Abstract
COVID-19 is now a severe threat to global health. Facing this pandemic, we developed a space-encoding microfluidic biochip for high-throughput, rapid, sensitive, simultaneous quantitative detection of SARS-CoV-2 antigen proteins and IgG/IgM antibodies in serum. The proposed immunoassay biochip integrates the advantages of graphene oxide quantum dots (GOQDs) and microfluidic chip and is capable of conducting multiple SARS-CoV-2 antigens or IgG/IgM antibodies of 60 serum samples simultaneously with only 2 μL sample volume of each patient. Fluorescence intensity of antigens and IgG antibody detection at emission wavelength of ~680 nm was used to quantify the target concentration at excitation wavelength of 632 nm, and emission wavelength of ~519 nm was used during the detection of IgM antibodies at excitation wavelength of 488 nm. The method developed has a large linear quantification detection regime of 5 orders of magnitude, an ultralow detection limit of ~0.3 pg/mL under optimized conditions, and less than 10-min qualitative detection time. The proposed biosensing platform will not only greatly facilitate the rapid diagnosis of COVID-19 patients, but also provide a valuable screening approach for infected patients, medical therapy, and vaccine recipients.
Graphical abstract

Similar content being viewed by others
Introduction
As of January 27, 2021, a cumulative total of over 98.2 million COVID -19 cases and 2.1 million deaths have been reported and is now a severe threat to global health [1]. Therefore, diagnosis and prognosis of COVID-19 are facing serious challenges for the whole world. The current common method to diagnose SARS-CoV-2 infection is based on the detection of viral RNA in nasal swab samples with RT-PCR [2,3,4,5,6,7]. PCR-based diagnosis is straightforward and reliable; however, it requires the manipulation detail in sample collection with long time of more than hours and is unable to track the immune response to the viral infection. Most recent studies on SARS-CoV-2 have shown that SARS-specific antibodies [8, 9], for instance, IgG, IgM, and IgA, appeared in serum after viral infection [10,11,12,13]. The researches have indicated that the neutralizing antibody dynamic levels at different time points for different persons present different clinical characteristics [14, 15]. Generally, the antibodies (IgG, IgM) appear from the 4th day after COVID-19 infection [16], and the seropositivity rate reached up to 100.0% within 20 days since onset and remained 100.0% until days 41–53 [14]. The patients aged over 31 years had a higher neutralizing antibody level than those aged 16–30 years [14]; the generation of IgG antibody in female tended to be stronger than male patients in disease early phase, and in severe status, the average IgG antibody level in female patients tended to be higher than that of in male patients [17]. Therefore, virus-specific antibodies in serum can be used as effective diagnostic biomarkers for viral infection screening and the evaluation of patient’s adaptive immune responses after patients are discharged and vaccine injection. In addition to SARS-CoV-2 specific antibodies, viral antigens (spike (S) protein [18,19,20] and nucleocapsid (N) proteins [21,22,23]) in circulating blood can be used for the prognosis investigation of COVID-19-related viremia. A number of groups reported the SARS-CoV-2 specific antibodies (IgG, IgM) and viral antigen detection and evaluation through different techniques [24,25,26]. Present detection methods of SARS-CoV-2 specific antibodies and viral antigens include electronic transistor-based biosensor [10], gold nanoparticle-based lateral flow assay [11], and enzyme-linked immunosorbent assays (ELISAs) [12]. Electronic transistor-based biosensor presents high sensitivity during SARS-CoV-2 antigen protein detection, and its practical applications are limited by very complex manufacturing processes [10]. Gold nanoparticle-based lateral flow assay is applied in rapid detection of SARS-CoV-2 IgG/IgM antibodies with limited sensitivity to conduct quantitative detection [27]. ELISA, on the other hand, can provide quantitative detection; however, it involves complicated labor manipulation process and long detection time [28]. Though the obvious progress of the SARS-CoV-2 antigen protein and antibody detection has been made through different techniques, the COVID19 pandemic still desires high-throughput, economic, highly-automatic, rapid, and sensitive platform to conduct simultaneous detection of SARS-CoV-2 IgG/IgM antibodies and SARS-CoV-2 antigen proteins for effective diagnosis, therapy, convalescent tracking, and future vaccination validation.
Carbon-based nanomaterials, for instance, graphene oxide (GO), reduced graphene oxide (rGO), and graphene quantum dots (GQDs) etc., were intensively explored in biosensors because of their fluorescence resonance energy transfer (FRET), biocompatible and economic properties, which have achieved essential progress in bio-molecular sensing [29,30,31]. However, most nanomaterial-based biosensors conducted detections in liquid phase, and it is still challenging to achieve ultrasensitive detection because of the limited factors including the distribution of complex substance in the reaction solution to the optical detection system, the influence of solution components on FRET efficiency, and relatively high background noise from the optical effects of nanoparticles in solution. Herein, we develop a high-throughput GOQDs-based immunoassay biochip with detection in solid phase to realize ultrasensitive and rapid simultaneous detection of COVID-19 specific antigens and immunoglobulin IgM/IgG antibodies including spike RBD protein (S-antigen), nucleocapsid protein (N-antigen), and SARS-CoV-2 spike RBD IgG/IgM (S-IgG/S-IgM) and nucleocapsid IgG/IgM (N-IgG/N-IgM). In solid phase detection, the immunoassay biochip integrated the advantages of GOQDs and microfluidics: (1) multiple parallel microchannels are integrated with the large surface/volume ratio and abundant functional groups of GOQDs to printed antibody/antigen barcode array and (2) the non-specifically adsorbed fluorescence molecule is quenched by the GOQDs substrate, which contributes to extremely low background noise. As a result, the proposed biochip is capable of the COVID-19 specific neutralizing antibodies and antigen biomarker detection using only 2 μL serum sample volume within as short as less than 10 min. The proposed immunoassay biochip will greatly facilitate rapid and economic screening of infected populations or vaccine response of vaccine recipients.
Materials and methods
Materials and reagents
Bull serum albumin (BSA) and 3-aminopropyltrimethoxisylane (APTES) were purchased from Sigma-Aldrich. The phosphate buffered saline (PBS) was purchased from Corning. The glass substrates (3″ × 1″) were purchased from Citotest. Silicon wafers were purchased from Meixin Electronic Technology Co. Ltd. SU-8 2025, AZ5214 photoresist and developer were ordered from Bynano Co. Ltd. Streptavidin-allophycocyanin (streptavidin-APC) was purchased from Biolegend (America). Treated chlorotrimethylsilane (TMCS) was purchased from Sinopharm Group Chemical Reagent Co. Ltd. Alexa Fluor® 488-conjugated goat anti-mouse IgG were purchased from Abcam. Anti-COVID-19 spike Mab (S-Ab), Anti-COVID-19 Nucleocapsid Mab (N-Ab), Recombinant COVID-19 Spike-RBD Protein (S-antigen), Recombinant COVID-19 Nucleocapsid Protein (N-antigen), Human IgM Anti-Covid-19 Spike (S-IgM), Human IgG Anti-COVID-19 Spike (S-IgG), Human IgM Anti-COVID-19 Nucleocapsid (N-IgM), Human IgG Anti-COVID-19 Nucleocapsid (N-IgG), and ELISA kit were purchased from Smart Life Sciences (Changzhou, China). Alexa Fluor™ 488-conjugated goat anti-human IgM and Alexa Fluor™ 633-conjugated goat anti-human IgG were purchased from Thermo Fisher Scientific. The ultrapure water (18.25 MΩ cm) used throughout all experiments was made by a Millipore system. All chemicals in this work were analytical grade or highest purity available for direct use without further purification.
