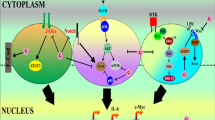
Abstract
A combination of omics techniques (transcriptomics and metabolomics) has been used to elucidate the mechanisms responsible for the antitumor action of a nanosystem based on a Ag core coated with mesoporous silica on which transferrin has been anchored as a targeting ligand against tumor cells (Ag@MSNs-Tf). Transcriptomics analysis has been carried out by gene microarrays and RT-qPCR, while high-resolution mass spectrometry has been used for metabolomics. This multi-omics strategy has enabled the discovery of the effect of this nanosystem on different key molecular pathways including the glycolysis, the pentose phosphate pathway, the oxidative phosphorylation and the synthesis of fatty acids, among others.
Graphical Abstract

Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The main drawbacks of traditional cancer treatments are the associated side effects and the presence of multidrug-resistant cancer cells, making it necessary to develop new approaches for cancer treatment [1,2,3]. In this sense, the use of nanosystems as drug delivery systems is a promising way for cancer treatment [4]. Mesoporous silica nanoparticle–based (MSNs) nanosystems have been widely employed with this aim thanks to their physico-chemical properties, such as high surface area, possibility of surface functionalization, or tunable size, among others, which confer these nanosystems a great versatility [5,6,7,8,9]. Apart from improving the pharmaceutical properties of therapeutic compounds, these nanoplatforms allow the co-delivery of different therapeutic agents, constituting an alternative to overcome cancer cell drug resistance [10,11,12] or even providing dual antimicrobial and osteogenic effects in the case of bone infection [13]. Taking this into account, the evaluation of the biomolecular mechanisms by which the nanosystems exert their antitumoral effects is crucial to identify possible cellular defense mechanisms activated after nanosystem exposure, thus allowing the improvement of the functionality of these nanoplatforms to increase their therapeutic action. In this context, the application of -omic techniques, such as proteomics, metabolomics, or transcriptomics, helps to evaluate impaired molecular pathways after exposure to cytotoxic agents [14, 15]. While metabolomics allows the identification of low molecular weight compounds involved in essential cellular functions in a wide variety of samples including cells, tissues, or body fluids [16, 17], transcriptomics profiling provides information about the expression levels of different genes comprising the transcriptome of a biological sample at a certain moment, since gene expression is very sensitive to external perturbations, such as the exposure to antitumoral agents [18]. The combination of these two powerful discovery tools (metabolomics and transcriptomics) allows for the identification of complete biochemical pathways involved in the biological process under study [19].
In our previous study, a hemocompatible transferrin-decorated core–shell nanosystem based on a silver nanoparticle core covered by a mesoporous silica coating (Ag@MSNs-Tf) was developed and extensively characterized. This nanosystem was proved to be selectively internalized by HepG2 cells, which overexpress their transferrin receptors; then, it was able to significantly reduce cellular viability and induce cellular death. In that work, it was demonstrated that Ag@MSNs-Tf was able to induce cellular apoptosis via the activation of the caspase cascade and through the interaction of different cell cycle components with membrane apoptotic receptors [20]. However, much of the biochemical mechanisms by which this nanosystem exerts its antitumor potential remain to be elucidated. In this work, in order to further reveal the molecular mechanisms involved, both targeted and untargeted metabolomics have been combined with transcriptomic profiling. The results obtained have allowed us to obtain a much more complete picture of how the Ag@MSNs-Tf nanosystem exerts its antitumor action, therefore providing useful information for the design of new, more complex, and more effective nanosystems.
Materials and methods
Ag@MSNs-Tf synthesis and characterization
Ag@MSNs-Tf nanosystem was synthesized as previously described [20]. Briefly, AgNPs were synthesized in a CTAB-containing medium prior to mesoporous silica shell formation. Then, Ag@MSNs were functionalized to display carboxylic acid groups (-COOH) onto their surface (Ag@MSNs-COOH). Finally, transferrin (Tf) was covalently grafted on the nanosystem via carbodiimide chemistry (Ag@MSNs-Tf). The nanosystem preparation was monitored after each synthetic step by thermogravimetric and differential thermal analysis (TGA and DTA) in a PerkinElmer Pyris Diamond TG/DTA analyzer, Fourier transform infrared (FTIR) spectroscopy using a Thermo Nicolet Nexus spectrometer, electrophoretic mobility measurements to calculate the values of the zeta-potential (ζ), and dynamic light scattering (DLS) performed in a Zetasizer Nano ZS Malvern instrument and transmission electron microscopy acquired in a JEOL JEM 1400 microscope.
Cell culture
Human hepatocellular carcinoma cell line (HepG2) was maintained in Dubelcco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum (FBS) and 1% penicillin/streptomycin at 37 °C and 5% CO2.
Cytotoxicity assays
For the cell viability assay, HepG2 cells were seeded on 96-well plates 24 h prior to the experiment. After cell attachment, cells were exposed to 10, 15, and 25 µg/mL of Ag@MSNs-Tf for 72 h. Then, 20 µL of MTT reagent (3-(4,5-dimethyl-thiazol-2-yl)2,5-diphenyl tetrazolium bromide) was added to each well and incubated for 4 h at 37 °C. The media was then removed, and the generated formazan crystals were dissolved in 100 mL of dimethyl sulfoxide. Absorbance was measured at 595 nm using a microplate reader (Sunrise, Tecan).
Targeted metabolomics
HepG2 cells were seeded into P100 plates and exposed to 10 µg/mL of the Ag@MSNs-Tf nanosystem for 72 h. After the exposure time, cells were placed on ice and washed with 10 mL of 0.9% NaCl. Before harvesting, 100 µL of methanol at − 20 °C was added. Then, 400 µL of an ice-cold 0.4% formic acid (v/v) solution was used to resuspend the cells, which were transferred to a 1.5-mL Eppendorf tube and vortexed for 1 min. After 3 min of incubation on ice, 45 µL of 15% NH4HCO3 (w/v) was added to neutralize the pH of the samples. Samples were then vortexed for 1 min and incubated for 20 min on ice. Prior to the LC–MS analysis, samples were centrifuged at 16,000 g and 4 °C for 10 min, and the supernatant was filtered through a PTFE membrane (pore size 0.22 µm). Total protein concentration of each sample was calculated by means of the Bradford assay, thus allowing further normalization of the concentration of each metabolite. The LC–MS analysis was carried out in multiple reaction monitoring (MRM) mode as previously described [21].
Differences in ATP, ADP, NADH, and NAD+ contents between control and cells exposed to Ag@MSNs-Tf were assessed by ANOVA statistical analysis at a 95% confidence level (p value < 0.05), followed by the Bonferroni test.
Untargeted metabolomics
Seven replicates (107 cells approx.) of each studied condition, HepG2 cells exposed to 10 µg/mL of Ag@MSNs-Tf nanosystem for 72 h and control untreated cells, were prepared. For the extraction of intracellular metabolites, cells were washed with 10 mL of 0.9% NaCl and placed on ice. The metabolic cycle was quenched by adding 400 µL of methanol at − 20 °C and 400 µL of ice-cold water. Then, cells were scraped and transferred into 1.5-mL tubes. Four hundred microliters of CHCl3 at − 20 °C was added to each sample; the tubes were incubated in a shaker for 20 min at 1400 rpm and 4 °C, and then centrifuged at 16,000 g and 4 °C for 5 min. After the separation of the polar and non-polar phases, 300 µL from each phase was transferred to different glass vials. The protein interphase was removed and the total protein concentration of each sample was estimated using the Bradford assay. 50 mg/L of 4-chloroplenylalanine was added to each sample as an internal standard. Then, the extracts were evaporated under a N2 stream. Prior to GC–MS analysis, the extracted metabolites were chemically derivatized by adding 30 µL of 50 mg/L methoxyamine hydrochloride in pyridine. The mixture was incubated at 500 rpm for 90 min at 37 °C. Subsequently, 60 µL of N,O-bis(trimethylsilyl)trifluoroacetamide (BTSFA) containing 1% trimethylsilyl chloride (TMCS) was added and incubated at 500 rpm for 1 h at 60 °C [22]. Samples were analyzed in a gas chromatograph (7890A, Agilent) coupled to a time-of-flight (TOF) high-resolution mass spectrometer (GCT premier Micromass, Waters) using the conditions previously described [21].
