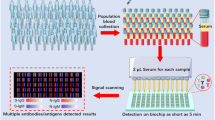
Abstract
A new detection strategy was developed to improve the sensitivity of a lateral flow immunoassay platform utilizing a delayed hydrophobic barrier fabricated with trimethylsilyl cellulose (TMSC). The SARS-CoV-2 spike receptor-binding domain (SARS-CoV-2 SP RBD) antigen was chosen as a model analyte to demonstrate the superior detectability of this scheme. The novel device consists of 2 separate layers, so-called delayed lateral flow immunoassay (d-LFIA). The upper layer is intended for the analyte or sample flow path, where the test solution flows freely straight to the detection zone to bind with the primary antibody. The lower layer, located just underneath, is designed for the SARS-CoV-2 spike receptor-binding domain-conjugated gold nanoparticles (SARS-CoV-2 SP RBD-AuNPs) used for producing a colorimetric signal. This layer is fabricated with a TMSC barrier to time-delay the movement of SARS-CoV-2 SP RBD-AuNPs, thus allowing the antigen to bind with the primary antibody more efficiently. This platform exhibited a 2.6-fold enhancement in the sensitivity and 9.1-fold improvement in the limit of detection (LOD) as compared with the conventional LFIA. In addition, this d-LFIA device was satisfactorily applied to accurate screening of COVID-19 patients.
Graphical abstract

Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
A newly emerging acute respiratory syndrome originated in Wuhan, China, and quickly spread into a pandemic publicly known as coronavirus disease 2019 (COVID-19) [1, 2]. The symptoms of SARS-CoV-2 infection were similar to those of influenzas, such as fever, cough, myalgia, and fatigue [3]. The gold standard detection method of coronavirus disease 2019 is based on the reverse transcription-polymerase chain reaction (RT-PCR), which typically provides high sensitivity and accuracy [4]. However, this approach still possesses some apparent disadvantages, such as high cost, lengthy analysis time, and limited accessibility for people from less developed countries [5]. Therefore, to complement the COVID-19 prompt diagnosis, the lateral flow immunoassay (LFIA) platform was adopted as a screening tool for SARS-CoV-2 antigen detection. This technique is rapid (with the result usually obtained in less than 15 min), inexpensive, uncomplicated, and suitable for point-of-care testing (POCT) [6].
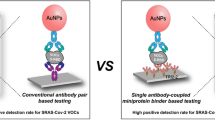
In the present work, we exploit the competitive immunoassay format to showcase the benefit that can be realized using our developed LFIA device. COVID-19 antigen was chosen as a model analyte in this research as it is now a global pandemic affecting millions of people worldwide. It is well-known that a major drawback of a typical immunoassay platform for COVID-19 antigen (i.e., antigen test kit or ATK) is its lack of sensitivity to detect trace amounts of analyte. This issue is primarily dedicated to the low efficiency of immunocomplex establishment. Therefore, increasing the binding efficiency between the target antigen and the antibody could be an effective way to overcome the aforementioned limitation. We expect that raising the binding efficiency of antigen could enhance the LFIA assaying performance.
In a competitive immunoassay format, the gold nanoparticles (AuNPs) are labeled with the analytes and dispensed on the conjugate pad. With a negative sample, the labeled analytes go through the strip and bind to both primary and secondary antibodies on the test and control lines, respectively, which results in the red color on both lines. In the positive case, the analyte (unlabeled) and the labeled analyte both move and competitively bind with the primary antibody on the test line. Afterward, the excess labeled analyte also binds to the secondary antibody on the control line [7], resulting in a full-scale color change on this end. The difference in the color on both lines is a preliminary indicative of the amount of target analytes that successfully occupy the antibody on the test line. Obviously, the analyte of lower concentration has far less chance to bind with the primary antibody than the more abundant labeled analyte, which can easily lead to a false negative interpretation. Hence, in this work, we intend to overcome this drawback by developing a new LFIA platform that efficiently increases the binding of the analyte with the primary antibody.