The detection antibodies of S-antigen and N-antigen were labeled with biotin using commercial kits (ab201795-Biotin, Abcam). Briefly, 100 μL of Biotin Modifier regent was added into lyophilized detection antibodies to obtain an antibody concentration of 1 mg/mL. After incubation for 15 min at room temperature, 10 μL biotin quencher reagent was added into the mixed solution and incubated for 4 min. Then the conjugates are ready for use without any purification. Biotinylated detection antibody is mixed with streptavidin-APC at a ratio of 100:1. The mixed solution was incubated for 10 min at room temperature and then is ready for use.
Immunoassay biochip fabrication
The biochips consist of the GOQDs functionalized substrate with detection barcode array, capturing antibody/antigen array microprinting PDMS chip and sample loading PDMS chip. The GOQDs functionalized substrate is assembled with high-density GOQDs through coupling agent APTES, on which capturing antibody/antigen array is imprinted through π – π stacking to produce repeatable and space-encoded antibody/antigen barcode array with the assistance of microprinting microfluidic chip. The capturing antibody/antigen array microprinting PDMS chip has 20 parallel microchannels of 20 μm in width to allow at most 20 kinds of capture antibodies/antigens microprinting. The sample loading PDMS chip is made of 1 mm thick PDMS with 60 detection chambers of 2 mm × 2 mm. Each chamber covers a complete antibody/antigen array and has a capacity of 4 μL sample volume.
GOQDs functionalization process
The glass substrates were cleaned thoroughly with ultrasonic in ethanol and deionized water, sequentially, and dried with N2 blowing. Then the substrate was treated with APTES solution and followed by ultrasound cleaning in deionized water. The APTES treated substrates were treated in GOQDs solution to assemble GOQDs on it. The properties of the GOQDs were characterized and analyzed as shown in supplementary material part and Fig. S1. Finally, extra GOQDs on the substrates were cleaned away by deionized water washing. The GOQDs functionalized substrates are ready for antibody/antigen detection barcode array microprinting as shown in Fig. S2a.
PDMS chip fabrication
The hard molds were designed and fabricated on silicon wafer through photolithography and etching process as shown in Fig. S3. To make antibody/antigen array microprinting and sample loading PDMS chip, silicon wafer molds were treated with chlorotrimethylsilane (TMCS) for 20 min. A ratio of Sylgard 184 A: B = 10:1 by weight was mixed thoroughly and loaded onto TMCS-treated silicon mold in a Petri dish. After all the bubbles in the PDMS were vacuumed out, the Petri dish containing PDMS mixer was cured at 80 °C for 60 min. Then the PDMS layers were peeled off from the silicon wafer molds. The antibody/antigen array microprinting PDMS layer (Fig. S2b) was punched to produce inlets and outlets for antibody/antigen barcode microprinting and then lined up and bonded with the GOQDs functionalized glass substrate forming spacing encoding microfluidic biochip. The sample loading PDMS chip was made of 1-mm-thick PDMS layer, and 60 chambers of 2 mm × 2 mm were punched according to the designed pattern (Fig. S2c).
Clinical sample preparation
First, 1–2 mL venous blood from the healthy person and vaccinated individual volunteers was collected into a non-anticoagulant tube and kept standing for 2 h. Then, the supernatant was collected into a new Eppendorf tube and centrifuged at 3000 rpm for 10 min. Finally, 100 μL supernatant was collected into a new tube and stored at −80 or − 20 °C for detection.
Antibody/antigen barcode array
To microprint detection barcode array, 2 μL capture antibody/antigen of 500 μg /mL was loaded into an inlet connected to a 20-μm-wide microchannel of the capturing antibody/antigen array microprinting PDMS chip. Loaded antibody/antigen was pumped through the microchannel and immobilized on the substrate through simple π – π stacking. Finally, the capturing antibody/antigen array microprinting PDMS chip was removed, and the substrate (Fig. S2d) was blocked with 3% BSA solution for 10 min to reduce non-specific adsorption.
SARS-CoV-2 IgG/IgM/antigen detection by the microfluidic biochip
The barcode array substrate was bonded with the sample loading PDMS chip. The target antibody/antigen samples (2 μL of each sample) were introduced into the chambers of the microfluidic biochip. After incubation at room temperature for 3–20 min, the sample loading PDMS chip was peeled off from the substrate and immersed in 1% BSA (BSA power diluted in PBS), and the substrate was flushed with 1% BSA. For antigen detection, 300 μL fluorescence-conjugated detection antibody complex of 10 μg/mL was loaded onto the substrate and dispersed all over the whole substrate. The reaction time between detection antibody and antigen is from 2 to 20 min. While for antibody detection, 300 μL goat-anti-human IgG and IgM secondary antibody labeled with fluorescence was loaded with concentration of 10 μg/ml and incubated for 2–20 min at room temperature. Finally, the substrate with detection fluorescence signal was washed by 1% BSA, PBS, and DI water and then span to dry. The substrate with detection fluorescence signal was scanned by GenePix 4400 to analyze target antibody/antigen concentration. Genepix 4400A scanner was used to obtain scanned fluorescence images in fluorescein isothiocyanate APC (635 nm), Alexa Fluor™ 633 (635 nm), and Alexa Fluor™ 488 (488 nm) channel. The image was analyzed using GenePix Pro software by aligning signals with array template. Fluorescence intensities were then extracted and exported into Excel files. The illustrated detection processes of SARS-CoV-2 IgM/ IgG/antigens on the biosensing chip are described in Fig. S4.
SARS-CoV-2 IgG/IgM/antigen detection by ELISA
The SARS-CoV-2 antibody or antigen is fixed on each well of ELISA plate. Then 100 μL of target antigen/antibody samples were introduced into each well of ELISA plate. After incubation at room temperature for 2 h, sample residue was removed from each well, and the substrate was cleaned with 1% BSA for five times. Then 100 μL of corresponding enzyme-linked antibody/antigen were added to bind the antigen/antibody immobilized on the solid carrier. After incubation for 2 h, the residue was removed from each well, and the substrate was cleaned with 1% BSA for five times. Finally, the substrate solution was added, and the catalysis led to a color change which can be measured for quantification of target antigen/antibody concentration.