Chromatographic data were analyzed with Mass Lynx software and the peak area of each metabolite was normalized with the corresponding internal standard area. Metabolites were identified based on both the mass spectra and the accurate mass value using the NIST MS search 2.0 library.
Transcriptome analysis
To evaluate potential alteration in the mRNA expression levels of HepG2 cells exposed to the proposed nanosystem, a microarray-based transcriptome profiling was carried out. HepG2 cells were seeded in P100 plates for 24 h, and then exposed to 10 µg/mL of Ag@MNSs-Tf for 72 h. The total mRNA was extracted from each sample using a commercial kit (PureLink® Invitrogen). Briefly, cells were harvested and centrifuged and the supernatant was removed. Cells were lysed with a buffer containing 2-mercaptoethanol. Lysates were then centrifuged and one volume of 70% ethanol was added to the samples. Seven hundred microliters of this volume was transferred to a spin cartridge and centrifuged. Afterwards, samples were rinsed with washing buffer and RNase-free water was added to the spin cartridge prior to centrifugation. The purified RNA was stored at − 80 °C for further analysis. Samples were processed with GeneChip® WT PLUS Reagent Kit (Applied Biosystems) hybridized with Clariom™ D array and Human (Applied Biosystems) and scanned with a GeneChip® Scanner 3000 7G (Applied Biosystems). Raw data were processed with RMA algorithm included in the Transcriptome Analysis Console (Applied Biosystems) for normalization and gene levels analysis. For each experimental condition (control and Ag@MSNs-Tf exposed cells), three microarray experiments corresponding to three independent RNA replicates were processed and analyzed. Fold changes between both experimental conditions were calculated as a quotient between the mean of the gene expression signals. Statistical analysis was performed with ebayes limma included in the Transcriptome Analysis Console.
RT-qPCR analysis of the electron transport chain complexes
The mRNA levels of the genes involved in the electron transport complexes were measured in HepG2 cells after exposure to 10 μg/mL of Ag@MSNs-Tf for 72 h. Total RNA was isolated using the TRIzol® reagent (Invitrogen) according to the manufacturer’s instructions. The quantity of extracted RNA was measured using a NanoDrop One (Thermo Fisher Scientific). The synthesis of cDNA with integrated removal of genomic DNA contamination was performed using a Quantitec reverse transcription kit (Qiagen) employing 1 μg of total RNA. RT-qPCR analysis was carried out using TaqMan gene expression assays (Thermo Fisher Scientific) and TaqMan Fast advance master mix (Thermo Fisher Scientific) according to the manufacturer’s instructions. The references of the TaqMan gene expression assays used are listed in Table S1 in Supplementary data. All reactions were performed in a final volume of 10 μL. The reaction protocol was 2 min at 50 °C, 10 min at 95 °C for activating the polymerase, and 40 cycles of 15 s at 95 °C and 1 min at 60 °C. Relative expression of genes was normalized using GAPDH as the endogenous control. Gene expression in each sample was calculated as 2−ΔΔCt.
Results
Effect of the nanosystem on the energy metabolism of tumoral cells
Before evaluating the effect of the proposed nanosystem on the energy metabolism of exposed tumoral cells, an analytical characterization of the material (Figs. S1 and S2 and Table S2 in Supplementary data) as well as assessment of its cytotoxicity (Fig. S3 in Supplementary data) has been carried out.
An important aspect when testing the anti-tumor potential of a drug is to check that it affects the energy metabolism of tumor cells, which is normally increased. For this reason, we evaluated the levels of the four key metabolites in energy metabolism (ATP, ADP, NADH, and NAD+) in cells exposed to the nanosystem (Ag@MSNs-Tf) in comparison to control cells. For this purpose, a targeted metabolomics method based on the MRM mode was employed. As shown in Fig. 1, the levels of both ATP and ADP were significantly increased after exposure to 10 µg/mL Ag@MSNs-Tf for 72 h, thus showing alteration of the energy-related machinery in treated cells. On the contrary, the levels of NADH and NAD+ remained unchanged at the tested exposure condition.
Since ATP levels were found to be significantly increased in cells exposed to the nanosystem, it was decided to evaluate whether this effect was related to an alteration in the expression levels of genes involved in the electron transport chain (ETC). For this purpose, four transcripts (NDUFS1, CYC1, COX5A, and MT-ATP6) were analyzed by RT-qPCR. CYC1 and MT-ATP6 were found significantly overexpressed in HepG2 cells after exposure to the nanosystem (Fig. 2); thus confirming the enhanced activity of the ETC. Furthermore, the overexpression of these two transcripts is also related to an increase in ATP synthesis, which can be well related to the results obtained by targeted metabolomics, where ATP levels were found to be significantly increased in cells treated with the nanosystem.
Untargeted metabolomics results
Metabolites with a NIST Rmatch higher than 700 were considered in order to guarantee the reliability of the identification. Based on this criterion, 26 metabolites present in both samples (control and cells exposed to the nanosystem) were quantified, including fatty acids, sugars, and amino acids, among other types of molecules (Table S3 in Supplementary material). To evaluate the similarity between the concentration levels of the metabolites, the Pearson correlation index (r) was used [23], which for 95% confidence would be 0.334 and 0.312 for n = 25 and n = 40, respectively. However, applying a stricter criterion, only correlations with an r value greater than 0.6 were taken into account (Table S4 in Supplementary material). Among these data, malic acid and isocitric acid (r = 0.8716); malic acid and ribitol (r = 0.8323); malic acid and phosphoric acid (r = 0.8149); malic acid and glutamine (r = 0.8026); isocitric acid and galactopyranose (r = 0.8232); D-ribofuranose and L-threonine (r = 0.8121); glutamine and D-ribofuranose (r = 0.8633); glutamine and erythritol (r = 0.8563); and L-threonine and erythritol (r = 0.8979) are worth noting since they showed a high positive correlation (> 0.8), which means that if the levels of one increased, the levels of the other increased similarly.
Subsequently, a principal component analysis (PCA) was performed on the quantified metabolites in order to detect metabolic alterations and patterns. This analysis allowed the identification of two distinct groups based on the first two principal components (Fig. 3A), which comprised 62.2% of the variance explained. On the other hand, principal component 1 (PC1), which explained 46% of the variance explained, was able to clearly separate the two experimental groups. The contribution of the variables (metabolites) represented by the loadings plot (Fig. 3B) showed the metabolites contributing the most to the separation of the groups: L- serine (23), rythonic acid (13), phosphoric acid (6), L-aspartic acid (5), and lauric acid (17).
PCA results from the GC–MS data. A 2D scores plot of the first principal component (PC1) versus the second principal component (PC2) for control (n = 7) (red area) and cells treated with 10 µg/mL of Ag@MSNs-Tf for 72 h (n = 7) (green area). B Loading plot of PC1 versus PC2 for the 26 quantified metabolites (the number of each metabolite correlates with the identified metabolites as shown in Supplemental Table S3)
Finally, using Student’s t-test (p-value < 0.05), it was found that 19 of the 26 quantified analytes showed significant differences in terms of concentration level when comparing control cells and cells treated with Ag@MSNs-Tf. Table 1 shows the 19 altered metabolites and includes their chromatographic retention time and Rmatch, confirming their correct identification. The table also includes the RM for each metabolite, which is defined as the ratio of the mean of the peak areas for each compound found in cells exposed to the nanosystem to the mean of the peak areas for the same compounds in control cells.