This strategy was accomplished using a delayed lateral flow immunoassay (d-LFIA) device, which comprises two separate layers. The first which is the upper layer, is used exclusively for the flow path of the analyte where they can move and bind freely to the primary antibody beforehand. The other layer underneath is designed for the flow path of labeled analyte and is printed with trimethylsilyl cellulose (TMSC) barrier downstream from the conjugate pad. TMSC is a hydrophobic material which makes the water wettability of the surface low [8, 9]. This hydrophobic barrier was used to delay the movement of the labeled analyte to the primary antibody on the test spot thus allowing the analyte to be captured more effectively in advance. Accordingly, the analyte has an increasing chance to bind with the primary antibody in the test spot completely. The excess of labeled analyte would then flow through the secondary antibody in the control spot as usual. The improvement in the analytical performances of this device was illustrated through the detection of SARS-CoV-2 antigen, a widely acknowledged biomarker for COVID-19. SARS-CoV-2 SP RBD-AuNPs, SARS-CoV-2 Spike S1 IgG antibody, and SARS-CoV-2 Spike S1 IgM antibody, were used as the labeled analyte, primary antibody, and secondary antibody, respectively. Various parameters including the delayed pattern of TMSC barrier and concentration of antibodies were optimized and the method was validated with clinical samples.
Experimental
Materials and reagents
SARS-CoV-2 spike receptor-binding domain (RBD, His Tag) (cat.: Z03483-100; Z03483-1), SARS-CoV-2 Spike S1 IgG antibody (cat no.: A02038-1), and SARS-CoV-2 Spike S1 IgM antibody (cat no.: A02046) were purchased from GenScript USA, Inc (NJ, USA). Sodium carbonate anhydrous (Na2CO3) and sodium hydroxide (NaOH) were obtained from Merck Millipore (Billerica, MA, USA). Sodium bicarbonate (NaHCO3) was purchased from May and Baker (England). Tween 20 was purchased from Glentham (UK). Trimethylsilyl cellulose (TMSC) was purchased from Thüringisches Institut für Textil- und Kunststoff-Forschung e.V. (TITK, Rudolstadt, Germany). Hexamethyldisiloxane (HMDSO, 98%), gold nanoparticles solution (20 nm, AuNPs), bovine serum albumin (BSA), phosphate-buffered saline (PBS) tablets, sucrose, and tris-hydrochloride (tris–HCl) were acquired from Sigma-Aldrich (St. Louis, MO). Sample pads (Fusion5) and absorbent pads were purchased from Ghealthcare Whatman (USA). Nitrocellulose membranes (unitcardsCN140 and unisartCN140 unback), glass fibers (GF33Glass) and plastic backing cards were purchased from SARTORU (USA). Aqueous solutions were freshly prepared by ultrapure deionized water (> 18.2 MΩ cm−1) obtained from a Milli-Q system (Millipore, USA).
Instruments
The TMSC delayed pattern was created using a Dimatix Materials Printer (Fujifilm DMP-2850, United State). The characterization was performed using the JEM-1400Flash series 120 kV transmission electron microscope (JEOL Ltd., Japan), and laser scanning confocal microscopy (LSCM) from LEXT OL55000 (Olympus, Japan). UV–visible absorption spectra were obtained with the HEWLETT-PACKARD 8453 UV–visible spectrometer (Agilent Technologies, UK). The lateral flow immunoassay was cut using the CM5000 Guillotine cutter from BioDot Inc. Infrared spectroscopy was performed with the Nicolet iS50 FTIR (ATR mode) Spectrometer (Thermo Scientific, United State).
Conjugation of the gold nanoparticles with SARS-CoV-2 spike receptor-binding domain (SARS-CoV-2 SP RBD)
The procedure for the conjugation of AuNPs and SARS-CoV-2 SP RBD was slightly modified from the previous report by P. Wang [10]. The detail of this procedure was provided in the Electronic Supplementary Material (ESM).
Preparation of the d-LFIA strip
The d-LFIA was assembled using components that have been separately prepared. The sample pads were treated with BSA (3% w/v in PBS) and tween-20 (1% v/v in PBS) to reduce non-specific adsorption of unwanted proteins and facilitate the solution flow. The conjugate pads were treated with sucrose (3% w/v in 1 M tris buffer pH 7.4) to consistently release conjugate and prevent the non-specific binding. These components were dried in an oven for 3–4 h at 60 °C and stored in a refrigerator. The d-LFIA strip was configured into 2 layers; LAYER 1 and LAYER 2, as depicted in Fig. 1. A nitrocellulose membrane (unitcardsCN140) containing a TMSC barrier was firstly affixed on a plastic backing card as the main body of LAYER 1. The TMSC was sonicated for 30 min and diluted with 98% HMDSO to the desired concentration (0.1% w/v) prior to printing on the nitrocellulose membrane. The TMSC solution was printed transversely onto the nitrocellulose membrane to create a 2 mm width × 3 mm length barrier using the Dimatix Materials printer (drop spacing = 10 μm, firing frequency = 17 V and droplet size = 10 pL). The TMSC-coated membrane was then left to dry at room temperature for 5 min. Next, the conjugate pad was assembled on this plastic backing card with 1-mm overlap on the front end of the unitcardsCN140. The sample and absorbent pads were then attached at each end of the plastic backing card, as shown in Fig. 1A.
