri M, Marchegiani E, Trozzi I, Guardigli M, Michelini E, Di Nardo F, Anfossi L, Baggiani C, Mirasoli M (2021) Recent advancements in enzyme-based lateral flow immunoassays. Sensors 21(10):3358. https://doi.org/10.3390/s21103358
Rodríguez MO, Covián LB, García AC, Blanco-López MC (2016) Silver and gold enhancement methods for lateral flow immunoassays. Talanta 148:272–278. https://doi.org/10.1016/j.talanta.2015.10.068
Wu KH, Huang WC, Chang SC, Shyu RH (2022) Colloidal silver-based lateral flow immunoassay for detection of profenofos pesticide residue in vegetables. RSC Adv 12:13035–13044. https://doi.org/10.1039/d2ra01654k
Anfossi L, di Nardo F, Giovannoli C et al (2013) Increased sensitivity of lateral flow immunoassay for ochratoxin A through silver enhancement. Anal Bioanal Chem 405:9859–9867. https://doi.org/10.1007/s00216-013-7428-6
Choi H, Kang T, Um K et al (2014) Reduction of silver ions in gold nanoparticle suspension for detection of dihydroxybenzene isomers. Colloids Surf A Physicochem Eng Asp 459:120–127. https://doi.org/10.1016/j.colsurfa.2014.06.050
Panferov VG, Safenkova I, Byzova NA et al (2018) Silver-enhanced lateral flow immunoassay for highly-sensitive detection of potato leafroll virus. Food Agric Immunol 29:445–457. https://doi.org/10.1080/09540105.2017.1401044
Panferov VG, Safenkova I, Varitsev YA et al (2016) Development of the sensitive lateral flow immunoassay with silver enhancement for the detection of Ralstonia solanacearum in potato tubers. Talanta 152:521–530. https://doi.org/10.1016/j.talanta.2016.02.050
Mirica AC, Stan D, Chelcea IC Mihailescu CM, Ofiteru A, Bocancia-Mateescu L.-A (2022) Latest trends in lateral flow immunoassay (LFIA) detection labels and conjugation process. Front Bioeng. Biotechnol 10. https://doi.org/10.3389/fbioe.2022.922772
Amina SJ, Guo B (2020) A review on the synthesis and functionalization of gold nanoparticles as a drug delivery vehicle. Int J Nanomedicine 15:9823–9857. https://doi.org/10.2147/IJN.S279094
Castrejón-Jiménez NS, García-Pérez BE, Reyes-Rodríguez Vega-Sánchez V, Martínez-Juárez VM, Hernández-González JC (2022) Challenges in the Detection of SARS-CoV-2: evolution of the Lateral Flow Immunoassay as a Valuable Tool for Viral Diagnosis. Biosensors (Basel) 12 (9):728. https://doi.org/10.3390/bios12090728
Reznickova A, Slavikova N, Kolska Z et al (2019) PEGylated gold nanoparticles: stability, cytotoxicity and antibacterial activity. Colloids Surf A Physicochem Eng Asp 560:26–34. https://doi.org/10.1016/j.colsurfa.2018.09.083
Qian W, Murakami M, Ichikawa Y, Che Y (2011) Highly efficient and controllable PEGylation of gold nanoparticles prepared by femtosecond laser ablation in water. J Phys Chem C 115:23293–23298. https://doi.org/10.1021/jp2079567
Haiss W, Thanh NTK, Aveyard J, Fernig DG (2007) Determination of size and concentration of gold nanoparticles from UV-Vis spectra. Anal Chem 79:4215–4221. https://doi.org/10.1021/ac0702084
Alfranca G, Artiga Á, Stepien G et al (2016) Gold nanoprism-nanorod face off: comparing the heating efficiency, cellular internalization and thermoablation capacity. Nanomedicine 11:2903–2916. https://doi.org/10.2217/nnm-2016-0257
Kim DS, Kim YT, Hong SB et al (2016) Development of lateral flow assay based on size-controlled gold nanoparticles for detection of hepatitis B surface antigen. Sensors (Switzerland) 16. https://doi.org/10.3390/s16122154
Khlebtsov BN, Tumskiy RS, Burov AM et al (2019) Quantifying the numbers of gold nanoparticles in the test zone of lateral flow immunoassay strips. ACS Appl Nano Mater 2:5020–5028. https://doi.org/10.1021/acsanm.9b00956
Tyagi H, Kushwaha A, Kumar A, Aslam M (2016) A facile pH controlled citrate-based reduction method for gold nanoparticle synthesis at room temperature. Nanoscale Res Lett 11. https://doi.org/10.1186/s11671-016-1576-5
Harimurti S, Rohiman A, Sulthoni MA, Idris I (2013) The effect of trisodium citrate concentration on the size of gold nanoparticles. In Proceeding in International Conference on Electronics Technology and Industrial Development, pp 282-284.