Transcriptome profiling
Transcriptomic analysis allowed the evaluation of more than 20,000 well-annotated human genes (Table S5 in Supplementary data). Table 2 lists the genes that were considered differentially expressed between control samples and cells exposed to Ag@MSNs-Tf. Genes with a log2 fold change above 1.2 (11 genes) or below 0.8 (11 genes) were considered altered at a significance level of 95%. The altered genes were found to be involved in different processes such as glycolysis, pentose phosphate pathway (PPP), electron transport chain (ETC), oxidative phosphorylation (OXPHOS), and amino acids transformation.
Discussion
In the present study, the biomolecular mechanisms responsible for the antitumor action observed for the new Ag@MSNs-Tf nanosystem have been studied in depth, using a multi-omic approach based on transcriptomics and both targeted and untargeted metabolomics.
In the first experiment, the state of the cellular energy machinery after treatment has been evaluated by means of targeted metabolomics, showing a deregulation based on a significant increase in ATP and ADP levels in the treated cells (Fig. 1). This alteration in ATP and ADP levels is also supported by the overexpression of CYC1 and MT-ATP6 genes found by RT-qPCR, since both genes are related to ATP synthesis. While ATP is the major source of energy for tumoral and non-tumoral cells for maintaining their viability [24], and it is known that ATP levels are increased in most tumors to satisfy the increased energy demand due to its fast growth, it has also been shown that ATP is also needed for apoptosis to occur, since processes as the apoptosome complex formation or the processing of pro-caspase 9 are ATP-dependent [25]. Such processes were previously proved by protein expression analysis to be induced in cells after treatment with Ag@MSNs-Tf [14, 20], which would explain the elevated levels of ATP found. As for the increased ADP levels induced by the nanosystem, it is known that the levels of this metabolite are usually increased as a consequence of ATP consumption, so higher ATP levels could lead to an increase in ADP levels. In addition, ADP levels have been reported to be related to the rate of oxidative phosphorylation (OXPHOS), since an increase in ADP levels produces a rise in the activity of this process [26]. However, since cancer cells reprogram their energetic metabolism to obtain ATP mostly from glycolysis, in a process known as the “Warburg effect” [27], it is still necessary to establish the source of the increase in ATP levels found in cells treated with Ag@MSNs-Tf nanosystem.
In this sense, the untargeted metabolomics analysis has allowed the acquisition of information about the status of the pathways involved in ATP production and other metabolic pathways. Metabolites of different natures such as carbohydrates, amino acids, tricarboxylic acid (TCA) cycle intermediates, fatty acids, and inorganic compounds have been identified. However, only 26 of these metabolites met the quantification criteria.
Out of these 26 metabolites, 19 were found to be significantly deregulated in HepG2 cells after exposure to the Ag@MSNs-Tf nanosystem (Table 1). Regarding the transcriptome profiling, 22 transcripts codifying enzymes involved in different metabolic pathways (Table 2) were found altered upon treatment with Ag@MSNs-Tf. Considering the set of metabolites and transcripts found by the multi-omics approach and that appear altered in HepG2 cells after treatment with the proposed nanosystem, it is possible to conclude that there are 4 main mechanisms by which this nanosystem exerts its anti-tumor action: the glycolysis, the pentose phosphate pathway (PPP), the oxidative phosphorylation, and the fatty acid biogenesis (Fig. 4). These mechanisms will be discussed below, including the pieces of evidence found in the omics analysis carried out.
Effect on glycolysis
As commented before, tumoral cells usually reprogram their energetic metabolism through a shift from OXPHOS to glycolysis due to the Warburg effect [28, 29]. Out of all the metabolites found deregulated, the increased levels of 2 isoforms of glucose (D-glucose (RM = 1.76) and D-glucopyranose (RM = 2.76)) are worth noting in this context. This high glucose level could be a symptom of the increased uptake of glucose and the activation of the glycolysis pathway as part of the Warburg effect, since it is known that cancer cells increase the uptake of glucose to generate glycolytic intermediates required for the synthesis of carbohydrates, fatty acids, and proteins, and to increase ATP production, as it has been observed after the exposure to Ag@MSNs-Tf (Fig. 1) [30, 31]. Taking this into account, the mRNA expression levels of all members of the glucose transporters SLC2A family were assessed after exposure to the nanosystem, finding 4 of them deregulated (SLC2A3, SLC2A5, SLC2A10, and SLC2A14). In the case of SLC2A5 (FC = 0.78) and SLC2A10 (FC = 0.79), these two transporters have low affinity for glucose as they are mainly involved in the transport of fructose in the case of SLC2A5 (also known as GLUT5) [32], and dehydroascorbic acid in the case of SLC2A10 (known as GLUT10) [33, 34]. Despite this fact, the downregulation of both transporters might have important effects on cancer cells, since the knockdown of GLUT5 has been proved to deplete fructose uptake and decrease cell proliferation [32]. As for GLUT10, this transporter is implicated in the maintenance of ascorbic acid homeostasis under oxidative stress conditions, so the downregulation of GLUT10 may promote the proliferation of ROS [33, 34]. Of special importance is the depletion of mRNA levels of SLC2A3 (or GLUT3) (FC = 0.50) because of the high affinity of this transporter towards glucose [35]. It has been demonstrated that hyperglycemia (as it has been observed in the study) induces the downregulation of GLUT3, and that the inhibition of this transporter was able to suppress the tumorigenic potential of hepatocarcinoma cells, since GLUT3 is involved in different pathways such as cAMP, NF-κβ, and p53 signaling [36, 37]. The transporter SLC2A14 (GLUT14), which has a strong similarity to GLUT3, was also found downregulated (FC = 0.54) [38, 39]. Thus, after analyzing the expression levels of the glucose transporters, it can be hypothesized that the observed accumulation of glucose was not due to an increase in the uptake of this metabolite, but because of an impairment in the glycolytic degradation of glucose. To support this hypothesis, the expression levels of all the transcripts codifying the different enzymes involved in the glycolysis pathway were additionally assessed. Among all the evaluated transcripts, two isoforms of the fructose-bisphosphate aldolase transcript, ALDOB (FC = 0.77) and ALDOC (FC = 0.79), which catalyzes the transformation of fructose 1,6-bisphosphate to glyceraldehyde 3-phosphate, were found downregulated. Nevertheless, the effect of the impairment of these isoforms on glycolysis is not clear since the downregulation of ALDOC produces contradictory effects on cell proliferation and migration in glioblastoma cells and in vivo [40]. Moreover, ALDOs have been demonstrated not to be the rate-controlling enzymes of the glycolysis pathway, since ALDOA downregulation does not affect intracellular ATP levels [41]. Another glycolytic transcript found deregulated after exposure to Ag@MSNs-Tf was the phosphoglycerate kinase 1 (PGK1, FC = 0.78) which catalyzes the transformation of 1,3-bisphosphoglycerate to 3-phosphoglycerate. Interestingly, overexpression of PGK1 has been observed in different types of tumors including liver cancer, and downregulation of PGK1 has been proved to reduce cell proliferation and tumorigenesis. Additionally, the knockdown of PGK1 has been demonstrated to inhibit glycolysis [42, 43]. In addition, the results pointed out a strong downregulation of the transcripts PFKFB3 (FC = 0.65) and PFKFB4 (FC = 0.50), which are two isoforms that catalyze the reversible transformation between fructose 2,6-bisphosphate (F2,6BP) and fructose-6-phosphate (F6P). PFKFB3 is a rate-limiting enzyme that has a strong kinase activity promoting the formation of F2,6BP and is usually upregulated in a wide variety of tumors, including hepatocellular carcinoma, where F2,6BP acts as an allosteric modulator promoting the activation of the glycolytic flux [44,45,46]. Thus, the downregulation of PFKFB3 leads to a decrease in the glycolytic flux resulting in a reduction in the cell growth of tumoral cells [44]. Moreover, PFKFB3 inhibition can be due to the presence of ROS (whose generation has been shown to be induced by exposure to AgNPs [45]) and redirects the glucose flux from glycolysis to the pentose phosphate pathway to increase nucleotide biosynthesis, regeneration of glutathione (GSH), and ROS scavenging [46, 47]. PFKFB4 has a more active kinase activity than phosphatase, and it is also overexpressed in several human cancers. PFKFB4 inhibition induces a decrease in F2,6BP levels and thus reduces the glycolytic flux and glucose uptake [48]. Other transcripts involved in the glycolysis pathway regulation were also found deregulated. Such is the case of glucose-6-phosphatase (G6PC) and the hexokinase 4 regulator (GCKR). The downregulation of G6PC as it was observed (FC = 0.42) permits tumoral cells to overcome glycolytic inhibition, acting as a survival mechanism [49]; although it has also been reported that loss of G6PC is implicated in the reduction of glucose levels [50]. In addition, the glucokinase regulator (GCKR) was found overexpressed (FC = 1.79) since its transcription is induced by high glucose levels to avoid the accumulation of glucose-6 phosphate [51].