For the assembly of LAYER 2, a double-sided adhesive tape was first attached onto LAYER 1 where the conjugate pad was underneath to be served as a linker between those two layers. Another piece of the nitrocellulose membrane (unisartCN140 unback) was subsequently placed on top of the adhesive tape (1-mm overlapped with the unitcardsCN140 of LAYER 1) to form a sandwich-like configuration, as schematically illustrated in Fig. 1A. Finally, another sample pad was assembled at the front end of the strip over the sample pad of LAYER 1. The dimensions of the respective elements are shown in Fig. 1B. The test (T) and control (C) spots were created further downstream from the TMSC barrier by dispensing with 0.25 μL of SARS-CoV-2 Spike S1 IgG (SARS-CoV-2 IgG) antibody (500 μL/mL in PBS) and 0.25 μL of SARS-CoV-2 Spike S1 IgM (SARS-CoV-2 IgM) antibody (1 mg/mL in PBS), respectively. The modified device was then incubated at 37 °C for 1 h to immobilize the capturing antibody onto the membrane. As the final step, 3 μL of SARS-CoV-2 SP RBD-conjugated AuNPs solution was dropped on the conjugate pad and dried at 37 °C for 20 min.
Detection procedures
To demonstrate the detection capability of the d-LFIA device proposed in this work, 8 μL of a sample solution containing SARS-CoV-2 SP RBD was first diluted with tris running buffer (pH 7.4) to obtain a final volume of 80 μL. This solution was subsequently dropped on the sample pad of the d-LFIA strip and allowed to develop for 15 min. A faded color would be observed on the test spot as a result of the target antigen successfully competing for the antibody, as opposed to the pure red color produced exclusively by the AuNP-conjugates on the control spot. On the contrary, a negative ( −) sample resulted in literally no or minimal changes of color on both spots as all the antibodies were captured solely by the AuNP-conjugates. A photograph of the detection zone was accurately taken by ManualShot, a smartphone application, under a light controlled box. The color intensity of the captured image was then analyzed by an Image J software using G color in the RGB mode.
Following the optimization, the developed d-LFIA was applied for the analysis of 12 throat swab samples, 8 of which were confirmed to be SARS-CoV-2-positive by the standard RT-PCR method. These clinical samples were obtained from the Faculty of Medicine Ramathibodi Hospital, Mahidol University, and have been certified by the Research Ethics Review Committee by the Faculty of Medicine (Certification of approval: COA. MURA2021/879).
Results and discussion
Characterization of SARS-CoV-2 SP RBD-AuNPs and d-LFIA device
In this section, the characterization of the AuNPs before and after the conjugation with SARS-CoV-2 SP RBD was carried out using UV–Vis spectroscopy and transmission electron microscopy (TEM) technique. Figure 2A illustrates the UV–Vis absorption spectra of the AuNPs and SARS-CoV-2 SP RBD-conjugated AuNPs. It is well-known that metallic nanoparticles (such as AgNPs or AuNPs) generally have a strong absorption band in the visible region. In this case, a characteristic peak of AuNPs was seen at 520 nm suggesting the size in the 20-nm range which conform to the supplier’s guideline [11]. In addition, TEM image (Fig. 2A) further showed homogenous distribution of AuNPs in aqueous solution. An average size of AuNPs dispersed in this solution was approximately 16 nm. Upon conjugation with SARS-CoV-2 SP RBD, the absorption peak shifted positively (red-shift) from 520 to 525 nm which can be attributed to an enlarged gold nanoparticle from such formation. All things considered; it was clearly indicated that the modification of the AuNPs with SARS-CoV-2 SP RBD was successful.