Material property characterization
Particle size and shape of GOQDs nanoparticles were characterized using high-resolution transmission electron microscopy TEM (FEI-G20, America). Surface morphologies of the substrate during fabrication process were examined using a SmartSPM AFM system. The UV-vis absorption of the GOQDs solution was obtained with a UV-vis absorption spectrophotometer (UV-2700, Shimadzu, Japan). A multifunctional microplate reader (CYTATION5, BioTek, USA) was utilized to perform the photoluminescence (PL) spectrum of GOQDs. Raman spectra of all the samples were conducted by a Renishaw inVia Raman microscope at room temperature excited by a laser of 785 nm. The fluorescence signals were collected by GenePix 4400 and analyzed with Genepix 4400 software.
Results and discussion
The construction of the biochip for SARS-CoV-2 antibody/antigen detection
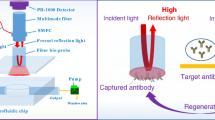
SARS-CoV-2 is a new kind of virus consisting of antigen protein (surface spike (S), membrane (M), and nucleocapsid (N) proteins) and RNA viral genome as shown in Fig. 1a, which invades humans through S protein interaction with human cells and duplicates itself through RNA viral genome in human cells. Once the person is infected by SARS-CoV-2, the adaptive immune system will develop SARS-CoV-2 antibodies (IgG/IgM) to prevent SARS-CoV-2 from invading. The schematic of designed microfluidic biochip is shown in Fig. 1b for the detection of the SARS-CoV-2 antigen proteins and IgG/IgM antibodies, which has the capability of 60 samples simultaneous detection consuming small sample volume of 2 μL. In order to microprint capture antibody/antigen barcodes on the substrate, GOQDs were assembled on amination-treated glass substrates and used to immobilize capture barcodes through simple π – π stacking. For SARS-CoV-2 antigen detection, the target antigen is sandwiched by capture and detection antibody specifically, as presented in Fig. 1c. To conduct anti-SARS-CoV-2 IgG/IgM detection, specific antigen is microprinted on the GOQDs functionalized substrate to capture target antibody firstly, and fluorescence-labeled goat-anti-human secondary antibody IgG and IgM are used for specific binding to IgG/IgM and output fluorescence signals as shown in Fig. 1d. The whole detection process was recorded in Video 1. To confirm the successful conjugation of each step of the detection process, the AFM and Raman (Fig. S5) characterizations were performed on the microfluidic biochip. The results showed slight changes in surface morphology after GOQDs functionalization, capture antibody immobilization, and target antigen capture, together with the appearance of representative Raman peaks for the typical material in each step, which indicated successful fabrication and sensing process of the microfluidic biochip for SARS-CoV-2 IgG/IgM/antigen detection. Finally, the detection conditions are optimized, and the results are shown in Fig. S6.
Schematic illustration of a the SARS-CoV-2 virus and b the immunoassay biochip: sample loading chip with reaction units (60 units per chip) is bonded with barcode array chip, and each unit covers an intact array of barcodes. Detection mechanism schematic of c SARS-CoV-2 antigen, the target antigen in sample is firstly captured by barcodes, and labeled detection antibody is conjugated with target. Detection mechanism schematic of d anti-SARS-CoV-2 IgM/IgG in samples, and labeled secondary antibody reacts with target antibody to present fluorescence signal
(MP4 285925 kb)
During practical applications, detection time is an extremely important parameter, and the time optimization of each detection step was performed, and the best conditions were presented in Fig. 2. Figure 2a shows the immobilization process of capture antibody/antigen barcode on the substrate, which took about 130 min. But these barcode substrates can be prepared and stored at 4 °C or − 20 °C ready for use. For SARS-CoV-2 antigen detection, as shown in Fig. 2b, it takes 20 min for capture antibody/antigen and target antigen/antibody to achieve adequate reaction and only 10 min for biotin and streptavidin conjugation. Also, it takes 20 min for adequate reaction of target antigen/antibody and detection antibody/secondary antibody. As a result, the best quantitative detection process consumes 40 min, since the conjugation of biotin labeled capture antibody and streptavidin-APC is prepared in advance before SARS-CoV-2 antigen detection, which is stored in the refrigerator ready for use anytime. For anti-SARS-CoV-2 IgG/IgM detection, as shown in Fig. 2c, it takes 20 min for sufficient incubation of microprinted antigen and IgG/IgM antibody and 10 min for adequate reaction of target IgG/IgM antibody and secondary antibodies. There is a tradeoff between detection time and detection limit. These optimized process time guarantees the lowest detection limit, and the total detection time could be reduced according to special requirement in practice, which increases the detection limit in some extent. The detection time can be shortened to be 10 min, as shown in Fig. 2 d and e by reducing sample incubation time to 6 min and fluorescent secondary antibody incubation to 4 min. In addition, the whole detection time can be even reduced to n 5 min, as shown in Fig. 2 f and g. However, the additional rinse steps during detection process could potentially take longer depending on the user without the assistance of automated equipment.
Graphical illustration for the assay protocols. a Procedure of the antibody/antigen array preparation processes. b Protocol for optimized antigen detection with detection time of 40 min. c Protocol for optimize IgG/IgM detection with detection time of 30 min. d Protocol for antigen detection with detection time of 10 min. e Protocol for IgG/IgM detection with detection time of 10 min. f Protocol for antigen detection with detection time of 5 min. g Protocol for IgG/IgM detection with detection time of 5 min
Specificity and sensitivity of SARS-CoV-2 antibody/antigen detection
To detect S-antigen and N-antigen proteins, 2 μL recombinant S-antigen and N-antigen samples with different concentrations from 1 to 105 pg/mL are loaded into the biochip followed by adding detection antibodies labeled with fluorescence. Both S-antigen and N-antigen detections appear excellent specificity, as in Fig. 3a. The fluorescence intensity dependence on target antigen concentration is presented in Fig. 3 b and c, together with their fitting equations, which show a similar binding affinity for both S-antigen and N-antigen. The results delivered that the biochip has a broad detection range, excellent specificity, and ultra-sensitivity of ~0.3 pg/mL for both SARS-CoV-2 S-antigen and N-antigen. The fitted linear relationship equations between fluorescence intensity and target concentration in Fig. 3 b and c provides standard curves to calculate the concentrations of targets in unknown samples. During practical applications, once the fluorescence intensity at different barcode area is detected, the concentration of corresponding target can be calculated by the fitted equations.