Effect on the pentose phosphate pathway (PPP) and other minor pathways
In addition to all the above, different metabolites implicated in the PPP were found deregulated in HepG2 cells after exposure to Ag@MSNs-Tf. Among all of them, the downregulation of D-ribose (RM = 0.19) should be pointed out since it is an essential component of key molecules such as DNA, RNA, acetyl-CoA, NADPH, and ATP [52, 53]. Ribose levels are increased in tumoral cells to support several cellular processes, so the depletion of this metabolite might have important effects on different metabolic processes [52, 54]. The transcript ribose 5-phosphate isomerase (RPIA), which was found slightly upregulated (FC = 1.24), is related to ribose metabolism. RPIA is usually upregulated after the activation of mTORC1 signaling, which redirects the glycolytic flux to generate ribose 5-phosphate and thus increases nucleotide synthesis [55, 56]. Apart from ribose, other byproducts of the PPP were found downregulated after treatment with Ag@MSNs-Tf such as ribitol (RM = 0.12), erythritol (RM = 0.12), or erythronic acid (RM = 0.02), what might be a symptom of the shift of the PPP towards the synthesis of ribose 5-phosphate to increase the nucleotide synthesis.
On another hand, the levels of myo-inositol (RM = 2.56) were found increased after exposure to the nanosystem. Since myo-inositol is endogenously synthesized from glucose, this result was expected due to the high levels of glucose found in the experiment as commented before [57]. Additionally, this result is particularly interesting because it has been described that myo-inositol might contribute to the inhibition of carcinogenesis in different organs such as the liver or lungs, since it is able to disrupt the PI3K/Akt survival pathway of tumoral cells and thus the cellular growth and cell cycle progression of such cells [57, 58]. Furthermore, galactose levels have been shown to be depleted in our study (RM = 0.33), what might involve the alteration of the galactose metabolism. The impairment of the galactose metabolism induces the downregulation of the PI3K/Akt pathway as well, and the reduction of galactose uptake might force the cells to use L-glutamine to obtain energy [59, 60].
Effect on amino acid–related pathways
Among others, the amino acid L-glutamine was found highly downregulated (RM = 0.30) after exposure to Ag@MSNs-Tf. It has been reported that glutamine depletion induces cell cycle arrest, as this metabolite plays an important role in cancer growth since, alongside glycine and aspartate, it is involved in the biosynthesis of purine and pyrimidine [28, 61]. Besides, glutamine is an important TCA anaplerosis metabolite, since it can generate α-ketoglutarate via its previous conversion to glutamate [61, 62]. The transformation of glutamine into glutamate is catalyzed by the enzyme glutaminase (GLS2), whose transcript was found somewhat upregulated (FC = 1.24). Thus, the overexpression of this transcript could respond to the need of the cells to replenish the levels of some metabolites involved in the TCA cycle. Moreover, the overexpression of GLS2 has been proved to have antiproliferative and tumor suppression properties in tumor cells since it also downregulates the PI3K/Akt signaling pathway [63, 64]. In addition, and besides its anti-proliferative activity, GLS2 overexpression is related to oxidative stress by contributing to the reduction of the intracellular levels of ROS through the production of glutathione (GSH) [63,64,65]. Apart from its role in TCA anaplerosis, glutamine is also involved in the biosynthesis of asparagine, acting as an amide group donor which is accepted by aspartate [61, 62]. This reaction is catalyzed by the enzyme asparaginase (ASNS), whose transcript was found upregulated after exposure to Ag@MSNs-Tf (FC = 1.33). Moreover, ASNS acts as a tumor growth suppressor and increases the sensibility of tumoral cells to apoptosis, since its depletion produces the accumulation of aspartate and glutamine [66]. Thus, the upregulation of GLS2 along with the overexpression of ASNS may explain the reduced levels of glutamine observed in the experiment. As in the case of glutamine, aspartate (RM = 0.05) also contributes to TCA cycle anaplerosis by generating citrate and malate, while it also takes part in nucleotide biosynthesis [67]. Although a strong decrease in aspartate levels was found in cells exposed to the nanosystem, none of the transcripts involved in neither the TCA cycle nor the nucleotide biosynthesis were found deregulated. This fact may mean that, like glutamine, aspartate is being consumed to produce arginine. Also, aspartate has been suggested to be a limiting metabolite for tumor growth and cell proliferation under hypoxic conditions, and reduced aspartate levels increase the dependence on glutaminase to replenish the TCA cycle [67, 68]. Despite the role of both glutamine and aspartate in TCA cycle anaplerosis, depleted levels of malate (RM = 0.03) and isocitrate (RM = 0.36) were found after exposure to the nanosystem, what can be due to the diversion of the glycolysis towards the PPP, since pyruvate may not be produced at the same level, or because of an increased activity of the TCA cycle and OXPHOS (pathway related to TCA cycle) to produce higher amounts of ATP.
Other amino acids such as threonine (RM = 0.17) or serine (RM = 0.09) were also found significantly deregulated. After exposure to Ag@MSNs-Tf, threonine levels were reduced, what may lead to cell death [69]. Moreover, it has been reported that depleted threonine levels affect S-adenosyl-homocysteine levels, which is a molecule necessary for tumor survival [61]. Even more interesting is the observed reduction in the serine levels which might have an impact on different processes. On one hand, serine is involved in the biosynthesis of GSH, acting as a substrate in the formation of cysteine, the limiting reagent in this process [61, 69, 70]. The reduction of serine levels has been related to the overexpression of cystathionine β-synthase (CBS), an enzyme that, together with cystathionine λ-lyase (CSE), is involved in the synthesis of GSH [29, 71, 72] and in cell proliferation and tumor growth [70]. Interestingly, the transcriptomics experiment confirmed this fact, since the transcripts encoding for both enzymes, CBS and CSE, were found upregulated (FC = 1.35 and FC = 1.39, respectively) upon Ag@MSNs-Tf exposure. On the other hand, serine is implicated in the biosynthesis of nucleotides (via the folate cycle) through its conversion to glycine in a reaction catalyzed by the enzyme serine hydroxymethyltransferase (SHMT2, FC = 1.08), and in the biosynthesis of other key biomolecules such as phospholipids. For this reason, a reduction in serine levels has been proved to inhibit tumor growth [73, 74]. Furthermore, upon serine starvation, a decrease in pyruvate kinase (PKM2) activity is induced, and consequently pyruvate is diverted to the TCA cycle [69, 75]. In the transcriptomics study, only the expression levels of the PKM transcript (FC = 0.88) were found slightly downregulated (below the established level). Under this situation, OXPHOS could acquire an important role in supporting cellular growth [75], what might explain the increased ATP levels observed in the targeted metabolomics assay (Fig. 1).