Park JW, Shumaker-Parry JS (2014) Structural study of citrate layers on gold nanoparticles: role of intermolecular interactions in stabilizing nanoparticles. J Am Chem Soc 136:1907–1921. https://doi.org/10.1021/ja4097384
Borse V, Konwar AN (2020) Synthesis and characterization of gold nanoparticles as a sensing tool for the lateral flow immunoassay development. Sensors Int 1. https://doi.org/10.1016/j.sintl.2020.100051
Ji X, Song X, Li J et al (2007) Size control of gold nanocrystals in citrate reduction: the third role of citrate. J Am Chem Soc 129:13939–13948. https://doi.org/10.1021/ja074447k
Harimurti S, Rohiman A, Sulthoni MA, et al (2013) The effect of trisodium citrate concentration on the size of gold nanoparticles embedded flash memory design for IoT SoC view project Stanas REE View project The Effect of Trisodium Citrate Concentration on the Size of Gold Nanoparticles as Catalyst in Growing Silicon Nanowire
Guerrini L, Alvarez-Puebla RA, Pazos-Perez N (2018) Surface modifications of nanoparticles for stability in biological fluids. Materials 11(7):1154. https://doi.org/10.3390/ma11071154
Hernandez Y, González-Pastor R, Belmar-Lopez C et al (2019) Gold nanoparticle coatings as efficient adenovirus carriers to non-infectable stem cells. RSC Adv 9:1327–1334. https://doi.org/10.1039/C8RA09088B
Chadha R, Sharma R, Maiti N et al (2015) Effect of SDS concentration on colloidal suspensions of Ag and Au nanoparticles. Spectrochim Acta A Mol Biomol Spectrosc 150:664–670. https://doi.org/10.1016/j.saa.2015.06.005
Shechtman O (2013) The coefficient of variation as an index of measurement reliability. In: Methods of Clinical Epidemiology, pp. 39-49. https://doi.org/10.1007/978-3-642-37131-8_4
Robson T, Shah DSH, Solovyova AS, Lakey JH (2018) Modular protein engineering approach to the functionalization of gold nanoparticles for use in clinical diagnostics. ACS Appl Nano Mater 1:3590–3599. https://doi.org/10.1021/acsanm.8b00737
Ramsey AV, Bischoff AJ, Francis MB (2021) Enzyme activated gold nanoparticles for versatile site-selective bioconjugation. J Am Chem Soc 143:7342–7350. https://doi.org/10.1021/jacs.0c11678
Kyrychenko A, Karpushina G, Svechkarev D et al (2012) Fluorescence probing of thiol-functionalized gold nanoparticles: is alkylthiol coating of a nanoparticle as hydrophobic as expected? J Phys Chem C 116:21059–21068. https://doi.org/10.1021/jp3060813
Milan J, Niemczyk K, Kus-Liśkiewicz M (2022) Treasure on the Earth—gold nanoparticles and their biomedical applications. Materials 15(9):3355. https://doi.org/10.3390/ma15093355
Ongo G (2014) Rapid detection of pathogenic bacteria using a silver enhancement sandwich assay, Master's thesis, McGill University
Li Q, Yang Y, Hu F et al (2019) Rapid detection of Escherichia coli O157:H7 by a fluorescent microsphere-based immunochromatographic assay and immunomagnetic separation. Anal Biochem 564–565:32–39. https://doi.org/10.1016/j.ab.2018.10.009
Shin JH, Hong J, Go H et al (2018) Multiplexed detection of foodborne pathogens from contaminated lettuces using a handheld multistep lateral flow assay device. J Agric Food Chem 66:290–297. https://doi.org/10.1021/acs.jafc.7b03582
Liu H, Chen C, Zhang C et al (2019) Functionalized AuMBA@Ag nanoparticles as an optical and SERS dual probe in a lateral flow strip for the quantitative detection of Escherichia coli O157:H7. J Food Sci 84:2916–2924. https://doi.org/10.1111/1750-3841.14766
Kong M, Shin JH, Heu S et al (2017) Lateral flow assay-based bacterial detection using engineered cell wall binding domains of a phage endolysin. Biosens Bioelectron 96:173–177. https://doi.org/10.1016/j.bios.2017.05.010
López-Soriano P, Noguera P, Gorris MT et al (2017) Lateral flow immunoassay for on-site detection of Xanthomonas arboricola pv. Pruni in symptomatic field samples. PLoS One 12. https://doi.org/10.1371/journal.pone.0176201
Hu J, Jiang YZ, Tang M et al (2019) Colorimetric-fluorescent-magnetic nanosphere-based multimodal assay platform for salmonella detection. Anal Chem 91:1178–1184. https://doi.org/10.1021/acs.analchem.8b05154
Huang H, Zhao G, Dou W (2018) Portable and quantitative point-of-care monitoring of Escherichia coli O157:H7 using a personal glucose meter based on immunochromatographic assay. Biosens Bioelectron 107:266–271. https://doi.org/10.1016/j.bios.2018.02.027
Acknowledgements
The authors thank Dr. Pilar García and Dr. Victor Ladero at the Dairy Research Institute of Asturias (IPLA-CSIC, Spain) for the preparation and inactivation of the bacterial cultures.
Funding
Open Access funding provided thanks to the CRUE-CSIC agreement with Springer Nature. This work is part of a project that has received funding from the European Union’s Horizon 2020 research and innovation program under the Marie Skłodowska-Curie Grant Agreement No. 813439. It was also funded by Consejería de Educación y Ciencia del Principado de Asturias (Ref. SV-PA-21-AYUD/2021/52132).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.






Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Bazsefidpar, S., Serrano-Pertierra, E., Gutiérrez, G. et al. Rapid and sensitive detection of E. coli O157:H7 by lateral flow immunoassay and silver enhancement. Microchim Acta 190, 264 (2023). https://doi.org/10.1007/s00604-023-05834-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00604-023-05834-8