SARS-CoV-2 antibody/antigen detection specificity and sensitivity of the biochip. Detection specificity of a S-antigen/N-antigen and d IgG/IgM antibodies. Fluorescence intensity dependence and fitted linear equation of c S-antigen, d N-antigen, e S-IgG, f N-IgG, g S-IgM, and h N-IgM. Note: Error bars are standard deviations in three parallel experiments. Note: Control sample in a and d is 3% BSA in PBS buffer, and the fluorescence signal is obtained at ~680 nm
To perform SARS-CoV-2 IgG/IgM antibody detection, the human S-IgM, S-IgG, N-IgM, and N-IgG with different concentrations from 1 to 105 pg/mL are loaded separately in each loading chamber with a full array of capture antigens on the biochip substrate to perform their specificity and sensitivity. Actually, S-antigen is used to capture both S-IgM and S-IgG, and N-antigen is used to capture both N-IgM and N-IgG, but their signals are separated by secondary IgM and IgG labeled with fluorescence. Figure 3d shows that they all specifically react with their corresponding capture antigen and secondary antibody, since paired capture antigen-antibody presents much higher fluorescence signal than mismatched pairs. The dependence of fluorescence intensity on antibody concentrations is shown in Fig. 3e–h, and further analysis demonstrates that fluorescence intensity has a similar linear relationship with target antibody concentrations in log scale for all four antibodies. The concentrations of targets in unknown samples can be calculated through the fitted equations in Fig. 3e–h based on the detected fluorescence intensity at different barcode strips. Detection limit of two target antigens and four target antibodies was calculated based on the representative equation LOD=3Sb1/S [32, 33], where S is the gradient of linear regression equation at low concentration (the low concentration range from 1 to 103 pg/mL in this manuscript) and Sb1 is the standard deviation of background. The calculation formula of standard deviation is \( {S}_{b1}=\sqrt{\frac{1}{N}\sum \limits_{i=1}^N{\left({x}_i- mean\right)}^2} \), where mean is the average of the background values (the valve is 60.12 in this article) and N = 20. The method equation in Table S1 is the linear regression equation at concentration range of 1 to 103 pg/mL, and the calculated detection limit is listed in Table S1, which are 0.23 pg/mL, 0.35 pg/mL, 0.21 pg/mL, 0.32 pg/mL, 0.31 pg/mL, and 0.29 pg/mL for S-antigen, N-antigen, S-IgM, S-IgG, N-IgM, and N-IgG, respectively. All the results indicate that the developed microfluidic biochip has good specificity and sensitivity for SARS-CoV-2 antibody/antigen detection. The negative control signal in Fig. 3 a and d is ~60 a.u., which means very low background noise contributed by the quenching function of GOQDs for fluorescence close to the substrate. The non-specifically bonded fluorescence-labeled detection antibody or secondary antibody does not show fluorescence intensity because of the short distance to the substrate. And only the detection antibody binding to the target has output fluorescence signal because of the enough long distance from the substrate.
Detection limit and detection time of SARS-CoV-2 antibodies/antigens
SARS-CoV-2 antibody/antigen detection time of the biochip is a very important factor during pandemic outbreak. At the same time, the critical detection limit is required to distinguish health and infected patients. To determine the effective detection time of SARS-CoV-2 antibody/antigen, we performed the target S-antigen, S-IgG, and S-IgM antibody detection with different detection time since the developed biochip has similar specificity and sensitivity for SARS-CoV-2 antibody/antigen detection demonstrated above. The fluorescence intensity results detected with 5 min, 10 min, 20 min, and 40 min for the S-antigen, S-IgG, and S-IgM antibody with concentrations from 5 to 500 ng/mL in serum are shown in Figs. S7-S9, which were used to determine the detection limit according to the representative formula LOD=3Sb1/S [32, 33]. The detection limit for S-antigen and S-IgG/IgM increases from ~0.3 pg/mL to ~4 ng/mL with detection time decreasing from 40 min to 5 min as shown in Fig. 4a–c, which result from the partial reaction between SARS-CoV-2 antibodies/antigen with the substrate and fluorescence labeled detection antibodies. To further validate the shortest effective detection time with reasonable sensitivity, the relative low S-antigen, S-IgG, and S-IgM sample concentration of 5 ng/ml was chosen to detect with 5–40 min for the microfluidic biochip. The results in Fig. 4d–f indicate that SARS-CoV-2 antibodies/antigen of 5 ng/mL could be distinguished basically in as short as 5 min and obviously in 40 min.
Detection limit and quantitative detection dependent on detection time of the microfluidic biochip. The detection limit of the biochip with 5, 10, 20, and 40 min detection time for a S-antigen, b S-IgG, and c S-IgM antibodies of SARS-CoV-2 antibody/antigen in serum. The detected fluorescence intensity of the biochip with 5, 10, 20, and 40 min for d S-antigen, e S-IgG, and f S-IgM antibodies with concentration of 5 ng/μL in serum. Note: The red-dotted lines represent fluorescence intensity of the background from 3 times detection. Error bars are standard deviations in three parallel experiments, and the fluorescence signal is obtained at ~680 nm in e–f
Reproducibility of SARS-CoV-2 antibody/antigen detection
The reproducibility of the biochip determines the detection accuracy of SARS-CoV-2 antibody/antigen. In order to perform the repeatability of proposed biosensing platform, 5 repeated tests were conducted for the same samples with SARS-CoV-2 antibodies/antigens of 100 ng/mL, and the detected fluorescence intensity is shown in Fig. S10. The detected concentration has small variations, and the relative standard deviation (RSD) of all six targets is less than 3%, as shown in Fig. 5a–f, which indicates excellent reproducibility of the proposed SARS-CoV-2 antibody/antigen detection microfluidic biochip.
Detection and analysis of simulated samples
During practical applications, all the targets are mixed in serum or other body fluids together with lots of bio-substance, and high-performance detection platform is desired to have the capability of handling clinical samples with excellent specification and sensitivity. Prior to performing serum sample analysis, it is necessary to test the platform using mixed target samples. First, S-antigen protein and N-antigen protein are mixed into 1% BSA or healthy serum to form 3 samples with different concentrations, and four kinds of target IgG/IgM antibodies are mixed into 1% BSA or healthy serum to form 3 samples, as listed in Table S2. Fluorescence quantitative analysis of target antigens is measured, as shown in Fig. 6a–c. Fluorescence quantitative analysis of target antibodies in mixed BSA and healthy serum is shown in Fig. 6d–f. The test concentrations derived from the relationship equation in Fig. 3 are described in Fig. 6g–l, which present recovery of 90.36–109.98% for all targets. In addition, detected concentrations of targets in spiked 1% BSA are consistent with those in spiked healthy serum, and it shows slight deviation less than 5%, as presented in Fig. S11. All the results delivered the practical application capability of the proposed biochip.