Effect on oxidative phosphorylation (OXPHOS)
As commented above, the glycolytic flux has been proved to be impaired in HepG2 cells exposed to Ag@MSNs-Tf; thus, the observed increase in the ATP levels might be due to an activation of oxidative phosphorylation (OXPHOS). Taking this into account, the expression levels of different transcripts involved in the ATP biosynthetic process and the OXPHOS were evaluated. Among all the deregulated transcripts, the upregulation of the ATP synthase subunit C lysine N-methyltransferase (FAM173B, FC = 1.44) should be highlighted, since it is involved in the assembly of the ATP synthase complex. Its expression is also related to the ATP synthesis process through the OXPHOS and to the mitochondrial respiration activity [76]. Additionally, the transcript encoding the DnaJ heat shock protein family (Hsp40) member C30 (DNAJC30), which was also found slightly upregulated (FC = 1.28), has been related to the ATP synthase complex. In this sense, it has been reported that the decreased expression of DNAJC30 results in reduced ATP levels [77, 78]. In addition, the gamma interacting protein 1 (GADD45GIP1), which is essential for the formation of the OXPHOS complexes, was found overexpressed (FC = 1.38) after exposure to Ag@MSNs-Tf, thus confirming the induced OXPHOS activity and the increased ATP production [79]. Interestingly, the copper-transporting ATPase 1 (ATP7A), which is implicated in the regulation of the OXPHOS, was also found upregulated (FC = 1.42). Although this ATPase is implicated in the transport of copper, it has been reported that ATP7A is able to carry silver ions and that the excess of both binding metals can increase the expression levels of this transporter [80, 81]. Thus, the increased ATP levels found in the experiment might also be a response of the cells to expel the silver ions generated from the Ag core of the nanosystem. In the same direction, we can highlight the downregulation found for the transcript encoding tafazzin (TAZ) (FC = 0.75), which is a protein involved in the maintenance of the mitochondrial membrane potential, thus indicating the impairment of the mitochondrial membrane potential after the exposure to the nanosystem [82].
Based on the above and to confirm the effect of Ag@MSNs-Tf on the electron transport chain (ETC) and the ATP production through the OXPHOS in treated cells, a RT-qPCR analysis was carried out to evaluate the expression levels of the following transcripts: NDUFS1, COX5A, CYC1, and MT-ATP6 (Fig. 2). The results showed that both CYC1 and MT-ATP6 were found significantly overexpressed in cells exposed to the nanosystem, confirming the enhanced activity of the ETC. Furthermore, the overexpression of these two transcripts is also related to an increase in ATP synthesis [83,84,85]. Thus, the previous hypothesis about the source of the increased ATP levels observed after exposure to the nanosystem can be confirmed and attributed to an enhanced activity of the OXPHOS, and specifically of the ETC complexes III and V.
Effect on fatty acid metabolism
Apart from all the above, we observed the accumulation of different fatty acids such as lauric acid (RM = 8.03), palmitic acid (RM = 2.00), and stearic acid (RM = 2.14) after exposure to Ag@MSNs-Tf. High levels of lauric acid have been proved to have toxic effects in different cell lines, including HepG2 cells [86]. The toxic properties of lauric acid are exerted through the induction of apoptosis due to the generation of ROS and the activation of the EGFR/ERK transduction pathway [87]. In the same way, the accumulation of palmitic and stearic acid is related to an increase in cellular apoptosis through different mechanisms in hepatocytes [88, 89]. In the case of palmitic acid, the apoptotic action is displayed through the activation of PPARα, the production of ceramide, the generation of NO and ROS, the induction of endoplasmic reticulum stress, or the activation of the factor κβ [90]. On another hand, stearic acid exerts its apoptotic action similarly to lauric acid, through the inhibition of the EGFR [91]. In order to explain the accumulation of these fatty acids, the expression levels of several transcripts involved in the biosynthesis and the β-oxidation of the fatty acids were evaluated. The most interesting transcript found slightly overexpressed was the acetyl-CoA carboxylase 1 (ACACA) (FC = 1.26), which is thought to be a fatty acid–controlling isoform. Its overexpression is related to the enhanced capacity of tumoral cells for de novo lipogenesis, what might explain the increased levels of the different fatty acids found upon Ag@MSNs-Tf exposure [92, 93]. Additionally, the levels of the coenzyme A precursor, the pantothenic acid, were depleted (RM = 0.63) after exposure to the nanosystem. This molecule leads to the formation of CoA with ATP, so it can be thought that the depleted levels of this precursor, along with the high levels of ATP found, mean a greater formation of CoA, needed for the synthesis of fatty acids and the operation of the TCA cycle [94, 95].
Conclusions
This study has unravelled some of the biomolecular mechanisms by which the hematocompatible Tf decorated core–shell Ag@MSNs hybrid nanosystem exerts its antitumor action. For this purpose, targeted and untargeted metabolomics analyses have been carried out in combination with a microarray transcriptomics analysis. The targeted metabolomics analysis has shown an alteration of the energy metabolism, with increased levels of ATP and ADP in cells exposed to the nanosystem. Additionally, the untargeted metabolomics analysis has identified a series of metabolites of different natures (carbohydrates, amino acids, TCA intermediates, fatty acids, and other minor molecules) significantly deregulated in hepatocarcinoma cells treated with the anti-tumoral nanosystem. The results of the metabolomics study were supported by the results derived from the transcriptomics assay. Overall, and considering the indications and targets found in both experiments, it has been deduced that the Ag@MSNs-Tf nanosystem exerts its antitumor action through different mechanisms. On one hand, exposure to the nanosystem impairs the glycolytic pathway, reducing the glycolytic flux and the uptake of glucose through the downregulation of some membrane glucose transporters. On the other hand, it has been confirmed that treated cells activate the oxidative phosphorylation pathway to increase the ATP levels as a defense mechanism to expel silver cations and to favor the formation of the apoptosome complex, necessary to induce cellular apoptosis. These findings have provided essential information on the biomolecular mechanisms of action of this potential antitumor nanosystem. Furthermore, these outcomes open the door to new research in which additional drugs loaded into the pores of the nanosystem could target the defense systems that were found to be activated in tumor cells exposed to the unloaded Ag@MSNs-Tf nanosystem, thereby increasing the effectiveness of the treatment. We are currently carrying out research in this direction.