UV–Vis absorption spectra (A) and TEM images (insert A) of the AuNP before (red line) and after (blue line) the conjugation with SARS-CoV-2 SP RBD. Laser scanning confocal microscopy (LSCM) of LFIA (B) and d-LFIA (C) membranes. Image of contact angle of the TMSC-modified nitrocellulose membrane (D). ATR-FTIR spectra of LFIA before (E) and after (F) TMSC printing on the nitrocellulose membrane
The existence of the TMSC barrier created on the nitrocellulose membranes was verified using laser scanning confocal microscopy (LSCM). Figure 2B shows the surface morphology of the unmodified (LFIA) nitrocellulose membrane while that of the TMSC-modified (d-LFIA) membrane is depicted in the Fig. 2C. The main difference between the two membranes was clearly observed by the dark zone on the d-LFIA surface which indicated a successful deposition of the TMSC barrier on the modified membrane. The LSCM technique also provides distinct colors to define the roughness of a surface. As displayed in Fig. 2B, the surface of the non-modified membrane differed significantly from its modified counterpart (Fig. 2C), particularly in the high-roughness area represented by the yellow color. The surface of d-LFIA membrane was interestingly smoother (blue color) after printing with the TMSC barrier. Moreover, the water contact angle (WCA) of both conventional and the TMSC-modified nitrocellulose membranes were investigated. The nitrocellulose membrane modified with TMSC produced an average static water contact angle (SCA) value of 124.6° as presented in Fig. 2D. This result signifies a superior hydrophobic property of d-LFIA membrane surface created by TMSC. On the contrary, the SCA could not be measured for the non-modified membrane due to the inherent surface hydrophilicity of a nitrocellulose membrane. Based on this evidence, it can be concluded that a hydrophobic property of the nitrocellulose membrane surface was successfully altered by modifying with the TMSC barrier.
In addition to those characterization techniques above, the attenuated total reflection infrared spectroscopy (ATR-IR) was carried out to further support this claim. The ATR-IR spectra of the non-modified and modified LFIA membranes are shown in Fig. 2E and F respectively. The non-modified LFIA revealed three intense bands of nitrate groups at around 1660 cm−1 and 1280 cm−1 for NO2, and 840 cm−1 for NO [12]. Meanwhile, the LFIA modified with TMSC, which comprises a trimethylsilyl [13], displayed new features of C-Si at around 757 cm−1, 840 cm−1, and 1250 cm−1 [9, 14,15,16] from TMSC. However, as observed in Fig. 2F, some of the nitrate peaks disappeared while the others were less intense. This might be ascribed to the fact that the nitrate groups were being concealed by the C-Si bonds introduced by TMSC barrier.
Design and detection principle of the d-LFIAs
In this section, these flow patterns were alternatively verified by the use of a red color dye. With the conventional LFIA device, the red dye solution flowed uninterruptedly through the test zone and reached the absorbent pad within 2 min as seen in Fig. 3A. Conversely, the red dye was briefly stopped when they reached the TMSC barrier for approximately 5–6 min (Fig. 3B), during which the barrier was gradually disintegrated as the TMSC continually dissolved in the running buffer. Following the dissolution, the red dye then continued to flow to the detection zone and reach the absorbent pad within roughly 10 min.
Flow behaviors of lateral flow immunoassay device without (A) and with (B) TMSC barrier at different times. Visual comparison of LFIA and d-LFIA devices for SARS-CoV-2 antigen testing (100 µg/mL) (C). Comparison of △intensity of test spots at various concentrations of SARS-CoV-2 antigen using LFIA and d-LFIA devices (D) (n = 3)
Hence, it can be conclusively said that, by using this d-LFIA device, a significant advantage could be gained by the target analyte on the top layer in terms of the interaction with antibody on the detection zone ahead of their competitors. Benefiting from this strategy, the resulting immunocomplexes between COVID-19 antigen and antibody can be predominantly achieved over its AuNP-conjugates. This action supposedly can lead to a significant enhancement of the sensitivity and in turn lower the detection limit of our platform. To validate this hypothesis, the detection performances (e.g., linearity, limit of detection and limit of quantification) toward SARS-CoV-2 antigen attained from both original and modified LFIAs were examined.