Detection results of mixed samples in 1% BSA and healthy serum. Fluorescence intensity of S-antigen and N-antigen in a sample 1, b sample 2, and c sample 3. Fluorescence intensity of IgG/IgM in mixed d sample 4, e sample 5, and f sample 6. The correlation of spiked and detected concentrations: g S-antigen, h N-antigen, i S-IgG, j N-IgG, k S-IgM, and l N-IgM. Error bars represent standard deviations in three parallel experiments. Note: the fluorescence signal is obtained at ~680 nm
High-throughput detection and analysis of the simulated samples
To demonstrate the high-throughput and statistical sensitivity of the biochip, we performed multiple SARS-CoV-2 antigen/antibody simultaneous detection with different concentrations. SARS-CoV-2 S-antigen, S-IgG, and S-IgM in serum were tested under 5 min in 2-μL sample volume. A total of 60 samples for each antigen/antibody were tested on one individual biochip including 12 negative control (0 ng/mL) and 16 positive simulated samples with concentration of 5, 50, and 100 ng/mL, respectively. The tested results were presented in Fig. 7a–c, which show apparent fluorescence signals for different samples and are consist with the concentrations of simulated samples. The quantitative fluorescence intensity from S-antigen, S-IgG, and S-IgM is shown in Figs. S12-S14, respectively. Figure 7d–f present the statistical analysis of the detected fluorescence intensity. The difference between negative control and positive simulated samples is significant, which delivers p < 0.0001 in Student t test. The detected concentrations of the simulated S-antigen, S-IgG, and S-IgM in serum samples were distributed in the range of 0.001–100 ng/mL, which had the recovery regime of 90.36–109.98%. Therefore, the developed microfluidic biochip provides a new high-throughput, rapid simultaneous SARS-CoV-2 antigen/antibody detection and analysis.
Detection of SARS-CoV-2 S-antigen, S-IgG, and S-IgM in spiked healthy serum. a–c Fluorescence pattern of detection chip. Fluorescence intensity dependence on concentration of SARS-CoV-2 S-antigen, S-IgG, and S-IgM a S-antigen, b S-IgG, and c S-IgM. d–f Statistical comparison between the negative samples and the positive samples. Since p < 0.05, the difference between these two groups is statistically significant. Assay results of spiked in healthy serum using the Proposed and Elisa Methods. Correlation between the two detection methods: g S-antigen and h S-IgG. Note: Control samples in a–c are serum sample from healthy persons. The fluorescence signal is obtained at ~680 nm in a and b, and the fluorescence signal is obtained at ~519 nm in c
To evaluate the detection accuracy of the proposed microfluidic biochip, a comparison of the spiked SARS-CoV-2 S-antigen and S-IgG in healthy serum and 1% BSA sample detection between our biochip and commercial Enzyme-Linked Immunosorbent Assay (ELISA) was performed, since ELISA is the gold standard for protein biomarkers analysis. The ELISA detection procedures of the SARS-CoV-2 S-antigen and S-IgG are illustrated in Fig. S15, which took ~4.5 h. The ELISA detected results of SARS-CoV-2 S-antigen and S-IgG are presented in Fig. S16. It has a linear range of 1–100 ng/mL and detection limit of 1 ng/ml for both simulated samples spiked in healthy serum and 1% BSA samples. In the detectable range of ELISA, the comparison of detected results between our biochip and commercial ELISA are presented in Fig. 7g–h, and the specific analysis data are shown in Table S3, which has recovery values of 97.19–103.01%. Thus, the proposed biochip is consistent with the commercial ELISA. Moreover, the microfluidic biochip has shorter detection time, lower detection limit, smaller reagent, and sample volume consuming than ELISA, indicating its potential use in rapid, economic, and high-throughput detection of SARS-CoV-2 antigens and IgG/IgM antibodies in clinical diagnosis and vaccine recipients’ evaluation.
Anti-SARS-CoV-2 IgG/IgM detection of clinical samples
In order to test the biochip in practical applications, 15 serum samples from vaccinated individuals of SARS-CoV-2 virus and 5 serum samples from healthy persons without vaccination were collected and detected. The results in Fig. 8a–d show that vaccinated individuals have much higher fluorescence intensity of all IgG/IgM antibodies than healthy persons without vaccination whose intensity is similar to background. Figure 8d–f presents the statistical analysis of the detected fluorescence intensity. The difference between healthy and vaccinated individuals is significant, which delivers p < 0.0019 in Student t test. The expression level of neutralizing antibodies, anti-S-RBD (S-IgG and S-IgM), represents the effectiveness of vaccination, because neutralizing antibodies could bind the S-RBD on SARS-CoV-2 virus and effectively protect human cells from virus infection by blocking the binding of SARS-CoV-2 virus with ACE2 on human cells. The level of anti- SARS-CoV-2 IgG/IgM may decrease with times goes after vaccination, and long-term tracking detections are needed to evaluate the protective induration of vaccination.
Detection of SARS-CoV-2 antigen/IgG/IgM antibodies in the serum from healthy and vaccinated individuals. Fluorescence intensity of a S-IgG, b N-IgG, c S-IgM, and d N-IgM; statistical comparison of e S-IgG, f N-IgG, g S-IgM, and h N-IgM between the healthy persons and vaccinated individuals. Note: The fluorescence signal of IgG in (a) and (b) is obtained at ~680 nm, and the fluorescence signal of IgM in (c) and (d) is obtained at ~519 nm
Discussion
The World Health Organization (WHO) has developed the ASSURED criteria as a benchmark to evaluate if the tests efficiently address the needs for disease control: affordable, sensitive, specific, user-friendly, rapid and robust, equipment-free, and deliverable to end users [34, 35]. However, none of the current methods fulfill all the criteria. For SARS-CoV-2 pandemics, currently, sensitive, rapid, high-throughput, and low-cost methods are essentially demanded to detect antibodies and antigens in practical applications including diagnostics, epidemiology, vaccine evaluation, disease understanding, and public health policies at the stage of large-scale outbreak all over the world [36]. Various immunosensors based on varied matrices have been compared on the basis of their performance and are summarized in Table S4. Most sensing platforms conduct single biomarker with narrow detection range, higher detection limit, and longer than 15-min qualitative detection time. The proposed microfluidic biochip platform has the capability of relatively large detection range of 5 orders of magnitude, simultaneous detection of 4 or even more (up 20 in current biochip design) biomarkers with 2 μL of each sample, high-throughput (60 samples for each biochip), as short as less than 10 min for qualitative detection, and 40 min for quantitative detection with ultralow detection limit of ~0.3 pg/mL, which essentially reduce SARS-CoV-2 antigen/antibody detection cost, time-consuming, and transmission probability. In addition, the main facilities to fabricate the proposed biochips are the photolithography process-related apparatus and a temperature-controlled furnace. The material cost of one biochip is about 1–2 dollar. Thus, in comparison to reported SARS-CoV-2 antigen/antibody detection methods, the proposed microfluidic biochip strategy could be more suitable for the current needs of SARS-CoV-2 pandemics.
Though the developed microfluidic biochip has the distinct merits mentioned above, some points and limitations still need to be discussed. Firstly, what is the best dilute factor for real SARS-CoV-2 antigen/antibody samples evaluation? As reported recently, the concentration of SARS-CoV-2 IgG/IgM antibodies in COVID-19 patients is in the range of tens ng/mL to several mg/mL [37, 38] And higher than 1 μg/mL, the neutralization of SARS-CoV-2 antibody concentration could ensure an effective protection (>50% protection) depending on the binding epitope and the affinity of the antibodies [15, 37, 39,40,41,42]. The proposed microfluidic biochip has detection limits of ~0.3 pg/mL under 40 min process and ~ 4 ng/mL under 5-min process. For quick qualitative evaluation of SARS-CoV-2 antibodies, direct detection of the samples without any dilution could be performed using the microfluidic biochip, which also takes the advantage of 2 μL small sample volume requirement of the microfluidic biochip. For quantitative evaluation of SARS-CoV-2 antibodies, the samples diluted in 1:10000 could still be distinguished if the patient sample is with lowest concentration of tens ng/mL [37, 38]. But in order to improve detection accuracy, the reasonable dilution rate should be determined based on the situation of the samples and applications for real practical tests. Secondly, the required multiple user steps need trained personnel at present, since the automated equipment is still in development. And the additional rinse steps during detection process could potentially take longer depending on the user without the assistance of automated equipment.