References
Surendan SP, Moon MJ, Park R, Jeong YY (2018) Bioactive nanoparticles for cancer immunotherapy. Int J Mol Sci 19:3877
Bu L-L, Yan J, Wang Z, Ruan H, Chen Q, Gunadhi V, Bell RB, Gu Z (2019) Advances in drug delivery for post-surgical cancer treatment. Biomaterials 219:119182
Iyer AK, Singh A, Ganta S, Amiji MM (2013) Role of integrated cancer nanomedicine in overcoming drug resistance. Adv Drug Deliv Rev 65:1784–1802
Gonzalez-Valdivieso J, Girotti A, Schneider J, Arias FJ (2021) Advanced nanomedicine and cancer: challenges and apportunities in clinical translation. Int J Pharm 599:120438
Vallet-Regí M, Rámila A, del Real RP, Pérez-Pariente J (2001) A new property of MCM-41: drug delivery system. Chem Mater 13:308–311
Manzano M, Vallet-Regí M (2020) “Mesoporous silica nanoparticles for drug delivery” Adv. Funct Mater 30:1902634
Castillo RR, Lozano D, Gonzalez B, Manzano M, Izquierdo-Barba I, Vallet-Regí M (2019) Advances in mesoporous silica nanoparticls for targeted stimuli-responsive drug delivery: an update. Expert Opin Drug Deliv 16:415–439
Vallet-Regí M (2022) Our contributions to applications of mesoporous silica nanoparticles. Acta Biomater 137:44–52
Vallet-Regí M, Schüth F, Lozano D, Colilla M, Manzano M (2022) Engineering mesoporous silica nanoparticles for drug delivery: where are we after two decades? Chem Soc Rev 51:5365–5451
Shi J, Kantoff PW, Wooster R, Farokhzad OC (2017) Cancer nanomedicine: progress, challenges and opportunities. Nat Rev Cancer 17:20–37
Paris JL, Villaverde G, Gómez S, Vallet-Regí M (2020) Nanoparticles for multimodal antivascular therapeutics: dual drug release, photothermal and photodynamic therapy. Acta Biomater 101:459–468
Paris JL, Vallet-Regí M (2020) Mesoporous silica nanoparticles for co-delivery of drugs and nucleic acids in oncology: a review. Pharmaceutics 12:526
Alvarez E, Izquierdo I, Colilla M, González B, Jimenez C, Estévez M, Vallet-Regí M (2021) A versatile multicomponent mesoporous silica nanosystem with dual antimicrobial and ostogenic effects. Acta Biomater 136:570–581
Montalvo-Quiros S, Aragoneses-Cazorla G, Garcia-Alcalde L, Vallet-Regí M, González B, Luque-Garcia JL (2019) Cancer cell targeting and therapeutic delivery of silver nanoparticles by transferrin decorated mesoporous silica nanocarriers: insights into the action mechanisms using quantitative proteomics. Nanoscale 11:4531–4545
Aragoneses-Cazorla G, Buendia-Nacarino MP, Mena ML, Luque-Garcia JL (2022) A multi-omics approach to evaluate the toxicity mechanisms associated with silver nanoparticles exposure. Nanomaterials 12:1762
Johnson CH, Ivanisevic J, Siuzdak G (2016) Metabolomics: beyond biomarkers and towards mechanisms. Nat Rev Mol Cell Biol 17:451–459
Roberts LD, Souza AL, Gerszten RE, Clish CB (2012) Targeted metabolomics. Curr Protoc Mol Biol 98:30.2.1–30.2.24
Machuca A, Garcia-Calvo E, Anunciacao DS, Luque-Garcia JL (2021) Integration of transcriptomics and metabolomics to reveal the molecular mechanisms underlying rhodium nanoparticles-based photodynamic cancer therapy. Pharmaceutics 13:1629
Ren S, Shao Y, Zhao X, Hong CS, Wang F, Lu X, Li J, Ye G, Yan M, Zhuang Z, Xu C, Xu G, Sun Y (2016) Integration of metabolomics and transcriptomics reveals major metabolic pathways and potential biomarker involved in prostate cancer. Mol Cell Proteomics 15:154–163
Aragoneses-Cazorla G, Serrano-Lopez J, Martinez-Alfonzo I, Vallet-Regí M, González B, Luque-Garcia JL (2021) A novel hemocompatible core@shell nanosystem for selective targeting and apoptosis induction in cancer cells. Inorg Chem Front 8:2697–2712
Garcia-Calvo E, Machuca A, Nerín C, Rosales-Conrado N, Anunciação DS, Luque-Garcia JL (2020) Integration of untargeted and targeted mass spectrometry-based metabolomics provides novel insights into the potential toxicity associated to surfynol. Food Chem Toxicol 146:111849
Sapcariu SC, Kanashova T, Weindl D, Ghelfi J, Dittmar G, Hiller K (2014) Simultaneous extraction of proteins and metabolites from cells in culture. MethodsX 1:74–80
Larson R, Farber B (2015) Elementary statistics: picturing the world, 6th edn. Pearson, Boston
Seyfried TM, Arismendi-Morillo G, Mukherjee P, Chinopoulos C (2020) On the origin of ATP synthesis in cancer. iScience 23:101761
Atlante A, Giannattasio S, Bobba A, Gagliardi S, Petragallo V, Calissano P, Marra E, Passarella S (2005) An increase in the ATP levels occurs in cerebellar cells en route to apoptosis in which ATP derives from both oxidative phosphorylation and anaerobic glycolysis. Biochim Biophys Acta 1708:50–62
Berg JM, Tymczko JL, Stryer L (2002) Biochemistry, 5th edn. W.H. Freeman, New York
Vaupel P, Schmidberger H, Mayer A (2019) The Warburg effect: essential part of metabolic reprogramming and central contributor to cancer progression. Int J Radiat Biol 95:912–919
Kodama M, Oshikawa K, Shimizu H, Yoshioka S, Takahashi M, Izumi Y, Bamba T, Tateishi C, Tomonaga T, Matsumoto M, Nakayama KI (2020) A shift in glutamine nitrogen metabolism contributes to the malignant progression of cancer. Nat Commun 11(1):1320
Phillips CM, Zatarain JR, Nicholls ME, Porter C, Widen SG, Thanki K, Johnson P, Jawad MU, Moyer MP, Randall JW, Hellmich JL, Maskey M, Qiu S, Wood TG, Druzhyna N, Szczesny B, Módis K, Szabo C, Chao C, Hellmich MR (2017) Upregulation of cystathionine-b-synthase in colonic epithelia reprograms metabolism and promotes carcinogenesis. Cancer Res 77:5741–5754
Li W, Zhang X, Sang H, Zhou Y, Shang C, Wang Y, Zhu H (2019) Effects of hyperglycemia on the progression of tumor diseases. J Exp Clin Cancer Res 38:327
Ramteke P, Deb A, Shepal V, Bhat MK (2019) Hyperglycemia associated metabolic and molecular alterations in cancer risk, progression, treatment and mortality. Cancers 11:1402
Weng Y, Fan X, Bai Y, Wang S, Hang H, Yang H, Zhu J, Zhang F (2018) SLC2A5 promotes lung adenocarcinoma cell growth and metastasis by enhancing fructose utilization. Cell Death Discov 4:38
Jiang C-L, Jen W-P, Tsao C-Y, Chang L-C, Chen C-H, Lee Y-C (2020) Glucose transporter 10 modulates adipogenesis via an ascorbic-mediated pathway to protect mice against diet-induced metabolic dysregulation. PLoS Genet 16:e31008823
Zoppi N, Chiarelli N, Cinquina V, Riteli M, Colombi M (2015) GLUT10 deficiency leads to oxidative stress and non-canonical avb3 integrin-mediated TGFb signaling associated with extracellular matrix disarray in arterial tortuosity syndrome skin fibroblasts. Hum Mol Genet 24:6769–6787
Dai W, Xu Y, Mo S, Li Q, Yu J, Wang R, Ma Y, Ni Y, Xiang W, Han L, Zhang L, Cai S, Qin J, Chen W-L, Jia W, Cai G (2020) GLUT3 induced by AMPK/CREB1 axis is key for withstanding energy stress and augments the efficacy of current colorectal cancer therapies. Signal Transduct Targeted Ther 5:177