The sensing capability of LFIA and d-LFIA devices for SARS-CoV-2 antigen were demonstrated herein. The SARS-CoV-2 IgG, which was immobilized on the test spot, was employed as a selective capturing element for COVID-19 antigen, SARS-CoV-2 SP RBD. The SARS-CoV-2 IgM was anchored on the control spot because it has multi-epitopes to ensure a definite binding with the SARS-CoV-2 SP RBD-AuNPs. When the blank (buffer) solution was applied, both devices (LFIA and d-LFIA) showed two distinctly colored spots on the test and control spots (Fig. 3C) clearly indicating that the immunocomplexes between SARS-CoV-2 SP RBD-AuNPs and SARS-CoV-2 IgG were thoroughly formed on both spots. Upon the addition of 100 µg/mL of SARS-CoV-2 SP RBD, the conventional LFIA device showed a slightly faint color and an intense red color on the test and control spots, respectively. The fading color on the test spot is an indicative of a competitive event between the SARS-CoV-2 SP RBD and its competitor, SARS-CoV-2 SP RBD-AuNPs. The difference between color intensity of the blank and analyte solution represents the amount of target antigen successfully occupying the antibodies. As for the d-LFIA device, the color was only observed on the control spot while the test spot was virtually invisible. The disappearing color on the test spot is presumably due to the fact that all the antibodies are fully captured by the target antigens that arrived earlier while the SARS-CoV-2 SP RBD-AuNPs are being delayed. Based on this strategy, the SARS-CoV-2 antigen has a major advantage in the immunocomplex formation over its AuNP-conjugate counterparts, and as a result, both sensitivity and detectability can be significantly improved. The remarkable enhancement of the proposed device can be illustrated through the differences of color intensity (Δintensity = intensitysample – intensityblank) produced by various concentrations of SARS-CoV-2 SP RBD as displayed in Fig. 3D.
d-LFIA optimization
Variable parameters affecting the d-LFIA for SARS-CoV-2 antigen detection including the concentration of SARS-CoV-2 IgG antibody and the drop spacing of TMSC were investigated to achieve the optimal sensing capability as shown in Fig. S1. The detail of optimization parameters was provided in the Electronic Supplementary Material (ESM).
Analytical performance
The analytical capability of the proposed d-LFIA device for SARS-CoV-2 antigen detection was assessed under its optimal conditions. A SARS-CoV-2 SP RBD stock solution was serially diluted to obtain the final concentrations in the range of 10 ng/mL to 105 ng/mL and subsequently subjected to the colorimetric analysis to establish a calibration curve. The test strips were photographed, after 15 min of reaction time, by a smartphone camera in a light controlled box and the G color intensity of the test spots were analyzed by the ImageJ software for quantification purpose. The △intensity values were calculated by subtracting the intensity acquired from the blank solution from that of the SARS-CoV-2 SP RBD sample. The calibration curves obtained from the modified d-LFIA and LFIA (unmodified) strips are exhibited in Fig. 4A and B respectively. The △intensity increased proportionally to the SARS-CoV-2 SP RBD concentration and the linearity were obtained within the studied range for both devices. Nevertheless, the △intensity obtained from the d-LFIA device was noticeably higher denoting that the sensitivity was significantly improved by the delaying process of the present platform. A sensitivity enhancement by approximately 2.6-fold was achieved as compared to that of the conventional LFIA device.
The limit of detection (LOD) and limit of quantification (LOQ) of the proposed device were calculated on the basis of 3SDblank/slope and 10SDblank/slope, and the values obtained were 0.11 ng/mL and 0.38 ng/mL, respectively. A roughly 9.1-fold lower detection limit was realized as compared to the conventional LFIA.
Table 1 compiles the detection methods and their analytical features for SARS-CoV-2 detection from the previous works and matches up against our study. It is undeniable that the best available option with high sensitivity and accuracy is RT-PCR; unfortunately, this method is very expensive, time-consuming, and possibly inaccessible for some users. Among the screening methods, LFIAs provide distinct advantages over the electrochemical methods in terms of reduced analysis time and superior detection limits. In comparison with previous colorimetric LFIA methods, our d-LFIA device offers a significant edge with regard to the sensitivity and detection limit. Although the LFIA modified with nanozyme chemiluminescence labeling [21] can produce a comparable LOD, the preparation of such modifying material is much more complicated.