Generally, a diagnostic technology development could be divided into four phases: Phase 1 refers to technologies that are at the proof-of-concept stage where researchers use synthetic targets to validate the concept. Phase 2 refers to technologies that have analyzed a small number of patient samples (i.e., <100 samples). Phase 3 typically refers to technologies that advance to clinical trials with a large patient cohort. Phase 4 is when the technology is commercialized and used in patients [5]. However, many approaches [43,44,45], for instance, ELISA, Fluorescence Immunoassays (FIAs), Lateral Flow Immunoassays (LFIAs), CRISPR Systems, and our microfluidic biochip, were developed for SARS-CoV-2 diagnosis and evaluation. Most of the methods are in phase 1 and 2, but they would still play essential roles in SARS-CoV-2 diagnostics, epidemiology, vaccine evaluation, disease understanding, and public health policies [5, 36]. Currently, ELISA, a gold standard of protein assay, is used to validate the accuracy of our microfluidic biochip, and remarkable results were obtained, indicating the potential applications in detection of the real SARS-CoV-2 samples. Limited by the lab environment qualification since clinical COVID-19 samples need to be done in authorized hospital or lab, the SARS-CoV-2 antigens of clinical samples have not been detected by our biochip, and only anti-SARS-CoV-2 IgG/IgM was tested in vaccinated individuals. Next, long-term tracking detection of vaccinated persons will be conducted to evaluate the vaccination effectiveness through collaborations.
Conclusion
In this work, we developed a microfluidic immunoassay to conduct rapid (less than 10 min), high-throughput (60 samples per chip), reliable, and ultrasensitive detection of multiple SARS-CoV-2 antigens and antibodies simultaneously. The platform integrates the advantages of nanomaterials to conjugate high-density biomolecules and microfluidics to realize high-efficient, low-volume, and high-integration biosensing platform. The proposed microfluidic biosensing platform not only has great potential benefits for the fast screening of SARS-CoV-2 infections, but also provides valuable tool for valid vaccination screening.
Placeholder TextPlaceholder TextPlaceholder TextPlaceholder Text
References
Coronavirus disease (covid-19) weekly epidemiological update. https://www.who.int/docs/default-source/coronaviruse/situation-reports
Chen LD, Li H, Ye YM, Wu Z, Huang YP, Zhang WL, Lin L (2020) A COVID-19 patient with multiple negative results for PCR assays outside Wuhan, China: a case report. BMC Infect Dis 20:517. https://doi.org/10.1186/s12879-020-05245-7
Corman VM, Landt O, Kaiser M, Molenkamp R, Meijer A, Chu DK, Bleicker T, Brunink S, Schneider J, Schmidt ML, Mulders DG, Haagmans BL, van der Veer B, van den Brink S, Wijsman L, Goderski G, Romette JL, Ellis J, Zambon M, Peiris M, Goossens H, Reusken C, Koopmans MP, Drosten C (2020) Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Euro Surveill 25:23–30. https://doi.org/10.2807/1560-7917.ES.2020.25.3.2000045
Loeffelholz MJ, Tang YW (2020) Laboratory diagnosis of emerging human coronavirus infections - the state of the art. Emerg Microbes Infect 9:747–756. https://doi.org/10.1080/22221751.2020.1745095
Udugama B, Kadhiresan P, Kozlowski HN, Malekjahani A, Osborne M, Li VYC, Chen H, Mubareka S, Gubbay JB, Chan WCW (2020) Diagnosing COVID-19: the disease and tools for detection. ACS Nano 14:3822–3835. https://doi.org/10.1021/acsnano.0c02624
Xu M, Wang D, Wang H, Zhang X, Liang T, Dai J, Li M, Zhang J, Zhang K, Xu D, Yu X (2020) COVID-19 diagnostic testing: technology perspective. Clinical and Translational Medicine 10:e158. https://doi.org/10.1002/ctm2.158
Chu DKW, Pan Y, Cheng SMS, Hui KPY, Krishnan P, Liu Y, Ng DYM, Wan CKC, Yang P, Wang Q, Peiris M, Poon LLM (2020) Molecular diagnosis of a novel coronavirus (2019-nCoV) causing an outbreak of pneumonia. Clin Chem 66:549–555. https://doi.org/10.1093/clinchem/hvaa029
Azer SA (2020) COVID-19: pathophysiology, diagnosis, complications and investigational therapeutics. New Microbes New Infect 37:100738. https://doi.org/10.1016/j.nmni.2020.100738
Fang B, Meng QH (2020) The laboratory's role in combating COVID-19. Crit Rev Clin Lab Sci 57:400–414. https://doi.org/10.1080/10408363.2020.1776675
Seo G, Lee G, Kim MJ, Baek SH, Choi M, Ku KB, Lee CS, Jun S, Park D, Kim HG, Kim SJ, Lee JO, Kim BT, Park EC, Kim SI (2020) Rapid detection of COVID-19 causative virus (SARS-CoV-2) in human nasopharyngeal swab specimens using field-effect transistor-based biosensor. ACS Nano 14:5135–5142. https://doi.org/10.1021/acsnano.0c02823
Li Z, Yi Y, Luo X, Xiong N, Liu Y, Li S, Sun R, Wang Y, Hu B, Chen W, Zhang Y, Wang J, Huang B, Lin Y, Yang J, Cai W, Wang X, Cheng J, Chen Z, Sun K, Pan W, Zhan Z, Chen L, Ye F (2020) Development and clinical application of a rapid IgM-IgG combined antibody test for SARS-CoV-2 infection diagnosis. J Med Virol 92:1518–1524. https://doi.org/10.1002/jmv.25727
Kohmer N, Westhaus S, Ruhl C, Ciesek S, Rabenau HF (2020) Clinical performance of different SARS-CoV-2 IgG antibody tests. J Med Virol 92:2243–2247. https://doi.org/10.1002/jmv.26145
Espejo AP, Akgun Y, Al Mana AF, Tjendra Y, Millan NC, Gomez-Fernandez C, Cray C (2020) Review of current advances in serologic testing for COVID-19. Am J Clin Pathol 154:293–304. https://doi.org/10.1093/ajcp/aqaa112