Shen X-H, Han Y-J, Yang B-C, Cui X-S, Kim N-H (2009) Hyperglycemia reduces mitochondrial content and glucose transporter expression in mouse embryos developing in vitro. J Reprod Dev 55:534–541
Zhuang Y, Zhao J, Xu X, Bi L (2018) Downregulation of GLUT3 promotes apoptosis and chemosensitivity of acute myeloid leukemia cells via EGFR signaling. Arch Iran Med 21:73–78
Augustin R (2010) The protein family of glucose transport facilitators: it’s not only about glucose after all. IUBMB Life 62:315–322
Wu X, Freeze HH (2002) GLUT14, a duplication of GLUT3, is specifically expressed in testis as alternative splice forms. Genomics 80:553–557
Kathagen-Buhmann A, Schulte A, Weller J, Holz M, Herold-Mende C, Glass R, Lamszus K (2016) Glycolysis and the pentose phosphate pathway are differentially associated with the dichotomous regulation of glioblastoma cell migration versus proliferation. Neuro Oncol 18:1219–1229
Tao Q-F, Yuan S-X, Yang F, Yang S, Yuan J-H, Wang Z-G, Xu Q-G, Ling K-Y, Cai J, Yu J, Huang W-L, Teng X-L, Zhou C-C, Wang F, Sun S-H, Zhou W-P (2015) Aldolase B inhibits metastasis through ten-eleven translocation 1 and serves as a prognostic biomarker in hepatocellular carcinoma. Mol Cancer 14:170
Cheng Y-J, Ding H, Du H-Q, Yan H, Zhao J-B, Zhang W-B, Zou Y-J, Liu H-Y, Xiao H (2014) Downregulation of phosphoglycerate kinase 1 by shRNA sensitizes U251 xenografts to radiotherapy. Oncol Rep 3(32):1513–1520
Hu H, Zhu W, Qin J, Chen M, Gong L, Li L, Liu X, Tao Y, Yin H, Zhou H, Zhou L, Ye D, Ye Q, Gao D (2017) Acetylation of PGK1 promotes liver cancer cell proliferation and tumorigenesis. Hepatology 65:515–528
Lu L, Chen Y, Zhu Y (2017) The molecular basis of targeting PFKFB3 as a therapeutic strategy against cancer. Oncotarget 8:62793–62802
Zhang X-F, Liu Z-G, Shen W, Gurunathan S (2016) Silver nanoparticles: synthesis, characterization, properties, applications, and therapeutic approaches. Int J Mol Sci 17:1534–1567
Franklin DA, He Y, Leslie PL, Tikunov AP, Fenger N, Macdonald JM, Zhang Y (2016) p53 coordinates DNA repair with nucleotide synthesis by suppressing PFKFB3 expression and promoting the pentose phosphate pathway. Sci Rep 6:38067
Yi M, Ban Y, Tan Y, Xiong W, Li G, Xiang B (2019) 6-Phosphofructo-2-kinase/fructose-2,6-biphosphatase 3 and 4: a pair of valves for fine-tuning of glucose metabolism in human cancer. Mol Metab 20:1–13
Chesney J, Clark J, Klarer AC, Imbert-Fernandez Y, Lane AN, Telang S (2014) Fructose-2,6-Bisphosphat synthesis by 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase 4 (PFKFB4) is required for the glycolytic response to hypoxia and tumor growth. Oncotarget 5:6670–6686
Abbadi S, Rodarte JJ, Abutaleb A, Lavell E, Smith CL, Ruff W, Schiller J, Olivi A, Levchenko A, Guerrero-Cazares H, Quinones-Hinojosa A (2014) Glucose-6-phosphatase is a key metabolic regulator of glioblastoma invasion. Mol Cancer Res 12:1547–1559
Wang Z, Dong C (2019) Gluconeogenesis in cancer: function and regulation of PEPCK, FBPase, and G6Pase. Trends Cancer 5:30–45
Rodríguez ML, Silva LF, Vangipurapu J, Modi S, Kuusisto J, Kaikkonen MU, Laakso M (2018) Functional variant in the GCKR gene affects lactate levels differentially in the fasting state and during hyperglycemia. Sci Rep 8:15989
Mahoney DE, Hiebert JB, Thimmesch A, Pierce JT, Vacek JL, Clancy RL, Sauer AJ, Pierce JD (2018) Understanding D-Ribose and mitochondrial function. Adv Biosci Clin Med 6:1–5
Gonzalez MJ, Seyfried T, Nicolson GL, Barclay BJ, Matta J, Vasquez A, D’Agostino D, Olalde J, Duconge J, Hunninghake R, Berdiel MJ, Cintrón A (2018) Mitochondrial correction: a new therapeutic paradigm for cancer and degenerative diseases. J Orthomol Med 33:1–20
Boros LG, Lee PWN, Brandes JL, Cascante M, Muscarlla P, Schirmer WJ, Melvin WS, Ellison EC (1998) Nonoxidative pentose phosphate pathways and their direct role in ribose synthesis in tumors: is cancer a disease of cellular glucose metabolism? Med Hypotheses 50:55–59
Ciou S-C, Chou Y-T, Liu Y-L, Nieh Y-C, Lu J-W, Huang S-F, Chou Y-T, Cheng L-H, Lo J-F, Chen M-J, Yang M-C, Yuh C-H, Wang H-D (2015) Ribose-5-phosphate isomerase A regulates hepatocarcinogenesis via PP2A and ERK signaling. Int J Cancer 137:104–115
Buj R, Chen C-W, Dahl ES, Leon KE, Kuskovsky R, Maglakelidze N, Navaratnarajah M, Zhang G, Doan MT, Jiang H, Zaleski M, Kutzler L, Lacko H, Lu Y, Mills GB, Gowda R, Robertson GP, Warrick JI, Herlyn M, Imamura Y, Kimball SR, Dgraff DJ, Snyder NW, Aird KM (2019) Suppression of p16 induces mTORC1-mediated nucleotide metabolic reprogramming. Cell Rep 28:1971–1980
Bizzarri M, Dinicola S, Bevilacqua A, Cucina A (2016) Broad spectrum anticancer activity of myo-inositol and inositol hexakisphosphate. Int J Endocrinol 2016:5616807
Chhetri DR (2019) Myo-Inositol and its derivatives: their emerging role in the treatment of human diseases. Front Pharmacol 10:1172
Tang M, Etokidem E, Lai K (2016) The Leloir pathway of galactose metabolism-a novel therapeutic target for hepatocellular carcinoma. Anticancer Res 36:6265–6272
Kase ET, Nikolic N, Bakke SS, Bogen KK, Aas V, Thoresen GH, Rustan AC (2013) Remodeling of oxidative energy metabolism by galactose improves glucose handling and metabolic switching in human skeletal muscle cells. PLoS One 8:e59972
Lieu EL, Nguyen T, Rhyne S, Kim J (2020) Amino acids in cancer. Exp Mol Med 52:15–30
Pavlova NN, Hui S, Ghergurovich JM, Fan J, Intlekofer AM, White RM, Rabinowitz JD, Thompson CB, Zhang J (2018) As extracellular glutamine levels decline, asparagine becomes an essential amino acid. Cell Metab 27:428–438
López de la Oliva AR, Campos-Sandoval JA, Gómez-García MC, Cardona C, Martín-Rufián MM, Sialana FJ, Castilla L, Bae N, Lobo C, Peñalver A, García-Frutos M, Carro D, Enrique V, Paz JC, Mirmira RG, Gutiérrez A, Alonso FJ, Segura JA, Matés JM, Lubeec G, Márquez J (2020) Nuclear translocation of glutaminase GLS2 in human cancer cells associates with proliferation arrest and differenciation. Sci Rep 10:2259–2275
Liu J, Zhang C, Lin M, Zhu W, Liang Y, Hong X, Zhao Yu, Young KH, Hu W, Feng Z (2014) Glutaminase 2 negatively regulates the PI3K/AKT signaling and shows tumor suppression activity in human hepatocellular carcinoma. Oncotarget 5:2635–2647
Suzuki S, Tanaka T, Poyurovsky MV, Nagano H, Mayama T, Ohkubo S, Lokshin M, Hosokawa H, Suzuki Y, Sugano S, Sato E, Nagao T, Yokote K, Tatsuno I, Prives C (2010) Phosphate-activated glutaminase (GLS2), a p53-inducible regulator of glutamine metabolism and reactive oxygen species. Proc Natl Acad Sci USA 107:7461–7466
Zhang B, Dong L-W, Tan Y-X, Zhang J, Pan Y-F, Yang C, Li M-H, Ding Z-W, Liu L-J, Jiang T-Y, Yang J-H, Wang H-Y (2013) Asparagine synthetase is an independent predictor of surgical survival and a potential therapeutic target in hepatocellular carcinoma. Br J Cancer 109:14–23