The analytical performance of the d-LFIA platform was further evaluated in the matter of selectivity toward SARS-CoV-2 antigen detection. Cross-reactivity analyses on other influenza viruses, including H1N1 virus, H3N2 virus, Colorado/06/2017-like virus and Phuket/3073/2013-like virus, human respiratory syncytial virus glycoprotein (RSV), human coxsackie adenovirus receptor (COX), human coronavirus MERS Spike glycoprotein (MERS), and SARS nucleocapsid protein (SARS-CoV-1) were thoroughly studied and their responses were exhibited in Fig. 5. The purpose of this cross-reactivity study was to see if any other virus antigens would react with the capturing antibodies used in this device. All test results were found to be negative with the exception of one, SARS-CoV-1, that was able to produce marginal response. The SARS-CoV-1 virus was known to induce the SARS-CoV-2 immune response [23] and hence not surprisingly discovered. A cutoff value of △intensity in the present work was established at 2.66 (A.U.) and any outturn over this value was considered to be COVID-19 positive.
Apart from the cross-reactivity study, the influence of common interferences potentially present in the test samples, including glucose, human serum albumin, and ascorbic acid, on the △intensity was also examined. The test samples containing 10 μg/mL of SARS-CoV-2 SP RBD were spiked with the above interfering species at the concentration similar to those reported previously [24, 25]. A change in the color intensity of less than 5% in comparison to its initial value is regarded as acceptable and interference-free. As expected, those interferences had literally no effect on the sensing capability of the d-LFIA as shown in Fig. 5. Therefore, it can readily be concluded that the developed d-LFIA device has satisfactorily high selectivity and specificity toward the SARS-CoV-2 antigen detection.
To meet a criteria of an effective sensing device, a long-term stability of the apparatus must be established. The d-LFIA strips were initially stored in a vacuum chamber and one was taken out for the measurement of 10 µg/mL SARS-CoV-2SP RBD every week. It was revealed that the proposed device was able to maintain the △intensity above 92% from its first reading for up to 5 weeks (Fig. S2) and thereby satisfying this requirement.
Clinical sample study
The d-LFIA strip was finally applied to detect SARS-CoV-2 SP RBD antigen in clinical samples to demonstrate its sensing efficiency. Twelve clinical throat swab samples were employed in this study, 8 of which were confirmed positive and 4 negative by the standard RT-PCR method. As shown in Table 2, seven out of eight positive samples and four negative samples were accurately determined by the d-LFIA strip. The only misinterpretation was a false negative derived from a sample which generated △intensity of 2.10, as tabulated in Table S1. Understandably, this particular sample likely contained a lower amount of antigen and the signal produced just failed the defined cutoff value (2.66). Overall, the sensitivity and specificity of the developed delayed LFIA strip for SARS-CoV-2 detection were found to be 87.5% and 100%, respectively.
Note: Sensitivity = the percentage of people who test positive for a disease that has that disease.
Specificity = the percentage of people without the disease who test negative for that disease.
Despite all the benefits, it is imperative to note there are certain drawbacks with the proposed method as well. The delaying strategy is particularly profitable to the competitive immunoassay where the interpretation of the antigen test results might be counterintuitive for the users already familiar with the currently used ATKs.
Conclusion
A novel delayed lateral flow immunoassay was successfully developed for rapid and enhanced detection of SARS-CoV-2 spike antigen. The d-LFIA device was modified using a trimethylsilyl cellulose barrier to delay the movement of RBD-conjugated AuNPs and allow the antigen to advantageously form immunocomplex with the antibody. As a result, the sensitivity and detectability of this platform were significantly improved as compared to the conventional LFIA. The applicability of the proposed method was demonstrated through a satisfactory screening of SARS-CoV-2 antigen in clinical samples. We believe that the proposed device can be used to gain analytical advantage in other competitive LFIA applications as well, particularly when higher sensitivity is required.