Wang X, Guo X, Xin Q, Pan Y, Hu Y, Li J, Chu Y, Feng Y, Wang Q (2020) Neutralizing antibody responses to severe acute respiratory syndrome coronavirus 2 in coronavirus disease 2019 inpatients and convalescent patients. Clin Infect Dis 71:2688–2694. https://doi.org/10.1093/cid/ciaa721
Ju B, Zhang Q, Ge J, Wang R, Sun J, Ge X, Yu J, Shan S, Zhou B, Song S, Tang X, Yu J, Lan J, Yuan J, Wang H, Zhao J, Zhang S, Wang Y, Shi X, Liu L, Zhao J, Wang X, Zhang Z, Zhang L (2020) Human neutralizing antibodies elicited by SARS-CoV-2 infection. Nature 584:115–119. https://doi.org/10.1038/s41586-020-2380-z
Xiang F, Wang X, He X, Peng Z, Yang B, Zhang J, Zhou Q, Ye H, Ma Y, Li H, Wei X, Cai P, Ma W-L (2020) Antibody detection and dynamic characteristics in patients with coronavirus disease 2019. Clin Infect Dis 71:1930–1934. https://doi.org/10.1093/cid/ciaa461
Zeng F, Dai C, Cai P, Wang J, Xu L, Li J, Hu G, Wang Z, Zheng F, Wang L (2020), A comparison study of SARS-CoV-2 IgG antibody between male and female COVID-19 patients: a possible reason underlying different outcome between sex. 92: 2050-2054. https://doi.org/10.1002/jmv.25989
Yan R, Zhang Y, Li Y, Xia L, Guo Y, Zhou Q (2020) Structural basis for the recognition of SARS-CoV-2 by full-length human ACE2. Science 367:1444–1448. https://doi.org/10.1126/science.abb2762
Liu Z, Xiao X, Wei X, Li J, Yang J, Tan H, Zhu J, Zhang Q, Wu J, Liu L (2020) Composition and divergence of coronavirus spike proteins and host ACE2 receptors predict potential intermediate hosts of SARS-CoV-2. J Med Virol 92:595–601. https://doi.org/10.1002/jmv.25726
Cui J, Li F, Shi ZL (2019) Origin and evolution of pathogenic coronaviruses. Nat Rev Microbiol 17:181–192. https://doi.org/10.1038/s41579-018-0118-9
Guo L, Ren L, Yang S, Xiao M, Chang YF, Dela Cruz CS, Wang Y, Wu C, Xiao Y, Zhang L, Han L, Dang S, Xu Y, Yang QW, Xu SY, Zhu HD, Xu YC, Jin Q, Sharma L, Wang L, Wang J (2020) Profiling early humoral response to diagnose novel coronavirus disease (COVID-19). Clin Infect Dis 71:778–785. https://doi.org/10.1093/cid/ciaa310
Chan-Yeung M, Xu RH (2003) SARS: epidemiology. Respirology 8:S9–S14. https://doi.org/10.1046/j.1440-1843.2003.00518.x
Liu Y, Eggo RM, Kucharski AJ (2020) Secondary attack rate and superspreading events for SARS-CoV-2. Lancet 395:e47. https://doi.org/10.1016/s0140-6736(20)30462-1
Seow J, Graham C, Merrick B, Acors S, Pickering S, Steel KJA, Hemmings O, O’Byrne A, Kouphou N, Galao RP, Betancor G, Wilson HD, Signell AW, Winstone H, Kerridge C, Huettner I, Jimenez-Guardeño JM, Lista MJ, Temperton N, Snell LB, Bisnauthsing K, Moore A, Green A, Martinez L, Stokes B, Honey J, Izquierdo-Barras A, Arbane G, Patel A, Tan MKI, O’Connell L, O’Hara G, MacMahon E, Douthwaite S, Nebbia G, Batra R, Martinez-Nunez R, Shankar-Hari M, Edgeworth JD, Neil SJD, Malim MH, Doores KJ (2020) Longitudinal observation and decline of neutralizing antibody responses in the three months following SARS-CoV-2 infection in humans. Nat Microbiol 5:1598–1607. https://doi.org/10.1038/s41564-020-00813-8
H-w J, Li Y, H-n Z, Wang W, Yang X, Qi H, Li H, Men D, Zhou J, S-c T (2020) SARS-CoV-2 proteome microarray for global profiling of COVID-19 specific IgG and IgM responses. Nat Commun 11:3581. https://doi.org/10.1038/s41467-020-17488-8
Muruato AE, Fontes-Garfias CR, Ren P, Garcia-Blanco MA, Menachery VD, Xie X, Shi P-Y (2020) A high-throughput neutralizing antibody assay for COVID-19 diagnosis and vaccine evaluation. Nat Commun 11:4059. https://doi.org/10.1038/s41467-020-17892-0
Huang C, Wen T, Shi F-J, Zeng X-Y, Jiao Y-J (2020) Rapid detection of IgM antibodies against the SARS-CoV-2 virus via colloidal gold nanoparticle-based lateral-flow assay. ACS Omega 5:12550–12556. https://doi.org/10.1021/acsomega.0c01554
Amanat F, Stadlbauer D, Strohmeier S, Nguyen THO, Chromikova V, McMahon M, Jiang K, Arunkumar GA, Jurczyszak D, Polanco J, Bermudez-Gonzalez M, Kleiner G, Aydillo T, Miorin L, Fierer DS, Lugo LA, Kojic EM, Stoever J, Liu STH, Cunningham-Rundles C, Felgner PL, Moran T, García-Sastre A, Caplivski D, Cheng AC, Kedzierska K, Vapalahti O, Hepojoki JM, Simon V, Krammer F (2020) A serological assay to detect SARS-CoV-2 seroconversion in humans. Nat Med 26:1033–1036. https://doi.org/10.1038/s41591-020-0913-5
Waiwijit U, Phokaratkul D, Kampeera J, Lomas T, Wisitsoraat A, Kiatpathomchai W, Tuantranont A (2015) Graphene oxide based fluorescence resonance energy transfer and loop-mediated isothermal amplification for white spot syndrome virus detection. J Biotechnol 212:44–49. https://doi.org/10.1016/j.jbiotec.2015.08.003
Tan S, Han R, Wu S, Liang H, Zhao Y, Zhao H, Li C-P (2019) A novel fluorescent sensing platform for insulin detection based on competitive recognition of cationic pillar[6]arene. Talanta 197:130–137. https://doi.org/10.1016/j.talanta.2019.01.004
Shi J, Chan C, Pang Y, Ye W, Tian F, Lyu J, Zhang Y, Yang M (2015) A fluorescence resonance energy transfer (FRET) biosensor based on graphene quantum dots (GQDs) and gold nanoparticles (AuNPs) for the detection of mecA gene sequence of Staphylococcus aureus. Biosens Bioelectron 67:595–600. https://doi.org/10.1016/j.bios.2014.09.059
Li M, Kang JW, Sukumar S, Dasari RR, Barman I (2015) Multiplexed detection of serological cancer markers with plasmon-enhanced Raman spectro-immunoassay. Chem Sci 6:3906–3914. https://doi.org/10.1039/C5SC01054C