Garcia-Bermudez J, Baudrler L, La K, Zhung XG, Fidelin J, Svlderskly VO, Papaglannakopoulos T, Molina H, Snuderl M, Lewis CA, Possemato RL, Blrsoy K (2018) Aspartate is a limiting metabolite for cancer cell proliferation under hypoxia and in tumors. Nat Cell Biol 20:775–781
Alkan HF, Walter KE, Luengo A, Madl T, Vander Heiden MG, Bogner-Strauss JG (2018) Cytosolic aspartate availability determines cell survival when glutamine is limiting. Cell Metab 28:706–728
Amelio I, Cutruzzolá F, Antonov A, Agostini M, Melino G (2014) Serine and glycine metabolism in cancer. Trends Biochem Sci 39:191–198
Niu W, Chen F, Wang J, Qian J, Yan S (2018) Antitumor effect of sikokianin C, a selective cystathionine b-synthase inhibitor, against human colon cancer in vitro and in vivo. Med Chem Commun 9:113–120
Jia H, Ye J, You J, Shi X, Kang W, Wang T (2017) Role of the cystathionine b-synthase/H2S system in liver cancer cells and the inhibitory effect of quinolone-indolone conjugate QIC2 on the system. Oncol Rep 37:3001–3009
Zhu H, Blake S, Chan KT, Pearson RB, Kang J (2018) Cystathionine b-synthase in physiology and cancer. BioMed Res Int 2018:3205125
Gao X, Lee K, Reid MA, Sanderson SM, Qiu C, Li S, Liu J, Locasale JW (2018) Serine availability influences mitochondrial dynamics and function through lipid metabolism. Cell Rep 22:3507–3520
Mattaini KR, Sullivan MR, Vander Heiden MG (2016) The importance of serine metabolism in cancer. JCell Biol 214:249–257
Maddocks ODK, Berkers CR, Mason SM, Zheng L, Blyth K, Gottlleb E, Vousden KH (2019) Serine starvation induces stress and p53 dependent metabolic remodeling in cancer cells. Nature 493:542–546
Malecki JM, Willemen HLDM, Pinto R, Ho AYY, Moen A, Kjonstad IF, Burgering BMT, Zwartkruis F, Eijkelkamp N, Falnes PO (2019) Lysine methylation by the mitochondrial methyltransferase FAM173B optimizes the function of mitochondrial ATP synthase. J Biol Chem 294:1128–1141
Tebbenkamp ATN, Varela L, Choi J, Paredes MI, Giani AM, Song JE, Sestan-Pesa M, Franjic D, Sousa AMM, Liu Z-W, Li M, Bichsel C, Koch M, Szigeti-Buck K, Liu F, Li Z, Kawasawa YI, Paspalas CD, Mineur YS, Prontera P, Merla G, Picciotto MR, Arnstern AFT, Horvath TL (2018) The 7q11.23 protein DNAJC30 interacts with ATP synthase and links mitochondria to brain development. Cell 175:1088–1104
Egea G, Jiménez-Altayó F, Campuzano V (2020) Reactive oxygen species and oxidative stress in the pathogenesis and progression of genetic diseases of the connective tissue. Antioxidants 9:1013–1051
Kim SJ, Kwon M-C, Ryu MJ, Chung HK, Tadi S, Kim YK, Kim JM, Lee SH, Park JH, Kweon GR, Ryu S-W, Jo YS, Lee C-H, Hatakeyama H, Goto Y-I, Yim Y-H, Chung J, Kong Y-Y, Shong M (2012) CRIF1 is essential for the synthesis and insertion of oxidative phosphorylation polypeptides in the mammalian mitochondrial membrane. Cell Metab 16:274–283
Chun H, Catterton T, Kim H, Lee J, Kim B-E (2017) Organ-specific regulation of ATP7A abundance is coordinated with systemic copper homeostasis. Sci Rep 7:12001
Monty J-F, Llanos RM, Mercer JFB, Kramer DR (2005) Copper exposure induces trafficking of the Menkes protein in intestinal epithelium of ATP7A transgenic mice. J Nutr 135:2762–2766
Ma W, Cui Y, Liu M, Tan Z, Jiang Y (2019) Downregulation of miR-125b promotes resistance of glioma cells to TRAIL through overexpression of Tafazzin which is a mitochondrial protein. Aging 11:2670–2680
Han Y, Sun S, Zhao M, Zhang Z, Gong S, Gao P, Liu J, Zhou J, Ma D, Gao Q, Wu P (2016) CYC1 predicts poor prognosis in patients with breast cancer. Dis Markers 2016:3528064
Wallac LaShanale, Mehrabi S, Bacanamwo M, Yao X, Aikhionbare FO (2016) Expression of mitochondrial genes MT-ND1, MT-ND6, MT-CYB, MT-COI, MT-ATP6 and 12S/MT-RNR1 in colorectal adenopolyps. Tumour Biol 37:12465–12475
Jonckheere AI, Smeitink JAM, Rodenburg RJT (2012) Mitochondrial ATP synthase: architecture, function and pathology. J Inherit Metab Dis 35:211–225
Sheela DL, Narayanankutty A, Nazeem PA, Raghavamenon AC, Muthangaparambil SR (2019) Lauric acid induce cell death in colon cancer cells mediated by thw wpidermal growth factor receptor downregulation: an in silico and in vitro study. Hum Exp Toxicol 38:753–761
Lappano R, Sebastiani A, Cirillo F, Rigiracciolo DC, Galli GR, Curcio R, Malaguamera R, Belfiore A, Cappello AR, Maggiolini M (2017) The lauric acid-activated signaling propts apoptosis in cancer cells. Cell Death Discov 3:17063–17071
Malhi H, Bronk SF, Wernerburg NW, Gores GJ (2006) Free fatty acids induce JNK-dependent hepatocyte lipoapoptosis. J Biol Chem 281:12093–12101
Harvey KA, Walker CL, Xu Z, Whitley P, Pavlina TM, Hise M, Zaloga GP, Siddiqui RA (2010) Oleic acid inhibits stearic acid-induced inhibition of cell growth and pro-inflammatory responses in human aortic endothelial cells. J Lipid Res 51:3470–3480
Ricchi M, Odoardi MR, Carulli L, Anzivino C, Ballestri S, Pinetti A, Fantoni LI, Marra F, Bertolotti M, Banni S, Lonardo A, Carulli N, Loria P (2009) Differential effect of oleic and palmitic acids on lipid accumulation and apoptosis in cultured hepatocytes. J Gastroenterol Hepatol 24:830–840
Evans LM, Cowey SL, Siegal GP, Hardy RW (2009) Stearate preferentially induces apoptosis in human breast cancer cells. Nutr Cancer 61:746–753
Bhattacharjee K, Nath M, Choudhury Y (2020) Fatty acid synthesis and cancer: aberrant expression of the ACACA and ACACB genes increases the risk for cancer. Meta Gene 26:100798–100808
Ye B, Yin L, Wang Q, Xu C (2019) ACC1 is overexpressed in liver cancers and contributes to the proliferation of human hepatoma HepG2 cells and the rat liver cell line BRL3A. Mol Med Rep 19:3431–3440
Kobayashi D, Kusama M, Onda M, Nakahata N (2011) The effect of pantothenic acid deficiency on keratinocyte proliferation and the synthesis of keratinocyte growth factor and collagen fibroblasts. J Pharmacol Sci 115:230–234
Leonardi R, Jackowski S (2007) Biosynthesis of pantothenic acid and coenzyme A. EcoSal Plus 2:10.1128
Funding
Open Access funding provided thanks to the CRUE-CSIC agreement with Springer Nature. This work was supported by Ministerio de Ciencia e Innovación (MINECO) grants PID2020-114529RB-I00 and PID2020-117091RB-I00 and the European Research Council ERC-2015-AdG (VERDI) Proposal No. 694160 (MV-R). CIBER is a public research consortium created by ISCIII whose actions are co-funded by the European Regional Development Fund. Guillermo Aragoneses-Cazorla received a pre-doctoral fellowship (FPU18/01176) from the Ministerio de Ciencia, Innovación y Universidades.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Aragoneses-Cazorla, G., Vallet-Regí, M., Gómez-Gómez, M.M. et al. Integrated transcriptomics and metabolomics analysis reveals the biomolecular mechanisms associated to the antitumoral potential of a novel silver-based core@shell nanosystem. Microchim Acta 190, 132 (2023). https://doi.org/10.1007/s00604-023-05712-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00604-023-05712-3