References
Cassaniti I, Novazzi F, Giardina F, Salinaro F, Sachs M, Perlini S, Bruno R, Mojoli F, Baldanti F, MotSMPC-T F (2020) Performance of VivaDiag COVID-19 IgM/IgG rapid test is inadequate for diagnosis of COVID-19 in acute patients referring to emergency room department. J Med Virol 92(10):1724–1727. https://doi.org/10.1002/jmv.25800
Huang C, Wen T, Shi F-J, Zeng X-Y, Jiao Y-J (2020) Rapid detection of IgM antibodies against the SARS-CoV-2 virus via colloidal gold nanoparticle-based lateral-flow assay. ACS Omega 5(21):12550–12556. https://doi.org/10.1021/acsomega.0c01554
Wasuwanich P, Thawillarp S, Ingviya T, Karnsakul W (2020) Coronavirus disease 2019 (COVID-19) and its gastrointestinal and hepatic manifestations. Siriraj Med J 72(4):272–282. https://doi.org/10.33192/Smj.2020.37
Yakoh A, Pimpitak U, Rengpipat S, Hirankarn N, Chailapakul O, Chaiyo S (2021) Paper-based electrochemical biosensor for diagnosing COVID-19: detection of SARS-CoV-2 antibodies and antigen. Biosens Bioelectron 176:112912. https://doi.org/10.1016/j.bios.2020.112912
Morales-Narváez E, Dincer C (2020) The impact of biosensing in a pandemic outbreak: COVID-19. Biosens Bioelectron 163:112274. https://doi.org/10.1016/j.bios.2020.112274
Nicol T, Lefeuvre C, Serri O, Pivert A, Joubaud F, Dubée V, Kouatchet A, Ducancelle A, Lunel-Fabiani F, Le Guillou-Guillemette H (2020) Assessment of SARS-CoV-2 serological tests for the diagnosis of COVID-19 through the evaluation of three immunoassays: two automated immunoassays (Euroimmun and Abbott) and one rapid lateral flow immunoassay (NG Biotech). J Clin Virol 129:104511. https://doi.org/10.1016/j.jcv.2020.104511
Bahadır EB, Sezgintürk MK (2016) Lateral flow assays: principles, designs and labels. TrAC Trends Anal Chem 82:286–306. https://doi.org/10.1016/j.trac.2016.06.006
Breitwieser D, Kriechbaum M, Ehmann HMA, Monkowius U, Coseri S, Sacarescu L, Spirk S (2015) Photoreductive generation of amorphous bismuth nanoparticles using polysaccharides – Bismuth–cellulose nanocomposites. Carbohydr Polym 116:261–266. https://doi.org/10.1016/j.carbpol.2014.06.017
Amornkitbamrung L, Mohan T, Hribernik S, Reichel V, Faivre D, Gregorova A, Engel P, Kargl R, Ribitsch V (2015) Polysaccharide stabilized nanoparticles for deacidification and strengthening of paper. RSC Adv 5(42):32950–32961. https://doi.org/10.1039/C4RA15153D
Wang P, Song J, Song R, Zhang M, Wu L, Li F, Yan Y, Zhou J, Chahan B, Liao M (2019) Preparation of monoclonal antibodies against Bc48 and development of a rapid detection assay for infection with Babesia caballi in China. Folia Parasitol (Praha) 66:005. https://doi.org/10.14411/fp.2019.005
Shawky SM, Awad AM, Allam W, Alkordi MH, El-Khamisy SF (2017) Gold aggregating gold: a novel nanoparticle biosensor approach for the direct quantification of hepatitis C virus RNA in clinical samples. Biosens Bioelectron 92:349–356. https://doi.org/10.1016/j.bios.2016.11.001
Costa MN, Veigas B, Jacob JM, Santos DS, Gomes J, Baptista PV, Martins R, Inácio J, Fortunato E (2014) A low cost, safe, disposable, rapid and self-sustainable paper-based platform for diagnostic testing: lab-on-paper. Nanotechnology 25(9):094006. https://doi.org/10.1088/0957-4484/25/9/094006
Fuchs P, Zhang K (2019) Efficient synthesis of organosoluble 6-azido-6-deoxy-2,3-O-trimethylsilyl cellulose for click reactions. Carbohydr Polym 206:174–178. https://doi.org/10.1016/j.carbpol.2018.11.003
Amornkitbamrung L, Marnul M-C, Palani T, Hribernik S, Kovalcik A, Kargl R, Stana-Kleinschek K, Mohan T (2018) Strengthening of paper by treatment with a suspension of alkaline nanoparticles stabilized by trimethylsilyl cellulose. Nano-Struct Nano-Objects 16:363–370. https://doi.org/10.1016/j.nanoso.2018.09.009