Wallace RA, Charlton JJ, Kirchner TB, Lavrik NV, Datskos PG, Sepaniak MJ (2014) Superhydrophobic analyte concentration utilizing colloid-pillar array SERS substrates. Anal Chem 86:11819–11825. https://doi.org/10.1021/ac5033947
Pokhrel P, Hu C, Mao H (2020) Detecting the coronavirus (COVID-19). ACS Sensors 5:2283–2296. https://doi.org/10.1021/acssensors.0c01153
Zhong J, Rösch EL, Viereck T, Schilling M, Ludwig F (2021) Toward rapid and sensitive detection of SARS-CoV-2 with functionalized magnetic nanoparticles. ACS Sensors 6:976–984. https://doi.org/10.1021/acssensors.0c02160
Rodriguez-Moncayo R, Cedillo-Alcantar DF, Guevara-Pantoja PE, Chavez-Pineda OG, Hernandez-Ortiz JA, Amador-Hernandez JU, Rojas-Velasco G, Sanchez-Muñoz F, Manzur-Sandoval D, Patino-Lopez LD, May-Arrioja DA, Posadas-Sanchez R, Vargas-Alarcon G, Garcia-Cordero JL (2021) A high-throughput multiplexed microfluidic device for COVID-19 serology assays. Lab Chip 21:93–104. https://doi.org/10.1039/D0LC01068E
Tan X, Krel M, Dolgov E, Park S, Li X, Wu W, Sun YL, Zhang J, Khaing Oo MK, Perlin DS, Fan X (2020) Rapid and quantitative detection of SARS-CoV-2 specific IgG for convalescent serum evaluation. Biosens Bioelectron 169:112572. https://doi.org/10.1016/j.bios.2020.112572
Iyer AS, Jones FK, Nodoushani A, Kelly M, Becker M, Slater D, Mills R, Teng E, Kamruzzaman M, Garcia-Beltran WF, Astudillo M, Yang D, Miller TE, Oliver E, Fischinger S, Atyeo C, Iafrate AJ, Calderwood SB, Lauer SA, Yu J, Li Z, Feldman J, Hauser BM, Caradonna TM, Branda JA, Turbett SE, La Rocque RC, Mellon G, Barouch DH, Schmidt AG, Azman AS, Alter G, Ryan ET, Harris JB, Charles RC (2020) Persistence and decay of human antibody responses to the receptor binding domain of SARS-CoV-2 spike protein in COVID-19 patients. Science. Immunology 5:eabe0367. https://doi.org/10.1126/sciimmunol.abe0367
Brouwer PJM, Caniels TG, van der Straten K, Snitselaar JL, Aldon Y, Bangaru S, Torres JL, Okba NMA, Claireaux M, Kerster G, Bentlage AEH, van Haaren MM, Guerra D, Burger JA, Schermer EE, Verheul KD, van der Velde N, van der Kooi A, van Schooten J, van Breemen MJ, Bijl TPL, Sliepen K, Aartse A, Derking R, Bontjer I, Kootstra NA, Wiersinga WJ, Vidarsson G, Haagmans BL, Ward AB, de Bree GJ, Sanders RW, van Gils MJ (2020) Potent neutralizing antibodies from COVID-19 patients define multiple targets of vulnerability. Science 369:643–650. https://doi.org/10.1126/science.abc5902
Cao Y, Su B, Guo X, Sun W, Deng Y, Bao L, Zhu Q, Zhang X, Zheng Y, Geng C, Chai X, He R, Li X, Lv Q, Zhu H, Deng W, Xu Y, Wang Y, Qiao L, Tan Y, Song L, Wang G, Du X, Gao N, Liu J, Xiao J, Su X-d, Du Z, Feng Y, Qin C, Qin C, Jin R, Xie XS (2020) Potent neutralizing antibodies against SARS-CoV-2 identified by high-throughput single-cell sequencing of convalescent patients’ B cells. Cell 182:73–84.e16. https://doi.org/10.1016/j.cell.2020.05.025
Pinto D, Park Y-J, Beltramello M, Walls AC, Tortorici MA, Bianchi S, Jaconi S, Culap K, Zatta F, De Marco A, Peter A, Guarino B, Spreafico R, Cameroni E, Case JB, Chen RE, Havenar-Daughton C, Snell G, Telenti A, Virgin HW, Lanzavecchia A, Diamond MS, Fink K, Veesler D, Corti D (2020) Cross-neutralization of SARS-CoV-2 by a human monoclonal SARS-CoV antibody. Nature 583:290–295. https://doi.org/10.1038/s41586-020-2349-y
Wu Y, Wang F, Shen C, Peng W, Li D, Zhao C, Li Z, Li S, Bi Y, Yang Y, Gong Y, Xiao H, Fan Z, Tan S, Wu G, Tan W, Lu X, Fan C, Wang Q, Liu Y, Zhang C, Qi J, Gao GF, Gao F, Liu L (2020) A noncompeting pair of human neutralizing antibodies block COVID-19 virus binding to its receptor ACE2. Science 368:1274–1278. https://doi.org/10.1126/science.abc2241
Mattioli IA, Hassan A, Oliveira ON, Crespilho FN (2020) On the challenges for the diagnosis of SARS-CoV-2 based on a review of current methodologies. ACS Sensors 5:3655–3677. https://doi.org/10.1021/acssensors.0c01382
Rahimi H, Salehiabar M, Barsbay M, Ghaffarlou M, Kavetskyy T, Sharafi A, Davaran S, Chauhan SC, Danafar H, Kaboli S, Nosrati H, Yallapu MM, Conde J (2021) CRISPR systems for COVID-19 diagnosis. ACS Sensors 6:1430–1445. https://doi.org/10.1021/acssensors.0c02312
Hartanto H, Wu M, Lam ML, Chen T-H (2020) Microfluidic immunoassay for detection of serological antibodies: a potential tool for rapid evaluation of immunity against SARS-CoV-2. Biomicrofluidics 14:061507. https://doi.org/10.1063/5.0031521
Acknowledgements
We thank Xiaoju Li from the State Key Laboratory of Microbial Technology, Shandong University, for the assistance in TEM.
Funding
This work was supported by the National Key R&D Plan of China (Grant No. 2017YFB0405400), the Major Innovation Projects in Shandong Province (2018YFJH0503), Natural Science Foundation for Distinguished Young Scientist of Shandong Province (Grant No. JQ201814), Qilu Young Scholar Funds (11500086963060), Shandong University multidisciplinary research and innovation team of young scholars, and Collaborative Innovation Center of Technology and Equipment for Biological Diagnosis and Therapy in Universities of Shandong.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
ESM 1
(DOCX 5070 kb)
Rights and permissions
About this article
Cite this article
Wang, C., Wang, C., Qiu, J. et al. Ultrasensitive, high-throughput, and rapid simultaneous detection of SARS-CoV-2 antigens and IgG/IgM antibodies within 10 min through an immunoassay biochip. Microchim Acta 188, 262 (2021). https://doi.org/10.1007/s00604-021-04896-w
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00604-021-04896-w