Puspasari T, Pradeep N, Peinemann K-V (2015) Crosslinked cellulose thin film composite nanofiltration membranes with zero salt rejection. J Membr Sci 491:132–137. https://doi.org/10.1016/j.memsci.2015.05.002
Reishofer D, Ehmann HM, Amenitsch H, Gspan C, Fischer R, Plank H, Trimmel G, Spirk S (2017) On the formation of Bi2S3-cellulose nanocomposite films from bismuth xanthates and trimethylsilyl-cellulose. Carbohydr Polym 164:294–300. https://doi.org/10.1016/j.carbpol.2017.02.008
Milosevic D, Moyer AM, Majumdar R, Kipp BR, Yao JD (2022) A reverse-transcription droplet digital PCR assay to detect and quantify SARS-CoV-2 RNA in upper respiratory tract specimens. J Clin Virol 153:105216. https://doi.org/10.1016/j.jcv.2022.105216
Grant BD, Anderson CE, Williford JR, Alonzo LF, Glukhova VA, Boyle DS, Weigl BH, Nichols KP (2020) SARS-CoV-2 coronavirus nucleocapsid antigen-detecting half-strip lateral flow assay toward the development of point of care tests using commercially available reagents. Anal Chem 92(16):11305–11309. https://doi.org/10.1021/acs.analchem.0c01975
Lee J-H, Choi M, Jung Y, Lee SK, Lee C-S, Kim J, Kim J, Kim NH, Kim B-T, Kim HG (2021) A novel rapid detection for SARS-CoV-2 spike 1 antigens using human angiotensin converting enzyme 2 (ACE2). Biosens Bioelectron 171:112715. https://doi.org/10.1016/j.bios.2020.112715
Roda A, Cavalera S, Di Nardo F, Calabria D, Rosati S, Simoni P, Colitti B, Baggiani C, Roda M, Anfossi L (2021) Dual lateral flow optical/chemiluminescence immunosensors for the rapid detection of salivary and serum IgA in patients with COVID-19 disease. Biosens Bioelectron 172:112765. https://doi.org/10.1016/j.bios.2020.112765
Liu D, Ju C, Han C, Shi R, Chen X, Duan D, Yan J, Yan X (2021) Nanozyme chemiluminescence paper test for rapid and sensitive detection of SARS-CoV-2 antigen. Biosens Bioelectron 173:112817. https://doi.org/10.1016/j.bios.2020.112817
Rashed MZ, Kopechek JA, Priddy MC, Hamorsky KT, Palmer KE, Mittal N, Valdez J, Flynn J, Williams SJ (2021) Rapid detection of SARS-CoV-2 antibodies using electrochemical impedance-based detector. Biosens Bioelectron 171:112709. https://doi.org/10.1016/j.bios.2020.112709
Gorkhali R, Koirala P, Rijal S, Mainali A, Baral A, Bhattarai HK (2021) Structure and function of major SARS-CoV-2 and SARS-CoV proteins. Bioinform Biol Insights 15:11779322211025876. https://doi.org/10.1177/11779322211025876
Noiphung J, Nguyen MP, Punyadeera C, Wan Y, Laiwattanapaisal W, Henry CS (2018) Development of paper-based analytical devices for minimizing the viscosity effect in human saliva. Theranostics 8(14):3797–3807. https://doi.org/10.7150/thno.24941
Mäkilä E, Kirveskari P (1969) A study of ascorbic acid in human saliva. Arch Oral Biol 14(11):1285–1292. https://doi.org/10.1016/0003-9969(69)90201-5
Acknowledgements
We thank Dr. Lunjakorn Amornkitbamrung for kindly providing the TMSC solution.
Funding
The authors gratefully thank financial supports from the Center of Excellence on Petrochemical and Materials Technology (PETROMAT), National Research Council of Thailand (NRCT) (N35A640020) and the Electrochemistry and Optical Spectroscopy Center of Excellence (EOSCE), Chulalongkorn University.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Ethics approval
The clinical samples were obtained from the Faculty of Medicine Ramathibodi Hospital, Mahidol University, and have been certified by the Research Ethics Review Committee by the Faculty of Medicine (Certification of approval: COA. MURA2021/879).
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Srithong, P., Chaiyo, S., Pasomsub, E. et al. A novel delayed lateral flow immunoassay for enhanced detection of SARS-CoV-2 spike antigen. Microchim Acta 189, 386 (2022). https://doi.org/10.1007/s00604-022-05467-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00604-022-05467-3